Abstract
Purpose
The prevalence of diabetes is increasing worldwide. The associations between the lipid profile and glycated hemoglobin (HbA1c), fasting glucose, and diabetes remain unclear, so we aimed to perform a cohort study and a two-sample Mendelian randomization (MR) study to investigate the causality between blood lipid profile and HbA1c, fasting glucose, and diabetes.
Methods
A total of 25,171 participants from the Taiwan Biobank were enrolled. We applied a cohort study and an MR study to assess the association between blood lipid profile and HbA1c, fasting glucose, and diabetes. The summary statistics were obtained from the Asian Genetic Epidemiology Network (AGEN), and the estimates between the instrumental variables (IVs) and outcomes were calculated using the inverse-variance weighted (IVW) method. A series of sensitivity analyses were performed.
Results
In the cohort study, high-density lipoprotein cholesterol (HDL-C) was negatively associated with HbA1c, fasting glucose, and diabetes, while the causal associations between HDL-C and HbA1c (βIVW = − 0.098, p = 0.003) and diabetes (βIVW = − 0.594, p < 0.001) were also observed. Furthermore, there was no pleiotropy effect in this study using the MR-Egger intercept test and MR-PRESSO global test.
Conclusions
Our results support the hypothesis that a genetically determined increase in HDL-C is causally related to a reduction in HbA1c and a lower risk of diabetes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hyperglycemia and diabetes are common diseases with an increasing prevalence worldwide. In 2021, the International Diabetes Federation (IDF) Diabetes Atlas Tenth edition reported approximately 537 million adults with diabetes increasing to 783 million by 2045 [1]. In addition, the estimated number of diabetes in South-East Asia will increase up to 152.8 million in 2045 [2]. It is a disease that often has complications due to chronic hyperglycemia, which causes long-term functional damage and failure of the eyes, kidneys and blood vessels [3]. With the increasing prevalence of diabetes, an increased use of medical resources and higher medical expenses will become the focus of attention in all countries.
Diabetes is a disease related to multiple risk factors including age, race, family history of diabetes, alcohol abuse, tobacco abuse, metabolic syndrome, genetic factors, and so on [4,5,6,7,8,9]. Diabetes is affected both by genes and environmental factors. Currently, several observational studies have suggested an association between the blood lipid profile and glycated hemoglobin (HbA1c) [10, 11], fasting glucose [12, 13] and diabetes [14, 15]. However, several Mendelian randomization (MR) studies have indicated inconsistent causal evidence between serum lipid profile and fasting glucose and diabetes [16,17,18,19,20,21,22,23,24,25,26]. Until now, no MR study has discussed the associations between high-density lipoprotein cholesterol (HDL-C) and low-density lipoprotein cholesterol (LDL-C) and HbA1c. A European MR study indicated that genetically increased HDL-C levels were associated with decreased fasting glucose levels [16]. One bidirectional two-sample MR study in India indicated that there was no causal relationship between LDL-C and fasting glucose and there was also no evidence for reverse causality of LDL-C affected by fasting glucose [17], while another European two-sample MR study showed lower LDL-C concentration was associated with higher fasting glucose based on PCSK9 variants [18]. Furthermore, several MR studies in European demonstrated that HDL-C levels had protective effects against diabetes [19,20,21,22,23,24], but a Copenhagen General Population Study showed that the levels of HDL-C were not causally associated with the risk of diabetes [25]; nevertheless, some European MR studies have indicated that a negative causal association between LDL-C and diabetes has been observed [18,19,20, 26], but a recent MR study in European demonstrated that genetically increased LDL-C level was associated with the risk of diabetes [23].
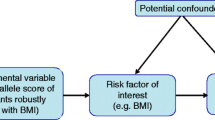
Mendelian randomization uses genetic variants as instrumental variables (IVs) to make inferences about causality based on observational studies [27]. The concept of MR is that when meiosis produces gametes, it is also random, not the result of human manipulation, so it is considered as natural randomized control trials (RCTs). In MR studies, researchers used IVs as a proxy for exposure and examined the causality between the exposure and outcome [28]. There are three assumptions of Mendelian randomization: first, the genetic instrument variables are related to exposure (relevance assumption); second, the genetic instrument variables are not associated with other factors (independence assumption); and third, the genetic instrument variables influence the outcome only through exposure (exclusion restriction assumption) [27].
Up to now, no Mendelian randomization (MR) study has been applied to investigate the causal associations between lipid profile and HbA1c while the causal relationship between lipid profile and fasting glucose and diabetes still remains unclear. Although there are many MR studies based on European ancestry populations, few are MR studies based on Asian populations. It is important because the frequency and distribution may have differences between each ethnicity, so it might not be appropriate to generalize the findings of European individuals to Asian individuals. Accordingly, there are three purposes of this study: first, we examined the association between HDL-C and LDL-C and HbA1c, fasting glucose, and diabetes based on a cross-sectional study and a cohort study; second, we evaluated the causal relationship between HDL-C and LDL-C and HbA1c, fasting glucose, and diabetes using one-sample Mendelian randomization and two-sample Mendelian randomization in Asian ancestry; and finally, we performed a series of sensitivity analyses to validate our results.
Methods
Observational analyses
Study population
The Taiwan Biobank (TWB) study conducted from 2012 to 2019 included 27,737 participants (13,834 men and 13,903 women) of Taiwan Han Chinese ethnicity without a history of cancer. The participants who enrolled in TWB were followed up within 2–4 years. The TWB collected questionnaires, physical examinations, genomic information, and urine and blood samples at the first recruitment and second follow-up [29]. In this study, individuals with missing lipid profiles and HbA1c data (N = 3) and failure in quality control (N = 2563) (Fig. 1) were excluded, so 25,171 participants were involved in cross-sectional and MR analysis. In addition, we also conducted a cohort study that excluded participants without a second follow-up, and a total of 10,570 participants were used in further analysis.
