Abstract
Purpose of the Review
Exposure to essential and non-essential metals is widespread. Metals exposure is linked to epigenetic, particularly DNA methylation, differences. The strength of evidence with respect to the metal exposure type, timing, and level, as well as the DNA methylation association magnitude, and reproducibility are not clear. Focusing on the most recent 3 years, we reviewed the human epidemiologic evidence (n = 26 studies) and the toxicologic animal model evidence (n = 18 studies) for associations between metals exposure and DNA methylation.
Recent Findings
In humans, the greatest number of studies focused on lead exposure, followed by studies examining cadmium and arsenic. Approximately half of studies considered metals exposure during the in utero period and measured DNA methylation with the genome-wide Illumina arrays in newborn blood or placenta. Few studies performed formal replication testing or meta-analyses. Toxicology studies of metals and epigenetics had diversity in model systems (mice, rats, drosophila, tilapia, and zebrafish were represented), high heterogeneity of tissues used for DNA methylation measure (liver, testis, ovary, heart, blood, brain, muscle, lung, kidney, whole embryo), and a variety of technologies used for DNA methylation assessment (global, gene specific, genome-wide). The most common metals tested in toxicologic studies were lead and cadmium.
Summary
Together, the recent studies reviewed provide the strongest evidence for DNA methylation signatures with prenatal metals exposures. There is also mounting epidemiologic evidence supporting lead, arsenic, and cadmium exposures with DNA methylation signatures in adults. The field of metals and DNA methylation is strengthened by the inclusion of both epidemiology and toxicology approaches, and further advancements can be made by coordinating efforts or integrating analyses across studies. Future advances in understanding the molecular basis of sequence specific epigenetic responses to metals exposures, methods for handling exposure mixtures in a genome-wide analytic framework, and pipelines to facilitate collaborative testing will continue to advance the field.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
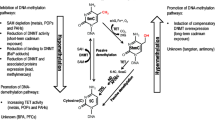
Environmental exposures to metals are an enduring public health issue. Non-essential metals, such as lead and cadmium, are those with no normal physiologic function in the body. At increasing levels, they have deleterious effects on multiple health endpoints, including neurodevelopment and neurodegenerative disorders, cancer, and cardiometabolic disorders [1,2,3]. Non-essential metal exposures arise through multiple sources and pathways, including contamination of food and water systems from industrial processes, as well as exposure through ambient air and tobacco smoke [4]. Other trace metals, such as manganese and selenium, are distinguished by their essential nature for normal biological processes, including serving as co-factors for enzymatic function or as a component of amino acids (selenocysteine) [5, 6]. Essential metals can exhibit non-linear dose–response curves with health endpoints, wherein toxicity is observed at both low and excess levels. Epidemiology studies can approximate human exposure to environmental trace metals through biomonitoring levels in human tissue (e.g., placenta, whole blood) and excretion samples (e.g. urine and fecal samples), but these studies are often observational and available tissues may be limited. Toxicology studies of trace metals are better able to assess causality and mechanisms of action; however, their findings may not always be able to be extrapolated to humans. Selection of model systems and relevance of dose for human exposures are important toxicologic study design factors. Across both epidemiology and toxicology studies of metals, the exposure timing (e.g., adulthood, in utero), route (e.g., inhalation, ingestion, injection), duration (e.g., acute, chronic), and source, and dose may influence the impacts. Altogether, the integration of complementary evidence from both epidemiology and toxicology studies are critical for advancing human health risk assessments of metals exposures.
Metals exposure levels related to health can have epigenetic marks as a biomarker or mechanism of that action. Upon human exposure, trace metals can interact with enzymes and interfere with intracellular gradients of micronutrients and reactive oxygen species [7]. These interactions are detectable via shifts in molecular signals, including epigenetic modifications [8]. There are several types of epigenetic modifications, and although their mechanisms are unique, they have important implications for regulating gene expression. DNA methylation is one epigenetic mechanism that can directly influence the magnitude of gene expression. Levels of epigenetic factors and responses to metals exposure can be tissue specific. For example, whole blood and saliva are less invasive source tissues in epidemiology studies, and they may be a proxy or surrogate of changes in the epigenome at hypothesized target organs where toxicological effects are occurring [9]. Studies conducted in animal models are better equipped to directly determine epigenetic mechanisms in target tissues, such as the brain or heart. The timing of epigenetic measurements is also important. When measuring epigenetics as a potential consequence of environmental exposures, prospective study designs with exposure prior to DNA methylation measurement are necessary. Biologically, there are major waves of epigenetic change throughout the life course, including the in utero or early life period for all tissues, or other time periods like puberty for specific tissues. Studies may monitor exposures during these windows of epigenetic susceptibility.
In this review article, we link together exciting recent research on metals and epigenetics. The field of metals and epigenetics has been active for at least two decades and previous review articles summarized early progress [10,11,12,13]. To bring the reader up to date, in this article, we focused on metals and epigenetics research published in the last 3 years. First, we cover the domain of human population-based epidemiology. Second, we review studies using model system-based toxicology. In both fields of research, we summarized their findings and evaluated the strength of the current evidence. Lastly, we reflect on the critical gaps and future directions of this field to make suggestions for strategies to make further advances.
Metals and Epigenetics: Epidemiologic Evidence
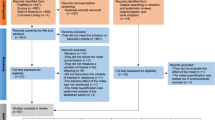
Many recent studies have associated metal exposures to changes in epigenetic endpoints. Through a PubMed search (Supplementary Methods) and subject matter expertise, we identified recent epidemiologic observational studies, published between January 1, 2019, and March 31, 2022. DNA methylation is the most common epigenetic endpoint used in epidemiologic studies that include epigenetic measurement. We included studies based on DNA methylation being the primary endpoint, metal exposure the primary exposure, and observational studies based on human subjects only (Supplemental Fig. 1). These epidemiological studies vary in design, sample characteristics, and timing of exposure and DNA methylation collection. Our findings included 26 observational studies, including three prospective studies, three meta-analyses, nineteen cross-sectional studies, and one case–control study (Table 1). We organized the results by study design and when possible, by metal exposure (Fig. 1).
DNA methylation is sequence specific and microarrays or sequencing technologies allow for measurement of DNA methylation at candidate locations or genome-wide to test for differentially methylated positions or regions. Epigenome-wide association studies (EWAS) are performed as a discovery analysis when all positions or regions are tested for differences. These methods often rely on bisulfite treatment of the DNA, which converts an un-modified cytosine to uracil and maintains modified cytosines in the sequence.
