Abstract
Cactaceae arose ca. 30–35 million years ago (Mya) and comprises keystone species of arid and semiarid biomes. Various specific features related to morphology, anatomy, and CAM metabolism have likely favored diversification and expansion under arid environments. The plastome of most cactus species also undergone several evolutionary features to adapt growth and development under adverse environmental conditions. Aiming to enhance the understanding of the plastid evolution in the subfamily Cactoideae, we sequenced and analyzed in detail the plastome of Rhipsalis teres (Vell.) Steud., an epiphytic species from the Atlantic forest. The plastome of R. teres contains the two main rearrangements found in other plastomes of the subfamily Cactoideae. Our structural analyses suggested that the ancestral plastome architecture served as the basic structure for homologous recombination-mediated diversification of the IR boundaries found in them. Cactoideae plastomes show several polymorphic RNA editing sites, which are mainly distributed in genes related to gene expression machinery. Interestingly, the loss of rpl33 and rpl36 genes suggests a relaxed plastid biosynthesis in these species. Also, the absence of the trnV-UAC gene indicates the emergence of a tRNA import mechanism given that, for steric reasons, all valine codons cannot be read by the tRNAVal(GAC). Finally, our data indicate that the environment imposed several evolutionary features in the plastomes of Cactoideae during evolution under arid and semiarid biomes allowing adaptation and diversification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cacti are appreciated around the world as ornamental plant species due to their varied shapes, sizes, and beautiful flowers. In addition, some species are valuable sources of food and new bioactive pharmaceutical compounds (Díaz et al. 2017; Inglese et al. 2017; Maciel et al. 2019; Ramírez-Rodríguez et al. 2020). Despite their high economic, cultural, and ecologic importance, they are among the most threatened taxonomic groups (Goettsch et al. 2015). The family Cactaceae comprises around 1400 species, including succulent and non-succulent plants that have a broad distribution across the Americas (Anderson 2001; Guerrero et al. 2019). It comprises keystone species of arid and semiarid biomes, and tropical rain forests (Anderson 2001). Several studies have supported the central Andes as the main center of origin of Cactaceae (Majure et al. 2012; Hernandez-Hernandez et al. 2014). Eastern Brazil is the second biodiversity hotspot derived from the Andean uplift (Ritz et al. 2007, 2012), and Mexico is another biodiversity hotspot of many cactus genera (Hernández-Hernández et al. 2014).
Cactaceae is the major succulent plant lineage that arose ca. 30–35 million years ago (Mya), with its fastest diversification occurring 10–5 Mya. Cactus radiation is a contemporaneous event if compared with the diversification of other succulent plant lineages and C4 plants (Arakaki et al. 2011). The emergence of succulent and C4 plants has probably favored the diversification and expansion of these plant lineages under arid environments (Arakaki et al. 2011). These evolutionary features involve several morphological, anatomic, and metabolic modifications including large cells for water storage, shallow root system, thick waxy cuticle, and CAM metabolism (Ogburn and Edwards 2010; Griffiths and Males 2017). Although Cactaceae contains early diverging lineages, poor in the number of species, and slight morphological succulence, the core cacti exhibit high diversity of species and pronounced morphological succulence (Arakaki et al. 2011; Guerrero et al. 2019).
Cactaceae comprises four subfamilies: Cactoideae, Maihuenioideae, Opuntioideae, and Pereskioideae. The last three subfamilies diverged early, whereas the former is the richest in the diversity of species. The species of Cactoideae are highly variable in growth habit and morphology, including treelike, shrubby, caespitose, climbing, or epiphytes. This subfamily has divided into two main clades: tribe Cacteae and Core Cactoideae (subdivided into Core Cactoideae I and II), which includes all the other tribes (Anderson 2001; Guerrero et al. 2019).