Ethical approval for the study was granted by the Institutional Review Board of Kaohsiung Medical University and Chung-Ho Memorial Hospital and the Ethics and Governance Committee (EGC) of the Taiwan Biobank. Written informed consent of participation was obtained from all participants when joining TWB and the personal information of each participant was fully encrypted for protection.
Covariates assessment
The covariates in our study included age, sex, education status (below high school graduate, college graduate or below, or above college graduate), marriage status (married or unmarried), sports habit (yes or no), drinking habit (yes or no), smoking habit (yes or no), body fat percentage, waist-hip ratio (WHR), family history of hyperlipidemia, and diabetes, and blood lipid profile (high-density lipoprotein cholesterol, low-density lipoprotein cholesterol).
BMI was calculated by dividing the body weight (kg) by the square of height (m2). WHR was the ratio of the waist circumference to the hip circumference.
Outcomes
HbA1c and fasting glucose were measured from blood samples after fasting for 8 h by using a Hitachi LST08 analyzer at Linkou Chang Gung Memorial Hospital. Diabetes was defined as fasting glucose levels higher than 126 mg/dL, HbA1c levels higher than 6.5% (48 mmol/mol), or a self-report of diabetes diagnosed by physicians [30,31,32].
Principle component analysis (PCA)
Population stratification (PS) is commonly caused by geographic isolation, which relates to low migration rates and gene flow over the course of several generations [33] and may operate as a confounder between genotypes and traits in genetic studies [34]. As principle component analysis (PCA) is a common dimension reduction method and is widely used to identify and adjust problems of PS in Genetics [35, 36], we visualized the results of PCA and presented these in Figure E1. In order to eliminate the effects of PS, the first eight ancestry principal components were adjusted in our study.
Mendelian randomization analyses
In this study, we performed a one-sample MR and a two-sample MR to examine the causality of the blood lipid profile and fasting glucose, HbA1c, and diabetes. One-sample MR was conducted by using individual data. The study design of two-sample MR analysis was established on two nonoverlapping populations [37]. The SNP-exposure summary effect size and standard error were obtained from the Asian Genetic Epidemiology Network, and we calculated the SNP-outcome effect size and standard error from the Taiwan Biobank.
Asian Genetic Epidemiology Network (AGEN)
AGEN is an Asian-based population consortium of genetic epidemiology studies of type 2 diabetes and cardiovascular disease-related phenotypes. This consortium aims to identify new loci by conducting genome-wide meta-analyses, replicate studies with similar phenotypes, and conduct other genotyping to discover novel loci. Variants were genotyped using Affymetrix or Illumina genome-wide genotyping arrays and imputed using a Hapmap Project Phase II reference panel and the 1000 Genomes Project Phase 3 of the University of Michigan imputation server reference panel [38]. Our study used summary data of HDL-C and LDL-C from two-stage GWAS meta-analyses based on 34,421 East Asian participants from 13 studies (CAGE-Network, CLHNS, GenSalt, KARE, KCPS II, SBCS, SCES, SiMES, SP2, SWMHS, TWSC, BES, and CHNS). The levels of blood lipid profiles were measured in mg/dL by standard biomedical methods. Participants with lowering cholesterol medication use were excluded from this study.
Genetic instrumental variables
In our research, summary data related to the blood lipid profile were obtained from AGEN. Firstly, we selected SNPs with a genome-wide significance threshold (P < 5 \(\times\) 10–8) associated with high-density lipoprotein cholesterol and low-density lipoprotein cholesterol; secondly, we removed the SNPs when they restricted linkage disequilibrium (R2 > 0.1); thirdly, when the F statistic was less than or equal to 10, it was treated as a weak instrumental variable and removed; fourthly, we excluded IVs directly associated with the outcome to meet assumption three of MR, so we included 161 SNPs as instrumental variables (IVs) for HDL-C on HbA1c, fasting glucose, and diabetes and included 81, 91, and 89 SNPs as IVs for LDL-C on HbA1c, fasting glucose and diabetes (criteria 2); and fifthly, according to independence between lipid profiles as exclusion of instrumental variables (IVs) associated with other lipid profiles based on the hypothesis of their independence, we searched the GWAS catalog and Phenoscanner to find the genome-wide significant variants that were related to other blood lipid profiles then we conducted MR analysis between HDL-C and HbA1c excluding 129 variants related to LDL-C, total cholesterol, or triglycerides. We also excluded 88 variants for MR analysis between LDL-C and HbA1c to reduce any possible pleiotropy effects for the Asian population [39, 40]. Finally, we included 40, 38, and 44 SNPs as IVs for HDL-C on HbA1c, fasting glucose, and diabetes to meet assumption three of MR where IVs did not directly associate with the outcome; thus, we included 8 SNPs as IVs for LDL-C on three outcomes (criteria 1) (Figure E2, Tables E1–E6) [41, 42].
Taiwan Biobank (TWB)
Genotyping was performed using a TWB chip based on an Axiom Genome-Wide CHB 1 Array plate (Affymetric, Inc., Santa Clara, CA, USA) and the Human Exome BeadChip (Illumina, Inc., San Diego, CA, USA) in TWB study [29].