Prospective Studies
Prospective studies are a strong epidemiologic study design as the exposures are measured prior to DNA methylation measures, limiting the potential for reverse causation. We identified three prospective studies testing prenatal maternal lead concentration levels and cord blood DNA methylation at birth. Although there were substantial differences in sample size, these studies were characterized by having repeated measures of lead concentrations during critical gestational periods with the purpose of unveiling (1) trimester-specific effects of maternal lead exposure on the offspring DNA methylation levels, and (2) sex-specific DNA methylation patterns in response to lead exposure. Specifically, in a cohort of Mexican mother–child pairs, first trimester blood lead concentrations were associated with DNA methylation levels at three sites, annotated to the genes RAB5A, EXT1, and a non-genic region, and sites were enriched for pathways related to neurodevelopment [24]. No associations were observed between second trimester maternal blood lead levels and DNA methylation, but third trimester blood lead concentrations were associated with one site in a non-genic region [24]. Additionally, in a study of mother–child pairs in Korea, the maternal blood lead levels measured during pregnancy weeks 12–20 were tested with infant sex-specific cord blood DNA methylation was tested [23]. Among males, 11 sites were differentially methylated by lead levels, and sites were enriched for endothelial cell development via CDH5 and axonogenesis via PLXNA4 [23]. Among females, no differentially methylated sites were observed [23]. Finally, a multi-metal epigenome-wide association study in the USA explored the relationship between prenatal exposure to essential and non-essential trace metals during first trimester of pregnancy and DNA methylation at two time points, in cord blood at birth and in whole blood at mid childhood [21]. Manganese concentrations were associated with higher DNA methylation at one site, mapping to the gene A2BP1, while lead exposure was associated with to lower DNA methylation of a site, mapping to the gene CASP8. In females, manganese exposure was associated with nine differentially methylated sites, seven of which persisted to mid-childhood [21]. Among males, manganese exposure was associated with higher DNA methylation of one site linked to the gene A2BP1, which persisted to mid-childhood [21]. Jointly, the results from these studies suggest that metal exposures during critical gestational periods, in particular during the first trimester of pregnancy, are associated with sex-dependent differences in DNA methylation in the offspring, which may persist until mid-childhood.
Meta-analyses
Discovery analyses through EWAS are often challenged by a greater number of DNA methylation positions being tested than number of participants in the study. Testing for replication or meta-analyses across samples can help reduce false positive associations. It has traditionally been difficult to perform meta-analyses of metal exposure epigenetic signatures due to heterogeneity in relevant design characteristics such as sample size, demographic composition (sex and genetic ancestry), timing of metal exposure (prenatal vs adulthood), and epigenetic tissues measured (cord blood, buccal cells, and blood). Three recent meta-analyses used standardized protocols to harmonize exposure assessment and DNA methylation datasets with the aim of identifying epigenetic signatures of copper [18•], arsenic [20•], and mercury exposure [22•], across different populations.
The first meta-analysis combined data from two mother-infant pair cohorts, the New Hampshire Birth Cohort Study and the Rhode Island Child Health Study, to explore the relationship between placental copper concentrations and genome-wide DNA methylation in placental tissue [18•]. These cohorts had similar collection of demographic and anthropometric measures from mother-infant pairs, assessment of metal concentrations, measurement of DNA methylation, and data analysis procedures. Through meta-analysis, placental copper concentrations were associated with DNA methylation at 15 sites and a region in the promoter of the gene GSTP1 [18•]. Sensitivity analyses indicated these results were robust to differences in the racial/ethnic composition of the two cohorts [18•]. With common investigators involved in the design of these two cohorts, harmonization of exposure assessment and DNA methylation measurement was facilitated.
In the meta-analysis of prenatal mercury exposure and DNA methylation, seven existing cohorts participated [22•], and this collaboration was facilitated through the Pregnancy and Childhood Epigenetics consortium [40]. Total mercury cord blood concentration was selected as the primary exposure variable. For cohorts with mercury assessment in different tissues (one in maternal hair and five in maternal whole blood), exposure levels were transformed to cord blood levels. The timing of DNA methylation measures varied across cohorts. Five cohorts measured cord blood DNA methylation at birth, four cohorts measured child blood DNA methylation at ages 7–8, and two cohorts had DNA methylation measures for both time points [22•]. By leveraging the increased power across cohorts, the meta-analysis identified mercury exposure was associated with higher DNA methylation levels at two sites mapped to the MED31 gene in both cord and childhood blood. These findings suggest prenatal exposure to mercury is associated with DNA methylation differences at birth that are sustained through childhood.
Third, the meta-analysis of arsenic and DNA methylation aimed at identifying arsenic related signatures of DNA methylation across two different studies (in Chile and Bangladesh) through standardized data pipelines, study design procedures, and common applications of statistical methods [20•]. The two study samples had differences in exposure assessment and selected tissues for DNA methylation analysis. The Chilean study assessed exposure based on recruitment from historical high or low arsenic exposure areas. The Bangladesh study measured baseline arsenic levels in water samples during recruitment [20•]. The Chilean sample used peripheral blood mononuclear cells (PBMCs) and buccal cells for DNA methylation measurement, while the Bangladesh study used blood samples only [20•]. Results were meta-analyzed using a combination of all PBMC samples from Chile and Bangladesh, and all PBMC samples (Chile and Bangladesh) plus buccal cells (Chile only). The meta-analysis from all PBMC samples revealed 11 differentially methylated regions, the meta-analysis from PBMC plus buccal cells identified 16 differentially methylated regions, and eight of the differentially methylated regions overlapped [20•]. Arsenic-associated sites were enriched for pathways related to fatty acid elongation, fatty acid metabolism, and lysosome activity [20•]. In general, these copper, mercury, and arsenic meta-analyses leverage large sample sizes to achieve statistical power to detect common epigenetic signatures from unique metal exposures across widely diverse populations. Standardized data procedures facilitated DNA methylation analysis from different tissues and time points to identify persistent epigenetic differences across the lifespan.
Cross-sectional Studies
Cross-sectional studies are a highly feasible epidemiologic study design because they only require one participant visit, though we should be cautious when interpreting findings. Across 19 cross-sectional studies identified, lead exposure was the most researched metal [16, 17, 25, 26]. These studies varied in sample size, geographical region, and target tissue. In general, lead blood levels were associated with gene specific DNA methylation patterns, higher average levels of DNA methylation, or differentially methylated sites. In Zambia, blood lead levels were correlated with differential patterns of DNA methylation of the ALAD and the p16 tumor suppressor gene promoter regions [16]. At least 84.3% of children with high lead exposure levels exhibited altered ALAD gen DNA methylation, in comparison to only 42.1% of children with low lead exposure levels [16]. In a study involving neonates from the USA, lead exposure was associated with lower cord blood DNA methylation at 30 sites and higher methylation at three sites [25]. These associations were attenuated after adjusting for blood cell type proportions, which is an important factor in DNA methylation profiles. A small study of occupational lead exposure in China analyzed DNA methylation associations with high blood lead levels (> 300 μg/L) versus low (< 100 μg/L), and lead concentrations were associated with 356 differentially methylated sites enriched for pathways associated with nervous system development [32]. Lastly, in Mexico, an epigenome-wide association study of prenatal lead exposure and cord blood DNA methylation found 47 differently methylated sites, 20 of which were previously identified to be associated with low birth weight [26]. These findings may suggest differences in DNA methylation may be a mechanism by which lead exposure contributes to low birth weight [26]. Altogether, these studies provide evidence of the variable effects of lead exposure on DNA methylation at early life stages and during adulthood, though a formal meta-analysis across studies has not yet been performed and is warranted.