Plastome evolutionary traits such as gene content, recombination events, loss of genes, gene transfer to the nucleus, RNA editing, and gene divergence are useful tools to understand processes of plant evolution, including species diversification and environmental adaptation (Greiner and Bock 2013; Vieira et al. 2016; Bock 2017; Pacheco et al. 2020a, b). Plastome sequences contain several molecular markers (Provan et al. 2001; Rogalski et al. 2015), useful for genetic studies, including genetic diversity, biogeography, structure of natural populations (Tsai et al. 2015; Rogalski et al. 2015; Roy et al. 2016; Stefenon et al. 2019), and phylogenetic studies (Nyffeler 2002; Lopes et al. 2019; Majure et al. 2019; Pacheco et al. 2019; 2020c).
The plastomes of most angiosperms normally show little size variation and encode a conserved set of genes (Bock 2007). Most genes are grouped into two categories: One comprises components of the photosynthetic machinery (i.e., photosystem I, photosystem II, cytochrome b6/f complex and ATP synthase) and the other includes the genes required for plastid gene expression (i.e., subunits of RNA polymerase, rRNAs, tRNAs, and ribosomal proteins) (Rogalski et al. 2015; Daniell et al. 2016). Moreover, plastomes contain other essential genes related to different cellular functions such protein import machinery (Kikuchi et al. 2013, 2018), plastid protein homeostasis (Kuroda and Maliga 2003), fatty acid biosynthesis (Kode et al. 2005), and chlorophyll biosynthesis (Agrawal et al. 2020). Loss of function of some protein complexes involved in photosynthesis displays different phenotypes from no discernible to white plants (Horváth et al. 2000; Albus et al. 2010; Krech et al. 2012). Disruption of plastid gene expression affects not only photosynthesis but also other essential cellular function, which can indeed inhibit cell division and lead to lethality (Drescher et al. 2000; Kode et al. 2005; Rogalski et al. 2006, 2008a; Agrawal et al. 2020). Plastid proteins encoded by the plastomes are translated on prokaryotic-type 70S ribosomes (Tiller and Bock 2014). Normally, all RNA components of these ribosomes (16S rRNA, 23S, 5S, and 4.5S), 30 tRNAs, and 21 ribosomal proteins (12 from the small ribosomal subunit and 9 from the large ribosomal subunit) are encoded by the plastomes (Tiller and Bock 2014; Daniell et al. 2016). Evolutionarily, lifestyle (i.e., non-photosynthetic organisms) and growth habit can be considered important factors to reduce the demand for protein biosynthesis in plastids, and consequently, a relaxed plastid translation containing a reduced set of ribosomal components could support protein biosynthesis in such situations (Rogalski et al. 2008b; Fleischmann et al. 2011; Alkatib et al. 2012b). Interestingly, Cactaceae is a photosynthetic lineage of angiosperms adapted to stress environmental conditions with slow growth and a notorious example of atypical evolution of plastomes (Sanderson et al. 2015; Solórzano et al. 2019).
Recently, Sanderson et al. (2015) and Solórzano et al. (2019) published plastid genomes (plastomes) of some cactus species belonging to the subfamily Cactoideae. These plastomes bear several unusual and interesting features if compared with the plastome organization found in most angiosperm lineages (Wicke et al. 2011). Currently, cactus plastomes of seven species of the genus Mammilaria (tribe Cacteae; Solórzano et al. 2019) and species of the tribe Echinocereeae (Core Cactoideae I), Carnegiea gigantea (Engelm.) Britton & Rose (Sanderson et al. 2015), and Lophocereus schottii (Engelm.) Britton & Rose are available in the plastid database. We wanted to compare representative species from the Core Cactoideae II with Rhipsalis teres (tribe Rhipsalideae) to answer the following question. Does the complete plastome sequence of a Neotropical Cactoideae species support the structural features and gene content found in the other few Cactaceae plastomes studied so far? The tribe Rhipsalideae includes epiphyte or lithophyte cacti (Anderson 2001). Its distribution occurs mainly in eastern South America, although some species occur in Central and North Americas. The epiphyte R. teres occurs in the Atlantic forest in Brazil (Zappi et al. 2011).