Quality control and strength of the instrumental variables
Quality control is an indispensable step in genetic research to avoid false-positive or false-negative results [43], so we had several exclusion criteria, as follows: For individuals, we checked for data integrity (missing rate > 5%), kinship less than second-grade relatives, out of the heterozygosity rate (without ± 3SD of mean), and gender mismatches. For SNPs, we excluded low minor allele frequency (< 0.05), low call rate (< 95%), and violations of Hardy–Weinberg (P values < 0.0001). After quality control, 25,171 participants and 602,723 variants remained for the final analysis (Fig. 1).
The strength of the instrument variable was evaluated by the F value. It represents the magnitude and quality of the effect on the SNP and exposure [44]. The calculation of the F value was a linear regression between GRS and exposure traits and it was adjusted for age, sex, and population stratification. To avoid weak instrumental variable bias, an F value above 10 was expected to provide sufficient strength for further analyses [45].
Data harmonization
In a two-sample MR study, inappropriate data harmonization can lead to distortion of the MR results. Harmonizing the data is necessary when combining two independent datasets according to the guidelines provided by Fortier et al. [37, 46]. First, we standardized the direction of all variants in the AGEN dataset, which meant that we adjusted all beta values of the effect size to be positive. Second, we matched the alleles in two datasets; that is, if the effect allele was different, we changed the effect allele and the reference allele in dataset 2 and multiplied the effect size by − 1, and the allele frequency was subtracted by 1. Finally, we removed the alleles that did not match in both datasets (n = 0).
One-sample Mendelian randomization
We conducted a one-sample Mendelian randomization (MR) using individual-level data to investigate the causal association between HDL-C and LDL-C and HbA1c, fasting glucose, and diabetes by 2-stage least-square regression [47]. In the first stage, we constructed the linear regression of the risk factors (e.g. HDL-C or LDL-C) on the IVs, resulting in the predicted value of risk factors by GRS, then in second stage, the HbA1c or fasting glucose was regressed on the predicted value by using linear regression, and diabetes was regressed on the predicted value by using logistic regression.
Two-sample Mendelian randomization
Compared to one-sample Mendelian randomization (MR), which uses individual-level data from the same sample for estimating the effect sizes of exposure and outcome, two-sample MR can be performed using not only individual data but also summary data, which can increase statistical power [48]. The effect size of exposure and outcome comes from separate datasets instead of using the same dataset, in order to avoid overfitting, and can also enlarge the study sample size [49]. Between two study designs, weak instruments bias can be avoided when using two independent samples (Fig. 2) [50].
The common approach is to examine this correlation through the inverse-variance weighted (IVW) method, which provides a causal estimate from summary data on multiple instrument variables [51]. The following is the formula of the estimate: \({\beta }_{IVW}=\frac{{\sum }_{k}{X}_{k}{Y}_{k}{\sigma }_{{Y}_{k}}^{-2}}{{\sum }_{k}{X}_{k}^{2}{\sigma }_{{Y}_{k}}^{-2}}\), and the standard error of the estimate is \(se\left({\widehat{\beta }}_{IVW}\right)=\sqrt{\frac{1}{{\sum }_{k}{X}_{k}^{2}{\sigma }_{{Y}_{k}}^{-2}}}\), where \({X}_{k}\) means the change in exposure per additional allele with the standard error \({\sigma }_{{X}_{k}}\), and \({Y}_{k}\) represents the change in outcome per additional allele with the standard error \({\sigma }_{{Y}_{k}}\) [52]. The estimates of SNP-exposure and SNP-outcome were obtained from the two separate datasets for each SNP. For each SNP-exposure, we extracted information, such as effect size and standard error, from the AGEN consortium. For each SNP-outcome, we conducted a multiple linear regression or multiple logistic regression adjusted for age, sex, one to eight population stratifications and other covariates including education status, marriage status, sport habit, drinking habit, smoking habit, body mass index, family history of hyperlipidemia and a family history of diabetes from the Taiwan Biobank to obtain the estimates.
Sensitivity analyses
We used a less strict standard (criteria 2) to select instrumental variables and conducted an MR analysis. Weighted median, MR-Egger, MR Pleiotropy Residual Sum and Outlier (MR-PRESSO), the mode-based estimate (MBE), and the contamination mixture method were used in our sensitivity analyses. Weighted median regression provided consistent causal estimates from the summary data, even if up to 50% of variants did not conform to the Instrument Strength Independent of Direct Effect (InSIDE) assumption [53]. MR-Egger also provided causal estimates, even though there were no valid instrumental variables, and the intercept term of this method is often used to assess pleiotropy [53]. The MBE method and the contamination mixture method were used when we included multiple instrumental variables, and it could provide a consistent estimate when we had some invalid instrumental variables. However, the contamination mixture method could further distinguish whether there were different mechanisms between the exposure and outcome from different genetic variant groups that had similar estimates [54, 55]. To test the directional pleiotropy and determine the outlier, we also used the MR-PRESSO method. Leave-one-out analysis was performed to detect whether a single SNP could affect the results [56]. Heterogeneity could be regarded as an index related to potential violation of MR assumption and was tested using Cochran’s Q statistics [57].
Statistical analyses
Continuous variables are represented by means and standard deviations, and categorical variables are expressed as values and percentages. The relationships between the lipid profiles, demographic characteristics, glycated hemoglobin (HbA1c), and fasting glucose are presented as beta values (\(\beta\)) and 95% confidence intervals (95% C.I.) analyzed by linear regression. We used logistic regression, odds ratios (ORs), and 95% confidence intervals (95% C.I.) to evaluate the risk of HDL-C and LDL-C on diabetes. We conducted multiple linear regression and multiple logistic regression adjusted for age, sex, one to eight principal components, education status, marriage status, sports habit, drinking habit, smoking habit, body mass index, family history of hyperlipidemia, and a family history of diabetes. Generalized estimating equations (GEE) were performed to analyze repeat measurement data and have robustness against deviations from normality. This method can also be used to analyze continuous or binary data, is insensitive to missing values, and considers the intraindividual correlation between measurements [58, 59]. Hence, we used multivariate generalized estimating equations (GEE) regression to examine longitudinal relationships between HDL-C and LDL-C and HbA1c, fasting glucose, and diabetes. A two-tailed test p value < 0.05 was regarded as statistically significant. Bonferroni correction was made (p value was multiplied by 6) for avoiding type 1 error resulting from multiple comparisons (2 traits × 3 outcomes = 6). Observational analyses were performed with SAS version 9.4 software, and Mendelian randomization analyses were performed on Plink version 1.90 and R version 3.5.1 using the “MendelianRandomization” and “MRPRESSO” packages.