After lead, the next most abundant metals examined in human cross-sectional studies are cadmium, arsenic, and chromium. Among 13 American Indian tribes, cadmium exposure in adults was associated with six DNA methylation sites, and sites were enriched for cancer pathways, cardiovascular disease risk factors, and inflammation [30]. A cross-sectional analysis of electroplating workers in China found chromium exposure was associated with eight DNA methylation sites, these results were confirmed in vitro and were associated with expression of SEMA4B, a gene involved in chromium-related carcinogenesis [31]. Chronic arsenic exposure has previously been associated with epigenetic dysregulation and carcinogenesis. A large US cross-sectional study observed arsenic exposure was associated with 20 differentially methylated sites and one differentially methylated region, which were consistent across sex groups and replicated findings from independent studies [28]. The most significant differentially methylated site mapped to the SLC7A11 gene, involved in transport of cysteine, an established mechanism related to arsenic [28]. In a cross-sectional study in Spain and Bangladesh, urinary arsenic levels were associated with six common differentially methylated sites and seven differentially hydroxymethylated sites, enriched for cardiometabolic disease, inflammation, and cancer [29]. Across these metals, there may be sufficient individual studies for a formal meta-analysis to examine consistency across populations.
Environmental exposure to metals occurs in mixtures, and a diverse set of cross-sectional studies have analyzed the associations between multiple metal exposures and DNA methylation. For example, among Chinese mothers exposed to e-waste recycling areas during pregnancy, concentrations of lead, cadmium, manganese, and chromium were tested for association with DNA methylation in their newborns’ cord blood [27]. Multiple metal exposures were associated with 125 differentially methylated sites, including 79 with higher DNA methylation and 46 with lower DNA methylation [27]. In Taiwan, a log unit increase in lead concentration was associated 0.315% higher global DNA methylation (p < 0.001), and a log unit increase in cadmium concentration was associated with 0.263% higher global DNA methylation (p < 0.001) [17]. One study evaluated the relationship between maternal concentrations of three non-essential metals (lead, cadmium, and mercury) and two essential trace metals (manganese and selenium), and whole blood DNA methylation levels during pregnancy [19•]. Cadmium and manganese exposure were associated with higher global DNA methylation [19•]. Lead was associated with 11 differentially methylated sites (false discovery rate < 0.1), enriched for neurology-related gene ontologies [19•], similar to previous observations [24]. These studies demonstrate that differences in DNA methylation may be observed at different life periods with concomitant exposure to multiple metals.
Case–control studies
Case–control studies can be leveraged for environmental epigenetic epidemiology to provide insights for populations with existing conditions. A matched case–control study conducted mediation analysis to explore the indirect effect of DNA methylation at the WNT3A gene on the relationship between prenatal lead exposure and non-syndromic cleft lip and/or palate (NSCL/P). In utero lead concentrations were associated (p < 0.05) with 0.52% higher DNA methylation at the WNT3A gene. They also observed that 9.2% of the lead exposure association with non-syndromic cleft lip and/or palate may be attributable to the mediating effect of WNT3A DNA methylation. Additional studies indirectly point to the role of metals in DNA methylation. For example, a matched case–control study explored the joint association between blood aluminum levels and DNA methylation of the beta-2 adrenergic receptor (ADRB2) gene in asthmatic children (n = 70) compared to those without asthma (n = 70) [41]. High aluminum levels were associated with asthma (OR = 11.6, 95%CI: 2.1, 63.4), but high ADRB2 DNA methylation was not associated with asthma (OR = 0.7, 95%CI: 0.2, 3.1), suggesting uncontrolled asthma may be affected by elevated blood aluminum concentrations rather than ADRB2 DNA methylation [41]. However, the study did not explore the relationship between aluminum concentrations and ADRB2 DNA methylation levels directly. In studies such as this, it will be possible to directly test for relationships between exposure and DNA methylation in the future.
Together, the recent epidemiology studies reviewed provide the strongest evidence for DNA methylation signatures with prenatal exposures to lead, mercury, arsenic, copper, and cadmium. Epidemiology studies of metals and DNA methylation are strengthened by prospective study designs and collaborative meta-analysis or replication testing. Common limitations of these studies include using surrogate tissues for both exposure and DNA methylation assessment, instead of the target tissues. DNA methylation measures area often collected at delivery, though critical periods in development may occur earlier. Complementary toxicology studies are able to overcome many of these challenges and provide controlled exposure doses and epigenetic measures in target tissues.
Metals and Epigenetics: Toxicologic Evidence
Overview of Metal Exposure Toxicology Studies with Epigenetic Endpoints
Metal exposure toxicology studies with epigenetic endpoints seek to investigate biological mechanisms of exposure toxicity and the role of changes in the epigenome. We searched for metal exposure toxicology studies with epigenetic endpoints published as current research between January 1, 2019, and March 21, 2022 (Supplementary Methods). We included studies based on epigenetic factors being the primary endpoint, metal exposure the primary exposure/treatment, and studies in animal models only (Supplemental Fig. 2). A total of 18 studies were identified (Table 2).
The studies profiled here were extremely diverse in every aspect. The metal exposures evaluated in these studies included: cadmium (7 studies), lead (5 studies), titanium dioxide (1 study), uranium (1 study), and arsenic (1 study), with an additional study that evaluated a mixture of lead and cadmium exposure (1 study) (Fig. 2). Exposure methods included drinking water, lactation, gavage, intraperitoneally injection and inhalation. Multiple in vivo experimental models were used including mice (C57BL/6 J, CD1, NIH/OlaHsd), rats (Sprague–Dawley, Wistar, Long-Evans), drosophila, zebrafish, and Nile tilapia fish. The target tissues evaluated for DNA methylation in the studies varied considerably. The liver (5) and testes (4) were the most common targets, with additional tissues including the spermatozoa (2), brain (2), blood (2), embryos (2), kidney (1), ovaries (1), heart (1), and muscle (1). DNA methylation was the principal epigenetic endpoint measured in all toxicology studies, though most of the studies evaluated multiple epigenetic endpoints. Methyl groups are added to DNA using DNA methyltransferase (DNMT) enzymes, and studies measure levels or activity of DNMT to understand regulation of DNA methylation. Expression DNMTs and ten-eleven translocation (TET) methylcytosine dioxygenases were often measured as secondary epigenetic endpoints.