Our data of R. teres plastome revealed new features compared with other cactus plastomes previously reported. We could identify among the plastomes sampled here evolutionary traits shared by them, including remarkable rearrangements, loss of introns, loss or pseudogenization of several genes, highly divergent genes, and several polymorphic RNA editing sites. Moreover, we inferred a phylogenetic analysis, based on concatenated plastid genes, in which the relationships were compared with structural data. Furthermore, we mapped several repetitive sequences in the plastome of R. teres, providing useful molecular markers for future genetic analyses in natural populations. Several evolutionary aspects related to plastome rearrangements, diversity of RNA editing sites, loss of genes and introns, plastid translation, and tRNA import in the subfamily Cactoideae are analyzed and discussed in detail.
2 Materials and method
Plant material, chloroplast isolation, and plastid DNA extraction
–Fresh and young cladodes of R. teres were collected from plants cultivated under greenhouse conditions. The plant material was maintained at 4ºC for 96 h to decrease starch levels (Lopes et al. 2018c). Posteriorly, chloroplast isolation and plastid DNA extraction were carried out according to Vieira et al. (2014).
Sequencing, assembling, and annotation, and data archiving statement
–Approximately 1 ng of chloroplast DNA was used to prepare sequencing libraries with Nextera XT DNA Sample Prep Kit (Illumina Inc., San Diego, CA, USA) according to the manufacturer's instructions. The obtained library was sequenced using Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA). The reads obtained (27,430 reads and an average length of 308.0 bp) were trimmed under the threshold with a probability of error < 0.05 and de novo assembled using CLC Genomics Workbench 8.0.2 software (CLC Bio, Aarhus, Denmark). The contigs used to assemble the R. teres plastome ranged from 658,56 to 258.47 of average coverage. The programs Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. 2004), Annotation of Organellar Genomes (GeSeq; Tillich et al. 2017), and BLAST were used for preliminary gene annotation. From the initial annotation, putative start and stop codons, and intron positions were determined by comparison with homologous genes from plastid genomes available in the organelle database (GenBank). All tRNA genes were meticulously checked by the server tRNAscan-SE (Lowe and Chan 2016). The physical circular map of the plastome was drawn using Organellar Genome DRAW (OGDRAW) (Greiner et al. 2019). The complete data (nucleotide sequences and gene features) of the R. teres plastome were deposited in the GenBank database under accession number MT387452.
Comparative analyses of plastome structure
–Structural features of the family Cactaceae were characterized by comparing the plastome of R. teres with the plastomes from other species of the family Cactaceae available in the Genebank database by multiple alignment analyses using the software Mauve Genome Alignment v2.3.1 (MAUVE) (Darling et al. 2004). As a reference, we used the plastome of Spinacia oleracea L. given that it shows the typical plastome organization found in most angiosperms. The linear maps of the plastid genes were drawn by OGDRAW (Greiner et al. 2019).
Identification and distribution of polymorphic SSRs, tandem repeats, and directed (D) and inverted (I) repeat loci
–Simple sequence repeats (SSRs) were mapped using the MIcroSAtellite (MISA) Perl script (Thiel et al. 2003), with thresholds of eight repeat units for mononucleotide SSRs, four repeat units for di- and trinucleotide SSRs, and three repeat units for tetra-, penta-, and hexanucleotide SSRs. Tandem repeats were identified using the program Tandem Repeats Finder (TRF) (Benson 1999). The parameters were set to 2, 7, and 7 for match, mismatch, and indel, respectively. The minimum alignment score to report repeats and maximum period size were set to 80 and 500, respectively. Subsequently, the repeats were manually verified and nested or redundant data were removed. The program REPuter (Kurtz et al. 2001) was used for inverted repeats (IRs) localization by forward vs. reverse complement (palindromic) alignment. The minimal repeat size was set to 30 bp and the identity of repeats ≥ 90% (hamming distance = 3).
Prediction of RNA editing sites
–Potential RNA editing sites in plastid protein-coding genes of the species belonging to the family Cactaceae, including R. teres, were predicted by the software Predictive RNA Editor for Plants (PREP) suite (Mower 2009). The cutoff value was set to 0.8. Absent genes, pseudogenes, and highly divergent genes found in different species of the family Cactaceae were excluded from this predictive analysis.