Data and resource availability
Summary data of HDL-C and LDL-C GWAS were used in our study is available at the AGEN consortium website (https://blog.nus.edu.sg/agen/summary-statistics/lipids/). Summary data (effect size and SE) of HbA1c, fasting glucose, and diabetes is available in Tables E1–E6.
Results
Participants
The distribution of the demographic characteristics for the study population is presented in Table 1. A total of 25,171 participants were enrolled from the Taiwan Biobank in a cross-sectional analysis and 10,570 participants were included in a cohort study. The median follow-up period was 3.35 years (range 2.85–3.88 years), and the number of incident diabetes was 976 in the cohort study. The average age of all participants in the cross-sectional study was 48.89 \(\pm\) 11.06 years old, men accounted for 50.32%, and the average BMI was 24.34 kg/m2. The average values of HDL-C, LDL-C, TC, and TG were 50.9, 120.9, 193.56, and 116.88 mg/dL, respectively. In the cohort study, participants were younger, had lower BMI, there were fewer males, and higher values of HDL-C, LDL-C, TC, and TG.
Cross-sectional study and cohort study
After adjusting for age, sex, first eight ancestry principal components, education status, marriage status, smoking habit, drinking habit, sports habit, BMI, family history of diabetes, and family history of hyperlipidemia, a negative association between HDL and HbA1c (βHDL-C = − 0.0051, p < 0.0001), and a positive association between LDL-C and HbA1c was observed (βLDL-C = 0.0004, p = 0.0046). When we adjusted for the same covariates, HDL-C (βHDL-C = − 0.131, p < 0.0001) and LDL-C (βLDL-C = − 0.01, p = 0.0136) were negatively associated with fasting glucose. Furthermore, we adjusted for the above covariates to examine the association between HDL-C, LDL-C, and diabetes. We found that HDL-C (ORHDL-C = 0.961, p < 0.0001) and LDL-C (ORLDL-C = 0.992, p < 0.0001) were negatively associated with diabetes. In Multivariate GEE models for the cohort study, we observed similar results, where HDL-C had a protective effect on HbA1c (β = − 0.006, p < 0.0001), fasting glucose (β = − 0.14, p < 0.0001) and diabetes (OR = 0.967, p < 0.0001). LDL-C revealed a negative relationship with diabetes (OR = 0.99, p < 0.0001), and had no association with HbA1c (β = 0.0002, p = 0.4814) and fasting glucose (β = − 0.011, p = 0.0587). The results are presented in Table 2.
Instrumental variables
In our study, we calculated the F value to evaluate the strength of the IVs. F values for IVs of HDL-C were 102.02 to 103.98 and F values for IVs of LDL-C were 13.29 to 13.96, respectively. The F values of all IVs were greater than 10, indicating that the strength of IVs was sufficient. The detailed information was shown in Tables E1–E6.
One sample Mendelian randomization analyses
In assessment of one-sample MR using individual-level data for causal association between HDL-C and LDL-C and HbA1c, fasting glucose and diabetes, we found that HDL-C was causally associated with HbA1c (βIVW = − 0.11, p = 0.05), fasting glucose (βIVW = − 3.67, p = 0.02) and diabetes (βIVW = − 0.02, p = 0.03), and there was no evidence between LDL-C and HbA1c, fasting glucose and diabetes.
Two sample Mendelian randomization analyses
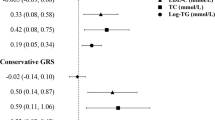
We conducted a two-sample MR to examine the causality between HDL-C and LDL-C and HbA1c, fasting glucose, and diabetes. We found that the increase in HDL-C caused a decrease in the HbA1c value (βIVW = − 0.098, p = 0.003), and the increase in LDL-C led to an increase in HbA1c (βIVW = 0.213, p = 0.034), as shown in Fig. 3 and Table E8. For fasting glucose, we found that neither HDL-C nor LDL-C had a significant causal effect on the value of fasting glucose, as shown in Fig. 4 and Table E9. In addition, we detected that higher HDL-C (βIVW = − 0.532, p = 0.002) reduced the risk of diabetes, but there was no genetic effect of LDL-C on the risk of diabetes in Fig. 5 and Table E10.
Sensitivity analyses
In our study, we applied the weighted median, MR-PRESSO, MBE, and contamination mixture method to check the robustness of the results from the IVW method and the intercept of the MR-Egger method to inspect the horizontal pleiotropy. We found that the effects of HDL-C on HbA1c in all methods had consistent negative directions, and all four sensitivity analyses were consistent. The effect of LDL-C on HbA1c in all methods had the same direction, but only the IVW, MR-PRESSO and contamination mixture methods had positive causal relationships (βIVW = 0.213, p = 0.034; βMR-PRESSO = 0.213, p < 0.001; βcontamination mixture method = 0.216, p = 0.034), as shown in Fig. 3 and Table E8. For fasting glucose, although HDL-C had the same direction in all MR methods, there was no causal association for IVW methods (βIVW = − 0.856, p = 0.387). In sensitivity analyses, the results of MBE and the contamination mixture method for the associations between HDL-C and fasting glucose were significant (βmode-based = − 5.331, p = 0.044; βcontamination mixture method = − 3.782, p = 0.004). The association between LDL-C and fasting glucose had no causal relationship, except for using the MR-PRESSO method (βMR-PRESSO = 2.718, p = 0.002). These results are shown in Fig. 4 and Table E9. The causal relationship between HDL-C and diabetes was estimated in the same direction in all methods, but LDL-C was not associated with diabetes in Fig. 5 and Table E10. After Bonferroni correction, we observed that HDL-C still has a causal association with diabetes.