Assessment Methods of DNA Methylation in Exposure Toxicology Studies
DNA methylation was the primary epigenetic endpoint measured in the exposure toxicology studies, though there were considerable differences in the assessment methods. Specific DNA methylation endpoints included global, genome-wide, and locus-specific. Approximately half of the studies reported results for global DNA methylation, which represents overall degree of methylated cytosine compared to total cytosine content [60]. Studies reporting changes in global DNA methylation used colorimetric, ELISA, or luminometric microplate–based assays, or bisulfite conversion/pyrosequencing of LINE-1, a repetitive DNA retrotransposon used as a proxy for global DNA methylation because it constitutes 17% of human genome [61]. The other half of included toxicology studies evaluated genome-wide DNA methylation with either whole genome bisulfite sequencing (WGBS), an expensive method that covers DNA methylation across the entire genome [62], or enhanced reduced representation bisulfite sequencing (EERBS), a more streamlined method that focuses coverage on a large number of biologically relevant loci. Lastly, a handful of profiled studies evaluated the DNA methylation at individual gene loci, primarily focusing on imprinted genes.
Differential DNA Methylation in the Liver in Metal Exposure Toxicology Studies
As a major organ, the liver is the primary site of xenobiotic metabolism in vivo, making it a prominent target for metal exposure toxicity [63]. In the studies profiled here, the liver was the most evaluated target tissue for changes in DNA methylation. Only two studies evaluated the same metal (lead) and target tissues (liver and blood), using the same model (mice) [54•, 57]. In both studies, maternal mice were exposed to lead through drinking water 2 weeks prior to pregnancy, during pregnancy, and through weaning [54•, 57]. Male and female base-pair resolution DNA methylation was measured in offspring at 3 weeks old in one study [57] and 5 months old in the other study [54•]. Although, both studies had hundreds of tissue- and sex-specific differentially methylated regions in lead-exposed tissues compared to controls, there was little in common between the results reported. For example, one of the goals of the two studies was to determine if DNA methylation changes in the liver corresponded to those in paired blood samples. In young mice, there were few differentially methylated regions in common between the blood and liver; however, in the adult mice, there were hundreds of differentially methylated regions in common between the two tissues. Moreover, each study had differentially methylated regions at different imprinted genes in common between blood and liver: young mice had Arid1b, Pde10a, Smoc2, Trappc9, and adult mice had Bargain, Peg12, Rasgrf1, Snrpn, and different enriched pathways. Together, the results of these two studies demonstrated that perinatal lead exposure caused changes in liver and blood DNA methylation in a dynamic manner over time, even after lead exposure had ceased.
In addition to the two studies, several other studies evaluated exposure induced differential DNA methylation in the liver. One study demonstrated that cadmium exposure in adult rats caused statically significant lower DNA methylation in liver gene promoter regions [52•], while a second study demonstrated that cadmium exposure in juvenile tilapia fish caused significant time- and dose-dependent lower global DNA methylation [46]. These findings suggest that cadmium exposure may cause a trend toward lower DNA methylation in functional regions of the genome, with potential impacts on gene expression.
Differential DNA Methylation in Brain and Embryotic Tissue in Metal Exposure Toxicology Studies
In addition to the liver, multiple toxicology evaluated exposure-induced differential DNA methylation in other target tissues. Neurotoxicity is one of the most studied adverse health outcomes of lead exposure. Two studies evaluated the effects of perinatal lead exposure on DNA methylation in different parts of the rodent brain [43, 45•]. One study evaluated neurons isolated from the mouse neuronal cortex [43], and the other study evaluated bulk tissue from the hippocampus [45•]. The neuron study reported a trend towards lower DNA methylation in gene promoter regions in the cortex [43]. In contrast, the hippocampus study reported higher DNA methylation in gene promoter regions of the hippocampus [45•]. The results of these two studies demonstrated that exposure to metals may cause variable changes in DNA methylation within different regions from the same organ.
Besides the brain, early embryonic target tissue was evaluated in several studies for exposure-induced differential DNA methylation. During early embryonic development, DNA methylation undergoes multiple windows of reprogramming, making embryos particularly vulnerable to exposure-induced alternations to DNA methylation, which can last into adulthood and impact chronic disease risk [64]. One study reported lower methylation in 12-h cadmium-exposed zebrafish embryos that returned back to baseline by 24-h exposure [42]. Another study reported no change to the global DNA methylation status of LINE-1 in preimplantation mouse embryos perinatally exposed to cadmium [58]. The findings of these two studies suggest that cadmium exposure may not significantly affect global DNA methylation; however, more comprehensive studies using base-pair resolution methods and cell type adjusted methods are needed to further understand the effects of cadmium on embryonic tissue DNA methylation changes.
Effects of Metal Exposure on Multi-generational Changes in DNA Methylation in Toxicology Studies
Multi-generational epigenetic studies are conducted with exposure in one generation and epigenetic markers measured in subsequent generations. Maternal (F0) exposure to toxicants during pregnancy can directly cause changes in DNA methylation and traits in the offspring (F1) generation [65]. The F2 generation was also directly exposed to the toxicant through germ cells, and changes have been observed [65]. Some research examines the F3 and subsequent generations that were never directly exposed to the toxicant. Reflected in current research, there has been considerable interest in studying effects of maternal metal exposure on DNA methylation patterns in adult offspring and subsequent generations. Half of the studies profiled here evaluated multigeneration effects of metal exposure. Although the variability in study designs makes it hard to draw specific conclusions about the findings, these studies reported some type of alterations to DNA methylation resulting from maternal exposure to metals. Most of the studies only involve evaluation of epigenetic changes in the F1 generation; however, several studies extend findings to the F2 and F3 generations. For example, in response to arsenic exposure in rats, lower global testes DNA methylation was observed in F0, higher testes DNA methylation was observed in F2 and F3, and higher ovary DNA methylation was observed in F3 [51]. Collectively, the current studies have demonstrated metal exposures are capable of causing multigenerational epigenetic alterations; however, the effects of such alterations on the health trajectory over the life course of individuals in future generations is not yet understood.
Summary, Critical Gaps, and Future Directions
Metals exposure and epigenetics research is in a period of rapid growth. A critical mass of studies and research groups are active in the area, which has generated breadth in the timing and type of exposure measures, as well as the timing and tissue of DNA methylation measures. Studies from the last 3 years offer compelling findings in their study population or model system. In recent human epidemiology studies, there is most evidence for lead and cadmium exposure during pregnancy and DNA methylation differences in tissues collected at birth (cord blood and placenta tissue). In recent toxicology studies, there is most evidence for prenatal lead and cadmium exposure causing differences in DNA methylation in liver from rodent models. Building on these successes, we identify areas of future development to advance the field.