When we used SNPs as IVs that were related to other lipid profiles (criteria 2), we observed similar results between HDL-C and HbA1c (βIVW = − 0.020, p = 0.002) and diabetes (βIVW = − 0.129, p < 0.001), but the effects were attenuated. The causal association between HDL-C and fasting glucose (βIVW = − 0.623, p = 0.006) was observed in this sensitivity analysis. The causal association between LDL-C and diabetes (βIVW = − 0.108, p = 0.03) was observed and the results were shown in Tables E11–E13.
In addition, we did not find any horizontal pleiotropy of IVs in any blood lipid profile by the intercept test of the MR-Egger regression test and the MR-PRESSO global test (p > 0.05). The results of pleiotropy and heterogeneity were shown in Table E14. Besides, there was no evidence that the causal estimation was affected by a single SNP in the leave-one-out analysis (Figure E4).
Discussion
To date, this is the first two-sample MR analysis to examine the causality between HDL-C, LDL-C, and HbA1c. Besides, few MR studies have explored the relationship between lipid profile and three outcomes of HbA1c, fasting glucose, and diabetes concurrently in Asian ancestry. In our study, we found that HDL-C had strongly negative causality with HbA1c and diabetes with the IVW method and all sensitivity analyses.
The results between observational studies and MR studies might have conflicting findings due to reverse causation and confounding. We aimed to overcome the limitation by using the MR approach and to provide evidence as to whether HDL-C and LDL-C were causally associated with HbA1c, fasting glucose, and diabetes. We observed evidence that the HDL-C levels had a causal association with HbA1c and diabetes in one-sample MR, and two-sample MR. Our findings were robust and consistent across different MR methods and various sensitivity analyses. LDL-C was not causally associated with HbA1c, fasting glucose, and diabetes in all methods and sensitivity analyses, and the results were not varying by statistical adjustment.
Up to now, few studies have explored genetically determined associations between lipid profiles and fasting glucose. A Dutch study found that higher levels of HDL-C were causally associated with lower levels of fasting glucose using two-sample MR analysis [16]. Inconsistent causality between LDL-C levels and fasting glucose had been found, where a European MR study indicated increased 1 mmol/L LDL-C would decrease 0.09 mmol/L fasting glucose [18]. Similar to our findings, an Indian study reported that the genetically predicted levels of LDL-C were not causally associated with the levels of fasting glucose [17]. Compared with other MR findings, we considered the diversity between each ethnic group, differences in SNP numbers, varying SNP selection criteria, or sample size might cause inconsistent results.
Previous MR studies have reported that higher levels of HDL-C and LDL-C were causally associated with a lower risk of diabetes using over one hundred IVs in European ancestry [19, 20]. An MR study using two large community-based studies (including UK Biobank) and independent case–control studies found a consistent result that demonstrated increased HDL-C and LDL-C levels would decrease the risk of diabetes [21]; furthermore, similar results using univariable MR analysis and multivariable MR analysis indicated that HDL-C was negatively associated with the risk of diabetes, but LDL-C did not have causal association with diabetes [22]. In line with our results, a recent MR study among Africans showed that genetically predicted HDL-C was associated with a decreased risk of diabetes, and a positive association between genetically predicted LDL-C and diabetes was also found [23]. An MR study from Danish populations demonstrated that the reduction of HDL-C levels would increase the risk of diabetes [24], but another Danish study showed no evidence between HDL-C and diabetes [25]. Further consistent results were observed between LDL-C and diabetes in previous MR studies where both studies found that decreased levels of LDL-C were causally associated with increased risk of diabetes [18, 26].
The development of diabetes involves several pathogenic processes, including \(\beta\)-cell dysfunction, insulin deficiency, and insulin resistance. These factors can cause an increase in insulin-mediated glucose production in the liver and a reduction in the uptake of insulin-mediated glucose production in skeletal muscle and adipose tissue, leading to hyperglycemia and then the development of diabetes [7, 60]. Prediabetes is defined as glycemic variables higher than a normal statement but lower than the diabetic threshold and plays an important role in the development of diabetes. Recent studies have also demonstrated the importance of the association between prediabetes, heart failure, and cardiovascular disease [61, 62]. Some studies have pointed out that HDL-C may directly influence glucose metabolism [13, 63]. There is evidence that HDL-C inhibits caspase 3 cleavage and blocks the harmful effect of LDL-C to directly protect \(\beta\)-cells [64]. Another study reported that HDL-C, LDL-C, and TC regulate the function and survival of \(\beta\)-cells and that HDL-C has antiobesity and insulin-sensitizing effects [65]. Moreover, a few studies have demonstrated that increasing HDL-C can be used as a treatment for diabetes [66, 67]. The influence of HDL-C on pancreatic \(\beta\)-cells and skeletal muscle cells in mice was discovered in this study, and an elevation of HDL-C in humans was associated with a reduction in blood glucose levels [67]. These associations might provide evidence for dyslipidemia contributing to prediabetes or diabetes. In addition, cholesterol ester transfer protein (CETP) is mainly related to the transfer of lipids in the plasma. It promotes the conversion of HDL to very low-density lipoprotein cholesterol (VLDL) and LDL in exchange for reverse-transported triglyceride (TG) [68]. In other words, CETP inhibitors not only decrease LDL-C but also increase HDL-C [66]. As mentioned above, the use of CETP inhibitors will increase HDL-C levels and decrease LDL-C levels, as well as affect glucose metabolism. Therefore, CETP inhibitors might not only treat dyslipidemia but regulate glucose metabolism as well. The underlying mechanism needs to be verified in the future.
We observed that the effect of HDL-C on HbA1c and fasting glucose was inconsistent in our study. The diagnose criteria for diabetes were based on fasting plasma glucose (FPG) or 2-h plasma glucose (2-h PG) value during a 75-g oral glucose tolerance test (OGTT), or HbA1c [69]. FPG is a relatively convenient and inexpensive method, but its disadvantages are that patients need to fast for 8 h, and FPG value will be affected by large diurnal variations, stress, or acute diseases [70]. HbA1c level is a stable and consistent indicator because it reflects blood glucose concentrations over the past eight to twelve weeks [71]. As a result, we speculated that these reasons might have led to our inconsistent results, and we believed that the result of HbA1c was more robust than fasting glucose.
Our study has several limitations. First, the Taiwan Biobank enrolls participants voluntarily, so there may be a problem of selection bias. However, the participants themselves were not informed of their genotypes and there was no significant difference in the ratio of men to women (men: 50.32%, women: 49.68%), so the problem of selection bias was reduced. Second, we did not perform multivariable MR because there were no common variants in HDL-C and LDL-C after strict exclusion criteria, however, we did not observe pleiotropy of IVs by MR-Egger intercept test and MR-PRESSO global test. Third, medication could not be taken into account in the analysis of our study, since the TWB did not collect information on drug use for diabetes (or high blood glucose, including insulin), so we were unable to obtain such information on treatment for patients. Finally, because the proportion of participants with type 1 diabetes was low (2.5%), we did not eliminate them from our study. We assumed that the small number of people with type 1 diabetes would not affect our findings.
There were several strengths in our study. First, there is no two-sample MR literature to discuss the causality between HDL-C, LDL-C and HbA1c in the Asian ancestry population. As genetic effects on disease should be different by race or ancestry, establishing such specific evidence is very important to understand the etiology by different genetic backgrounds. Second, the results of Mendelian randomization studies are often influenced by linkage disequilibrium (LD), horizontal pleiotropy, and population stratification. We removed SNPs with a strict criterion of LD (\({r}^{2}\ge 0.1\)) in our study. Third, we selected the genetic variants as instrumental variables (IVs) with strict criteria (excluded variants associated with other lipid traits) so that there was no pleiotropy effect in our study. Fourth, we adjusted the population stratification in all analyses by principal component analysis. Fifth, we used the large GWAS consortium from Asian countries and an F value > 10 when selecting the IVs. Sixth, we used five sensitivity analyses and the Bonferroni correction method to elevate the robustness of our results. Finally, we conducted a cohort study to enhance our findings.
Conclusion
Our results support the hypothesis that an increase in HDL-C is causally related to a lower level of HbA1c and a lower risk of diabetes in Asian ancestry. It is necessary to investigate the mechanism of HDL-C’s effects on HbA1c in the future.
Data availability
Summary data of blood lipid profiles GWAS were used in our study is available at the AGEN consortium website (https://blog.nus.edu.sg/agen/summary-statistics/lipids/).
Abbreviations
- MR:
-
Mendelian randomization
- IVW:
-
Inverse-variance weighted
- HbA1c:
-
Glycated hemoglobin
- HDL-C:
-
High-density lipoprotein cholesterol
- LDL-C:
-
Low-density lipoprotein cholesterol
- RCTs:
-
Randomized controlled trials
- IVs:
-
Instrumental variables
- SNPs:
-
Single nucleotide polymorphisms
- WHR:
-
Waist-hip ratio
- BMI:
-
Body mass index
- PRESSO:
-
Pleiotropy residual sum and outliers
- MBE:
-
Mode-based estimate
References
International Diabetes Federation diabetes atlas Tenth edition. 2021
Saeedi P, Petersohn I, Salpea P, Malanda B, Karuranga S, Unwin N et al (2019) Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: results from the International Diabetes Federation Diabetes Atlas, 9(th) edition. Diabetes Res Clin Pract 157:107843
Diagnosis and classification of diabetes mellitus (2013) Diabetes Care 36(Suppl 1):S67-74
Siqueira ISL, Alves Guimarães R, Mamed SN, Santos TAP, Rocha SD, Pagotto V et al (2020) Prevalence and risk factors for self-report diabetes mellitus: a population-based study. Int J Environ Res Public Health 17:6497
Malta DC, Bernal RTI, Iser BPM, Szwarcwald CL, Duncan BB, Schmidt MI (2017) Factors associated with self-reported diabetes according to the 2013 National Health Survey. Rev Saude Publ 51:12s
Bertoldi AD, Kanavos P, França GV, Carraro A, Tejada CA, Hallal PC et al (2013) Epidemiology, management, complications and costs associated with type 2 diabetes in Brazil: a comprehensive literature review. Global Health 9:62