The most pressing and perplexing question in metals exposure epigenetics, and indeed in environmental epigenetics more broadly, is the molecular basis for highly reproducible, sequence-specific epigenetic differences. Metals may have a generalized response on epigenetic modifying enzymes. For examples, exposures, including some metals, that generate reactive oxygen species can deplete available methyl groups [66] or oxidize DNA which alters methyl binding domains [67]. Metals can also influence levels of DNMT enzymes [68] and cadmium can non-competitively bind DNMT, changing enzyme function [69]. Metabolism of metals, particularly arsenic, may also influence the availability of the methyl substrate for DNA methylation [12]. These types of epigenetic enzyme-based mechanisms would be expected to produce widespread or global differences in DNA methylation, which have been observed [68]. However, they fail to account for the highly sequence specific differences in DNA methylation that are also observed with metals exposures. The availability of sequences for action by epigenetic enzymes (based on the presence or absence of transcription factors or histone occupancy) varies by developmental timing and tissue, and this has been hypothesized to dictate the exposure-epigenetic specificity [70, 71]. Similarly, while groups of environment-related DNA methylation differences are concurrently associated with gene expression differences [72], and new findings suggest epigenetic factors may influence cellular cytoskeletal structures [73], the range of potential consequences of environment-related DNA methylation differences have not been fully explored. Early life epigenetic programming may have lagged effects in adulthood [74], requiring longitudinal investigations of consequences. Targeted, sequence-specific epigenetic editing techniques [75, 76], such as using piRNA to edit DNA methylation [77], CRISPR to modulate chromatin marks or perturb DNA [78], or the DNA binding proteins zinc finger nucleases and transcriptional activator like effector nucleases [79] are emerging. Experimental toxicologic work using these types of techniques will revolutionize the biologic understanding of the consequences of altered DNA methylation. Understanding the biologic basis for reproducible, sequence-specific signatures of metals exposure and their consequences will catapult the field forward.
There are numerous complementary areas of future development in the fields of metals exposure modeling and in epigenetic measurement. With respect to metals exposure, especially for essential metals where adverse effects are expected at both low and high exposures, we need to incorporate non-linear dose–response curves. In epidemiology studies, the choice of biological media for exposure assessment is a function of important considerations, including metabolism and half-life of metals, directness of inference to biological processes according to tissue specificity, and invasiveness towards human subjects sampling. Additionally, some metals, such as arsenic may be conjugated or modified upon metabolism within the human body. As such, different species of metals may be quantified and afford inference of the metal at different stages of the metabolism process. Metals exposures occur in combination and can influence absorption and metabolism of each other. Advanced methods for modeling exposure mixtures has been applied [21], though will be challenged by features of complex exposure matrices, including sparsity of effect estimate signals within small sample sizes, collinearity between exposures, and potential high-dimensional interactions between exposures. Inclusion of multiple offspring generations of model systems may yield important insight into potential transgenerational effects attributable to metals exposures. Multigenerational studies should take particular care when interpreting potential effects, as up to three generations may be directly exposed to the parent chemical. Future work may also incorporate genetic polymorphisms that influence metals metabolism, and in turn, may modify the relationship between metals exposure and epigenetics.
In the area of epigenetic markers, repeated longitudinal epigenetic measures will help answer questions of the persistence and timing of exposure and epigenetic associations. The development of human DNA methylation microarrays enabled standardization of measurement across populations, and the new mouse DNA methylation microarray and the new custom mammalian array [80] should enable new opportunities for cross-toxicology study investigation. Notably, standard laboratory methods collapse all DNA modifications (methylation, hydroxymethylation, formylation, carboxylation), though findings are often attributed to the most abundant DNA modification, which is DNA methylation. Studies highlighted here have largely focused on DNA methylation for utilitarian purposes (stability of the marker in archived samples), though biologically DNA methylation is expected to follow histone modifications [81]. Histone modifications are expected to be rapid responses to environmental conditions with more immediate impacts on gene expression, while DNA methylation provides longer-term maintenance of environmental signals [82]. Earlier environmental epigenetic influences may be captured by using additional epigenetic markers, including histone modifications. Measures of hydroxymethylation may also be considered, particularly early in development or in brain or placenta tissues where the marker is most abundant [83]. Rapid expansion in technology to measure multi-omics data frames in combination with enhanced exposure assessment will yield new opportunities. Improvements in the areas of metals exposure modeling and expansion on epigenetic markers under consideration or standardization of epigenetic measures will build on the existing foundational research.
Additionally, advances in the field of metals and epigenetics together will continue. The rigor of associations between metals exposures and epigenetics is enhanced by testing triangulation using diverse study designs and is a major current strength. This includes testing findings across toxicologic model systems for evolutionary conservation and across diverse human study samples with wide ranges of exposure for assessing generalizability or specificity of signals. Statistically, we have noted that several the current studies lack replication testing and are underpowered for epigenome-wide assessment. As with any genome-wide analysis with relatively small effect sizes and small sample sizes, false positive associations are expected. It is time for partnerships and replication testing to increase the impact and rigor of the research. Larger and more collaborative epidemiologic studies which involve replication testing or meta-analysis, akin to the recent mercury study in the PACE consortium [22•], are critically needed. There is also high potential for these collaborative meta-analyses of metals and epigenetics through the Toxicant Exposures and Responses by Genomic and Epigenomic Regulators of Transcription (TaRGET II) consortium [84], the Environmental Influences on Child Health Outcomes (ECHO) consortium [85], and the Cohorts of Heart and Aging Research in Genetic Epidemiology (CHARGE) consortium [86]. When testing associations between metals exposures and epigenetics, we need to be clear about the limits of the inferences we can make (association, biomarker, mechanism, cell composition marker) [87]. For example, reproducible epigenetic signatures of metals may be assessed as biomarker of exposure, as has been effectively demonstrated with smoking epigenetic signatures [88] and follow-up questions can include the persistence and cross-tissue applicability of the biomarker signal. Currently, human studies lack evidence of a causal relationship between metals exposure and epigenetic signatures, but toxicologic studies are advancing this area. Epigenetic signatures may be a marker of metals exposure-induced altered cell type composition, particularly in toxicology studies where cell type has generally not been adjusted for, which could be an important biologic effect of exposures [89]. Continued and expanded collaboration across metals and epigenetics studies will enable the assessment of reproducible findings, which will open opportunities for using these signatures in biomarker or mechanistic studies.
Conclusion
In summary, metals exposures that are common in populations are associated with epigenetic markers, specifically DNA methylation. To date, epigenome-wide studies have largely investigated cadmium and lead exposure measured in blood and observed sequence specific differences in DNA methylation, though replication testing is needed. It is an exciting era for metals and epigenetics studies with emerging methods for multi-metal exposure assessment, reproducible epigenome-wide DNA methylation measures, and pipelines to facilitate collaborative testing. The next several years are expected to bring further rapid advancements.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance
Bakulski KM, Seo YA, Hickman RC, Brandt D, Vadari HS, Hu H, et al. Heavy metals exposure and Alzheimer’s disease and related dementias. J Alzheimers Dis. 2020;76:1215–42.