Zheng Y, Ley SH, Hu FB (2018) Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol 14:88–98
Gudjinu HY, Sarfo B (2017) Risk factors for type 2 diabetes mellitus among out-patients in Ho, the Volta regional capital of Ghana: a case–control study. BMC Res Notes 10:324
Temneanu OR, Trandafir LM, Purcarea MR (2016) Type 2 diabetes mellitus in children and adolescents: a relatively new clinical problem within pediatric practice. J Med Life 9:235–239
Khan HA, Sobki SH, Khan SA (2007) Association between glycaemic control and serum lipids profile in type 2 diabetic patients: HbA1c predicts dyslipidaemia. Clin Exp Med 7:24–29
Laverdy OG, Hueb WA, Sprandel MC, Kalil-Filho R, Maranhão RC (2015) Effects of glycemic control upon serum lipids and lipid transfers to HDL in patients with type 2 diabetes mellitus: novel findings in unesterified cholesterol status. Exp Clin Endocrinol Diabetes 123:232–239
Wang S, Ji X, Zhang Z, Xue F (2020) Relationship between lipid profiles and glycemic control among patients with type 2 diabetes in Qingdao, China. Int J Environ Res Public Health 17:5317
Drew BG, Duffy SJ, Formosa MF, Natoli AK, Henstridge DC, Penfold SA et al (2009) High-density lipoprotein modulates glucose metabolism in patients with type 2 diabetes mellitus. Circulation 119:2103–2111
Zhu XW, Deng FY, Lei SF (2015) Meta-analysis of Atherogenic Index of Plasma and other lipid parameters in relation to risk of type 2 diabetes mellitus. Prim Care Diabetes 9:60–67
Davis PJ, Liu M, Sherman S, Natarajan S, Alemi F, Jensen A et al (2018) HbA1c, lipid profiles and risk of incident type 2 Diabetes in United States Veterans. PLoS ONE 13:e0203484
Liu J, van Klinken JB, Semiz S, van Dijk KW, Verhoeven A, Hankemeier T et al (2017) A Mendelian randomization study of metabolite profiles, fasting glucose, and type 2 diabetes. Diabetes 66:2915–2926
Agarwal T, Lyngdoh T, Dudbridge F, Chandak GR, Kinra S, Prabhakaran D et al (2020) Causal relationships between lipid and glycemic levels in an Indian population: a bidirectional Mendelian randomization approach. PLoS ONE 15:e0228269
Schmidt AF, Swerdlow DI, Holmes MV, Patel RS, Fairhurst-Hunter Z, Lyall DM et al (2017) PCSK9 genetic variants and risk of type 2 diabetes: a mendelian randomisation study. Lancet Diabetes Endocrinol 5:97–105
Fall T, Xie W, Poon W, Yaghootkar H, Mägi R, Knowles JW et al (2015) Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes 64:2676–2684
White J, Swerdlow DI, Preiss D, Fairhurst-Hunter Z, Keating BJ, Asselbergs FW et al (2016) Association of lipid fractions with risks for coronary artery disease and diabetes. JAMA Cardiol 1:692–699
Zhu Z, Zheng Z, Zhang F, Wu Y, Trzaskowski M, Maier R et al (2018) Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun 9:224
Yuan S, Larsson SC (2020) An atlas on risk factors for type 2 diabetes: a wide-angled Mendelian randomisation study. Diabetologia 63:2359–2371
Soremekun O, Karhunen V, He Y, Rajasundaram S, Liu B, Gkatzionis A et al (2022) Lipid traits and type 2 diabetes risk in African ancestry individuals: a Mendelian Randomization study. EBioMedicine 78:103953
Haase CL, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R (2015) HDL cholesterol and risk of type 2 diabetes: a mendelian randomization study. Diabetes 64:3328–3333
Marott SC, Nordestgaard BG, Tybjærg-Hansen A, Benn M (2016) Components of the Metabolic syndrome and risk of type 2 diabetes. J Clin Endocrinol Metab 101:3212–3221
Pan W, Sun W, Yang S, Zhuang H, Jiang H, Ju H et al (2020) LDL-C plays a causal role on T2DM: a Mendelian randomization analysis. Aging (Albany NY) 12:2584–2594
Lee K, Lim CY (2019) Mendelian randomization analysis in observational epidemiology. J Lipid Atheroscler 8:67–77
Smith GD, Ebrahim S (2003) “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32:1–22
Chen CH, Yang JH, Chiang CWK, Hsiung CN, Wu PE, Chang LC et al (2016) Population structure of Han Chinese in the modern Taiwanese population based on 10,000 participants in the Taiwan Biobank project. Hum Mol Genet 25:5321–5331
Lin WY, Liu YL, Yang AC, Tsai SJ, Kuo PH (2020) Active cigarette smoking is associated with an exacerbation of genetic susceptibility to diabetes. Diabetes 69:2819–2829
Diagnosis and classification of diabetes mellitus (2006) Diabetes Care 29(Suppl 1):S43–S48
Gillett MJ (2009) International Expert Committee report on the role of the A1c assay in the diagnosis of diabetes. Diabetes Care 32(7):1327–1334
Daniel L. Hartl AGC (2007) Principles of population genetics. 4th edn
Hellwege JN, Keaton JM, Giri A, Gao X, Velez Edwards DR, Edwards TL (2017) Population stratification in genetic association studies. Curr Protoc Hum Genet. 95:1.22.1-3.3
Zhang F, Zhang L, Deng HW (2009) A PCA-based method for ancestral informative markers selection in structured populations. Sci China C Life Sci 52:972–976
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Hartwig FP, Davies NM, Hemani G, Davey SG (2016) Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol 45:1717–1726
Network AGE. 2017.
Nowak C, Ärnlöv J (2018) A Mendelian randomization study of the effects of blood lipids on breast cancer risk. Nat Commun 9:3957
Beeghly-Fadiel A, Khankari NK, Delahanty RJ, Shu XO, Lu Y, Schmidt MK et al (2020) A Mendelian randomization analysis of circulating lipid traits and breast cancer risk. Int J Epidemiol 49:1117–1131
Luo M, Sun M, Wang T, Zhang S, Song X, Liu X et al (2023) Gut microbiota and type 1 diabetes: a two-sample bidirectional Mendelian randomization study. Front Cell Infect Microbiol 13:1163898
Zhang LP, Zhang XX (2022) Relationship between lipids and sleep apnea: Mendelian randomization analysis. World J Clin Cases 10:11403–11410
Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT (2010) Data quality control in genetic case–control association studies. Nat Protoc 5:1564–1573
Burgess S, Thompson SG (2011) Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 40:755–764
Pierce BL, Ahsan H, Vanderweele TJ (2011) Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 40:740–752
Fortier I, Raina P, Van den Heuvel ER, Griffith LE, Craig C, Saliba M et al (2017) Maelstrom research guidelines for rigorous retrospective data harmonization. Int J Epidemiol 46:103–105
Jung S (2013) Structural equation modeling with small sample sizes using two-stage ridge least-squares estimation. Behav Res Methods 45:75–81
Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG (2015) Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 30:543–552
Burgess S, Davies NM, Thompson SG (2016) Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 40:597–608
Burgess S, Davey Smith G, Davies NM, Dudbridge F, Gill D, Glymour MM et al (2019) Guidelines for performing Mendelian randomization investigations. Wellcome Open Res 4:186
Burgess S, Dudbridge F, Thompson SG (2016) Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med 35:1880–1906
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525
Hartwig FP, Davey Smith G, Bowden J (2017) Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46:1985–1998
Burgess S, Foley CN, Allara E, Staley JR, Howson JMM (2020) A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun 11:376
Huang D, Lin S, He J, Wang Q, Zhan Y (2022) Association between COVID-19 and telomere length: a bidirectional Mendelian randomization study. J Med Virol 94:5345–5353
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife 7
Dragioti E, Gerdle B, Larsson B (2019) Longitudinal associations between anatomical regions of pain and work conditions: a study from The SwePain Cohort. Int J Environ Res Public Health 16:2167
Westerlund H, Kivimäki M, Singh-Manoux A, Melchior M, Ferrie JE, Pentti J et al (2009) Self-rated health before and after retirement in France (GAZEL): a cohort study. Lancet 374:1889–1896
Diagnosis and classification of diabetes mellitus (2014) Diabetes Care 37(Suppl 1):S81-90
Cai X, Zhang Y, Li M, Wu JH, Mai L, Li J et al (2020) Association between prediabetes and risk of all cause mortality and cardiovascular disease: updated meta-analysis. BMJ 370:m2297
Cai X, Liu X, Sun L, He Y, Zheng S, Zhang Y et al (2021) Prediabetes and the risk of heart failure: a meta-analysis. Diabetes Obes Metab 23:1746–1753
Gordon SM, Hofmann S, Askew DS, Davidson WS (2011) High density lipoprotein: it’s not just about lipid transport anymore. Trends Endocrinol Metab 22:9–15
Rahmoun MN, Ghembaza CE, El-Amine GM (2019) Lipid profile in type 2 patients with diabetes from Tlemcen: A Western Algerian population. Diabetes Metab Syndr 13:1347–1351
von Eckardstein A, Sibler RA (2011) Possible contributions of lipoproteins and cholesterol to the pathogenesis of diabetes mellitus type 2. Curr Opin Lipidol 22:26–32
Fazio S, Linton MF (2013) Killing two birds with one stone, maybe: CETP inhibition increases both high-density lipoprotein levels and insulin secretion. Circ Res 113:94–96
Drew BG, Rye KA, Duffy SJ, Barter P, Kingwell BA (2012) The emerging role of HDL in glucose metabolism. Nat Rev Endocrinol 8:237–245
Chapman MJ, Le Goff W, Guerin M, Kontush A (2010) Cholesteryl ester transfer protein: at the heart of the action of lipid-modulating therapy with statins, fibrates, niacin, and cholesteryl ester transfer protein inhibitors. Eur Heart J 31:149–164
Association AD (2020) 2. Classification and diagnosis of diabetes: standards of medical care in diabetes—2021. Diabetes Care 44:S15–S33
Sacks DB (2011) A1C versus glucose testing: a comparison. Diabetes Care 34:518–523
Little RR, Sacks DB (2009) HbA1c: how do we measure it and what does it mean? Curr Opin Endocrinol Diabetes Obes 16:113–118
Funding
This work was supported by grants from the Ministry of Science and Technology, Taiwan [MOST 107-2314-B-037-086-MY3 and MOST 110-2314-B-037-048 MY3]. The funding agencies had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
Author information
Authors and Affiliations
Contributions
YCL and TNW contributed to the conception and design. TNW contributed to the acquisition of data. YCL and HPT contributed to statistical analysis and interpretation. YCL and TNW drafted the manuscript for important intellectual content. TNW is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Conflict of interests
The authors have declared that they have no conflict of interest.
Ethical approval
Ethical approval for the study was granted by the Institutional Review Board of Kaohsiung Medical University and Chung-Ho Memorial Hospital and the Ethics and Governance Committee (EGC) of the Taiwan Biobank.
Informed consent
Written informed consent of participation was obtained from all participants when joining TWB and the personal information of each participant was fully encrypted for protection.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, YC., Tu, HP. & Wang, TN. Blood lipid profile, HbA1c, fasting glucose, and diabetes: a cohort study and a two-sample Mendelian randomization analysis. J Endocrinol Invest 47, 913–925 (2024). https://doi.org/10.1007/s40618-023-02209-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40618-023-02209-x