Cosselman KE, Navas-Acien A, Kaufman JD. Environmental factors in cardiovascular disease. Nat Rev Cardiol. 2015;12:627–42.
Vrijheid M, Casas M, Gascon M, Valvi D, Nieuwenhuijsen M. Environmental pollutants and child health—a review of recent concerns. Int J Hyg Environ Health. 2016;219:331–42.
Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ. Heavy metal toxicity and the environment. Exp Suppl. 2012;101:133–64.
Argüello JM, Raimunda D, González-Guerrero M. Metal transport across biomembranes: emerging models for a distinct chemistry. J Biol Chem. 2012;287:13510–7.
Lewicka I, Kocyłowski R, Grzesiak M, Gaj Z, Oszukowski P, Suliburska J. Selected trace elements concentrations in pregnancy and their possible role - literature review. Ginekol Pol. 2017;88:509–14.
Milnerowicz H, Ściskalska M, Dul M. Pro-inflammatory effects of metals in persons and animals exposed to tobacco smoke. J Trace Elem Med Biol. 2015;29:1–10.
Ruiz-Hernandez A, Kuo C-C, Rentero-Garrido P, Tang W-Y, Redon J, Ordovas JM, et al. Environmental chemicals and DNA methylation in adults: a systematic review of the epidemiologic evidence. Clin Epigenetics. 2015;7:55.
Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD. Epigenetic research in neuropsychiatric disorders: the “tissue issue.” Curr Behav Neurosci Rep. 2016;3:264–74.
Cheng T-F, Choudhuri S, Muldoon-Jacobs K. Epigenetic targets of some toxicologically relevant metals: a review of the literature. J Appl Toxicol. 2012;32:643–53.
Fragou D, Fragou A, Kouidou S, Njau S, Kovatsi L. Epigenetic mechanisms in metal toxicity. Toxicol Mech Methods. 2011;21:343–52.
Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6:820–7.
Ryu H-W, Lee DH, Won H-R, Kim KH, Seong YJ, Kwon SH. Influence of toxicologically relevant metals on human epigenetic regulation. Toxicol Res. 2015;31:1–9.
Yang W, Guo Y, Ni W, Tian T, Jin L, Liu J, et al. Hypermethylation of WNT3A gene and non-syndromic cleft lip and/or palate in association with in utero exposure to lead: a mediation analysis. Ecotoxicol Environ Saf. 2021;208: 111415.
Yu M, Zhou X, Ju L, Yu M, Gao X, Zhang M, et al. Characteristics of iron status, oxidation response, and DNA methylation profile in response to occupational iron oxide nanoparticles exposure. Toxicol Ind Health. 2020;36:170–80.
Yohannes YB, Nakayama SM, Yabe J, Nakata H, Toyomaki H, Kataba A, et al. Blood lead levels and aberrant DNA methylation of the ALAD and p16 gene promoters in children exposed to environmental-lead. Environ Res. 2020;188: 109759.
Lin C-Y, Lee H-L, Hwang Y-T, Huang P-C, Wang C, Sung F-C, et al. Urinary heavy metals, DNA methylation, and subclinical atherosclerosis. Ecotoxicol Environ Saf. 2020;204: 111039.
• Kennedy E, Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, et al. Copper associates with differential methylation in placentae from two US birth cohorts. Epigenetics. 2020;15:215–30. This recent epidemiology study is notable because it uses a meta-analysis approach to identify replicated associations between copper exposure and placenta DNA methylation.
• Aung MT, Bakulski KM, Feinberg JI, Dou JF, Meeker JD, Mukherjee B, et al. Maternal blood metal concentrations and whole blood DNA methylation during pregnancy in the early autism risk longitudinal investigation (EARLI). Epigenetics. 2021;17;253–68.
• Bozack AK, Boileau P, Wei L, Hubbard AE, Sille FCM, Ferreccio C, et al. Exposure to arsenic at different life-stages and DNA methylation meta-analysis in buccal cells and leukocytes. Environ Health. 2021;20:79. This recent epidemiology study is notable because it uses a meta-analysis approach to identify replicated associations between arsenic exposure and DNA methylation in two tissues.
Bozack AK, Rifas-Shiman SL, Coull BA, Baccarelli AA, Wright RO, Amarasiriwardena C, et al. Prenatal metal exposure, cord blood DNA methylation and persistence in childhood: an epigenome-wide association study of 12 metals. Clin Epigenetics. 2021;13:208.
• Lozano M, Yousefi P, Broberg K, Soler-Blasco R, Miyashita C, Pesce G, et al. DNA methylation changes associated with prenatal mercury exposure: a meta-analysis of prospective cohort studies from PACE consortium. Environ Res. 2022;204: 112093. (This recent epidemiology study is notable because it uses a meta-analysis approach to identify replicated associations between mercury exposure and newborn blood DNA methylation.)
Park J, Kim J, Kim E, Kim WJ, Won S. Prenatal lead exposure and cord blood DNA methylation in the Korean Exposome Study. Environ Res. 2021;2021(195):110767.
Rygiel CA, Dolinoy DC, Perng W, Jones TR, Solano M, Hu H, et al. Trimester-specific associations of prenatal lead exposure with infant cord blood DNA methylation at birth. Epigenet Insights. 2020;2020(13):2516865720938669.
Montrose L, Goodrich JM, Morishita M, Kochmanski J, Klaver Z, Cavalcante R, et al. Neonatal lead (Pb) exposure and DNA methylation profiles in dried bloodspots. Int J Environ Res Public Health. 2020;17:6775.
Heiss JA, Téllez-Rojo MM, Estrada-Gutiérrez G, Schnaas L, Amarasiriwardena C, Baccarelli AA, et al. Prenatal lead exposure and cord blood DNA methylation in PROGRESS: an epigenome-wide association study. Environ Epigenet. 2020;6:dvaa014.
Zeng Z, Huo X, Zhang Y, Hylkema MN, Wu Y, Xu X. Differential DNA methylation in newborns with maternal exposure to heavy metals from an e-waste recycling area. Environ Res. 2019;171:536–45.
Bozack AK, Domingo-Relloso A, Haack K, Gamble MV, Tellez-Plaza M, Umans JG, et al. Locus-specific differential DNA methylation and urinary arsenic: an epigenome-wide association study in blood among adults with low-to-moderate arsenic exposure. Environ Health Perspect. 2020;128:067015.
Domingo-Relloso A, Bozack A, Kiihl S, Rodriguez-Hernandez Z, Rentero-Garrido P, Casasnovas JA, et al. Arsenic exposure and human blood DNA methylation and hydroxymethylation profiles in two diverse populations from Bangladesh and Spain. Environ Res. 2022;204: 112021.
Domingo-Relloso A, Riffo-Campos AL, Haack K, Rentero-Garrido P, Ladd-Acosta C, Fallin DM, et al. Cadmium, smoking, and human blood DNA methylation profiles in adults from the strong heart study. Environ Health Perspect. 2020;128:067005.
Feng L, Guo X, Li T, Yao C, Xia H, Jiang Z, et al. Novel DNA methylation biomarkers for hexavalent chromium exposure: an epigenome-wide analysis. Epigenomics. 2020;12:221–33.
Zhang X-X, He Z, Feng B, Shao H. An epigenome-wide DNA methylation study of workers with an occupational exposure to lead. J Appl Toxicol. 2019;39:1311–9.
Ha F, Li N, Long C, Zheng P, Hu G, Jia G, et al. The effect of global DNA methylation on PDCD5 expression in the PBMC of occupational chromate exposed workers. J Occup Environ Med. 2021;63:600–8.
Issah I, Arko-Mensah J, Rozek LS, Zarins KR, Agyekum TP, Dwomoh D, et al. Global DNA (LINE-1) methylation is associated with lead exposure and certain job tasks performed by electronic waste workers. Int Arch Occup Environ Health. 2021;94:1931–44.
Childebayeva A, Goodrich JM, Chesterman N, Leon-Velarde F, Rivera-Ch M, Kiyamu M, et al. Blood lead levels in Peruvian adults are associated with proximity to mining and DNA methylation. Environ Int. 2021;155: 106587.
Lee J-E, Kim H-R, Lee M, Kim N-H, Wang K-M, Lee S, et al. Smoking-related DNA methylation is differentially associated with cadmium concentration in blood. Biochem Genet. 2020;58:617–30.
Nishizawa-Jotaki S, Sakurai K, Eguchi A, Tanabe H, Watanabe M, Mori C. Association between mercury in cord serum and sex-specific DNA methylation in cord tissues. J Dev Orig Health and Disease. 2021;12:124–31.
Li Z, Guo C, Li X, Wang Z, Wu J, Qian Y, et al. Associations between metal exposure and global DNA methylation in potentially affected people in E-waste recycling sites in Taizhou City, China. Sci Total Environ. 2020;711: 135100.
Montes-Castro N, Alvarado-Cruz I, Torres-Sánchez L, García-Aguiar I, Barrera-Hernández A, Escamilla-Núñez C, et al. Prenatal exposure to metals modified DNA methylation and the expression of antioxidant- and DNA defense-related genes in newborns in an urban area. J Trace Elem Med Biol. 2019;55:110–20.
Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, et al. Cohort profile: pregnancy and childhood epigenetics (PACE) consortium. Int J Epidemiol. 2018;47:22–23u.
Nafea OE, El-Korashi LA, Gehad MH, Yousif YM, Zake LG. Association between blood aluminum and beta-2 receptor gene methylation with childhood asthma control. Hum Exp Toxicol. 2020;39:1301–9.
Bian X, Gao Y. DNA methylation and gene expression alterations in zebrafish embryos exposed to cadmium. Environ Sci Pollut Res Int. 2021;28:30101–10.
Dou JF, Farooqui Z, Faulk CD, Barks AK, Jones T, Dolinoy DC, et al. Perinatal lead (Pb) exposure and cortical neuron-specific DNA methylation in male mice. Genes (Basel). 2019;10.
• Guan D-L, Ding R-R, Hu X-Y, Yang X-R, Xu S-Q, Gu W, et al. Cadmium-induced genome-wide DNA methylation changes in growth and oxidative metabolism in Drosophila melanogaster. BMC Genomics. 2019;20:356. https://doi.org/10.1186/s12864-019-5688-z. This recent toxicology study is notable because it used whole-genome bisulfite sequencing, the gold standard method. This study also directly connected gene-specific DNA methylation changes to measured gene expression changes.
• Hong T, Li S-M, Jia B, Huang Y, Shu K, Yuan K-W, et al. DNA methylation changes in the hippocampus of learning and memory disorder offspring rats of lead exposure during pregnant and lactation period. Ann Palliat Med. 2021;10:1059–69. This recent toxicology study is notable because in addition to evaluating changes in DNA methylation in the hippocampus, functional learning and memory tests were also carried out, directly connecting DNA methylation changes with functional outcomes specific to the relevant tissue.
Hu F, Yin L, Dong F, Zheng M, Zhao Y, Fu S, et al. Effects of long-term cadmium exposure on growth, antioxidant defense and DNA methylation in juvenile Nile tilapia (Oreochromis niloticus). Aquat Toxicol. 2021;241: 106014.
Legendre A, Elmhiri G, Gloaguen C, Magneron V, Kereselidze D, Saci N, et al. Multigenerational exposure to uranium changes morphometric parameters and global DNA methylation in rat sperm. C R Biol. 2019;342:175–85.
Ma Y, Guo Y, Ye H, Huang K, Lv Z, Ke Y. Different effects of titanium dioxide nanoparticles instillation in young and adult mice on DNA methylation related with lung inflammation and fibrosis. Ecotoxicol Environ Saf. 2019;176:1–10.
Tachibana K, Kawazoe S, Onoda A, Umezawa M, Takeda K. Effects of prenatal exposure to titanium dioxide nanoparticles on DNA methylation and gene expression profile in the mouse brain. Front Toxicol. 2021;3: 705910.
• Nakayama SMM, Nakata H, Ikenaka Y, Yabe J, Oroszlany B, Yohannes YB, et al. One year exposure to Cd- and Pb-contaminated soil causes metal accumulation and alteration of global DNA methylation in rats. Environ Pollut. 2019;252:1267–76. https://doi.org/10.1016/j.envpol.2019.05.038. This recent toxicology study is notable because it used the unique exposure method of contaminated soil for a long period of time (1 year). It also evaluated a mixture of lead and cadmium.
Nava-Rivera LE, Betancourt-Martínez ND, Lozoya-Martínez R, Carranza-Rosales P, Guzmán-Delgado NE, Carranza-Torres IE, et al. Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure. Sci Rep. 2021;11:8276.
• Ren C, Ren L, Yan J, Bai Z, Zhang L, Zhang H, et al. Cadmium causes hepatopathy by changing the status of DNA methylation in the metabolic pathway. Toxicol Lett. 2021;340:101–13. https://doi.org/10.1016/j.toxlet.2020.12.009. This recent toxicology study is notable because it used whole-genome bisulfite sequencing, the gold standard method. This study also evaluated visual and functional changes in the liver corresponding with DNA methylation changes in the same tissue.
Saintilnord WN, Tenlep SYN, Preston JD, Duregon E, DeRouchey JE, Unrine JM, et al. Chronic exposure to cadmium induces differential methylation in mice spermatozoa. Toxicol Sci. 2021;180:262–76.
• Svoboda LK, Neier K, Wang K, Cavalcante RG, Rygiel CA, Tsai Z, et al. Tissue and sex-specific programming of DNA methylation by perinatal lead exposure: implications for environmental epigenetics studies. Epigenetics. 2021;16:1102–22. This recent toxicology study is notable because it details a coordinated evaluation of changes in DNA methylation resulting from perinatal in two tissues, liver, and blood. The purpose of this coordinated evaluation is to test weather DNA methylation in blood can be used as a proxy for changes in DNA methylation in liver, allowing for a potentially less invasive biomarker for liver epigenetic changes. The study also evaluated sex-specific DNA methylation.
Svoboda LK, Wang K, Jones TR, Colacino JA, Sartor MA, Dolinoy DC. Sex-specific alterations in cardiac DNA methylation in adult mice by perinatal lead exposure. Int J Environ Res Public Health. 2021;18:E577.
Wang X, Wang M, Zeng L, Su P. Hypomethylation of LINE-1 retrotransposons is associated with cadmium-induced testicular injury. Environ Sci Pollut Res Int. 2020;27:40749–56.
Wang K, Liu S, Svoboda LK, Rygiel CA, Neier K, Jones TR, et al. Tissue- and sex-specific DNA methylation changes in mice perinatally exposed to lead (Pb). Front Genet. 2020;11:840.
Zhu J, Huang Z, Yang F, Zhu M, Cao J, Chen J, et al. Cadmium disturbs epigenetic modification and induces DNA damage in mouse preimplantation embryos. Ecotoxicol Environ Saf. 2021;219: 112306.
Tai Z, Guan P, Zhang T, Liu W, Li L, Wu Y, et al. Effects of parental environmental copper stress on offspring development: DNA methylation modification and responses of differentially methylated region-related genes in transcriptional expression. J Hazard Mater. 2022;424: 127600.
Vryer R, Saffery R. What’s in a name? Context-dependent significance of “global” methylation measures in human health and disease. Clin Epigenet. 2017;9:2.
Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703.
Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–105.
Gu X, Manautou JE. Molecular mechanisms underlying chemical liver injury. Expert Rev Mol Med. 2012;14: e4.
Breton-Larrivée M, Elder E, McGraw S. DNA methylation, environmental exposures and early embryo development. Anim Reprod. 2019;16:465–74.
Tuscher JJ, Day JJ. Multigenerational epigenetic inheritance: One step forward, two generations back. Neurobiol Dis. 2019;132: 104591.
Kreuz S, Fischle W. Oxidative stress signaling to chromatin in health and disease. Epigenomics. 2016;8:843–62.
Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 2004;32:4100–8.
Sanchez OF, Lee J, Yu King Hing N, Kim S-E, Freeman JL, Yuan C. Lead (Pb) exposure reduces global DNA methylation level by non-competitive inhibition and alteration of dnmt expression. Metallomics. 2017;9:149–60.
Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286:355–65.
Collings CK, Anderson JN. Links between DNA methylation and nucleosome occupancy in the human genome. Epigenet Chromatin. 2017;10:18.
Martin EM, Fry RC. Environmental Influences on the epigenome: exposure- associated DNA methylation in human populations. Annu Rev Public Health. 2018;39:309–33.
Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet. 2016;98:680–96.
Walker C, Burggren W. Remodeling the epigenome and (epi)cytoskeleton: a new paradigm for co-regulation by methylation. J Exp Biol. 2020;223:jeb220632.
Treviño LS, Dong J, Kaushal A, Katz TA, Jangid RK, Robertson MJ, et al. Epigenome environment interactions accelerate epigenomic aging and unlock metabolically restricted epigenetic reprogramming in adulthood. Nat Commun. 2020;11:2316.
de Groote ML, Verschure PJ, Rots MG. Epigenetic Editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012;40:10596–613.
Gjaltema RAF, Rots MG. Advances of epigenetic editing. Curr Opin Chem Biol. 2020;57:75–81.
Perera BPU, Svoboda L, Dolinoy DC. Genomic tools for environmental epigenetics and implications for public health. Curr Opin Toxicol. 2019;18:27–33.
Nakamura M, Gao Y, Dominguez AA, Qi LS. CRISPR technologies for precise epigenome editing. Nat Cell Biol. 2021;23:11–22.
Yim YY, Teague CD, Nestler EJ. In vivo locus-specific editing of the neuroepigenome. Nat Rev Neurosci. 2020;21:471–84.
Arneson A, Haghani A, Thompson MJ, Pellegrini M, Kwon SB, Vu H, et al. A mammalian methylation array for profiling methylation levels at conserved sequences. Nat Commun. 2022;13:783.
Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, et al. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev. 2015;29:2449–62.
Przybilla J, Buske P, Binder H, Galle J. Histone modifications control DNA methylation profiles during ageing and tumour expansion. Front Life Sci. 2013;7:31–43.
Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104:341–6.
Wang T, Pehrsson EC, Purushotham D, Li D, Zhuo X, Zhang B, et al. The NIEHS TaRGET II Consortium and environmental epigenomics. Nat Biotechnol. 2018;36:225–7.
Breton CV, Landon R, Kahn LG, Enlow MB, Peterson AK, Bastain T, et al. Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun Biol. 2021;4:1–15.
Colicino E, Marioni R, Ward-Caviness C, Gondalia R, Guan W, Chen B, et al. Blood DNA methylation sites predict death risk in a longitudinal study of 12, 300 individuals. Aging (Albany NY). 2020;12:14092–124.
Bakulski KM, Fallin MD. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen. 2014;55:171–83.
Reese SE, Zhao S, Wu MC, Joubert BR, Parr CL, Håberg SE, et al. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ Health Perspect. 2017;125:760–6.
Campbell KA, Colacino JA, Park SK, Bakulski KM. Cell types in environmental epigenetic studies: biological and epidemiological frameworks. Curr Environ Health Reports. 2020;7:185–97.
Funding
Support for Dr. Elkin was provided by the NIH award number T32 DK071212. Mr. Higgins was supported by the NIH award number R01 AG067592-01S1. Support for Dr. Aung was provided by the NIH award number P30 ES030284. Dr. Bakulski was supported by the NIH awards: R01 AG070897, R01 AG067592, R35 ES031686, R01 MD013299, and R01 ES025531.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Human and Animal Rights and Informed Consent
This article did not involve human participants or animal models.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Metals and Health
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elkin, E.R., Higgins, C., Aung, M.T. et al. Metals Exposures and DNA Methylation: Current Evidence and Future Directions. Curr Envir Health Rpt 9, 673–696 (2022). https://doi.org/10.1007/s40572-022-00382-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40572-022-00382-4