Phylogenetic inference
–To infer the R. teres phylogenetic position within the suborder Cactineae, we used the maximum likelihood (ML) method for the phylogenetic reconstruction based on 58 concatenated plastid genes. The taxa sampled for this phylogenetic analysis encompass all species belonging to the suborder Cactineae containing complete plastome sequences available in the GenBank, which included representative species of the families Cactaceae, Basellaceae, Montiaceae, Halophytaceae, Portulacaceae, and Talinaceae. Also, Spinacia oleracea (Chenopodiaceae: Caryophyllales) was used as an outgroup species to root the tree. All plastomes sampled and analyzed here are listed in Supplementary Table S1. For this purpose, plastid genes were firstly extracted from the GenBank database and aligned individually using the software MUSCLE (Edgar 2004) implemented in MEGA 6.0 (Tamura et al. 2013). Gene sequences were concatenated using DnaSP v.6 software (Rozas et al. 2017). Posteriorly, a maximum likelihood tree was constructed using IQTREE v1.6.6 (Nguyen et al. 2015). Five hundred nonparametric bootstrap replications were used to assess branch supports, which generated five partitions grouping the genes in the best substitution models: GTR + F + G4 (atpA, atpB, atpF, petA, psaC, psbH, rbcL, rpl2, rpl14, rpl16, rpoB, rpoC1, rps4, rps7, rps12, rps14, and ycf3); TVM + F + I (atpH, atpI, petB, petD, petG, petN, psaA, psaB, psbA, psbB, psbC, psbD, psbE, psbF, psbJ, psbL, psbN, and psbZ); TVM + F + G4 (atpE, cemA, psaI, psbK, psbM, psbT, rpl36, rpoA rpoC2, rps2, rps8, rps11, rps16, ccsA, matK, and rpl22); TIM3 + F + G4 (clpP); TPM3 + F + G4 (infA, rpl20, rps3, rps15, rps18, and rps19). Lastly, the consensus tree generated in this analysis was visualized using the software FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Gene divergence analysis in plastomes of Cactaceae
–We aligned 60 protein-coding genes found in all 11 plastomes of the family Cactaceae, including R. teres, and other six genes absent in one or more of these species using the software Muscle implemented in Mega 6.0 (Edgar 2004; Tamura et al. 2013). The phylogenetic reconstruction of each gene was performed to assess the gene divergence. The phylogenies were inferred based on the ML method following the same steps abovementioned for the phylogenetic inference using concatenated plastid genes as the dataset. The gene divergence was estimated by the sum of total branch lengths that link the operational taxonomical units to the common ancestor of the species sampled here.
Sliding window analysis
– Hotspots of nucleotide polymorphism of R. teres and other plastomes of the family Cactaceae were inferred by sliding window analysis. Firstly, the sequences of interest were aligned using ClustalW implemented in Mega 6.0 (Tamura et al. 2013). Posteriorly, the sliding window analysis was carried out by the software DnaSP v.6 (Rozas et al. 2017). The window length and the step size were set to 20 and 5 bp, respectively.
3 Results
General features of the R. teres plastome
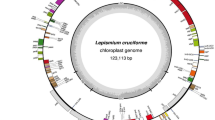
–The R. teres plastome is a circular DNA molecule of 122,389 bp in length with the typical quadripartite structure constituted of two inverted repeat regions, IRA and IRB (IRs), of 8,488 bp each separated by a large single copy (LSC) of 81,397 bp and a small single copy (SSC) of 24,016 bp (Fig. 1). We identified 99 unique genes, which are 66 protein-coding genes, 29 tRNA genes, and four rRNA genes (Table 1). Additionally, ycf2, trnI-CAU, and trnL-CAA genes are completely duplicated in the IRs, while the ycf1 is partially duplicated in these regions. Seven protein-coding genes and five tRNA genes harbor one intron, and the ycf3 gene contains two introns. On the other hand, clpP, rpl2, and rpoC1 genes, which typically harbor introns, lost their introns in the R. teres plastome. Concerning the plastid genes commonly found in most angiosperms, seven of them are absent (trnV-UAC, rpl33, ndhA, ndhE, ndhG, ndhH, and ndhI) and other seven are pseudogenes (ndhB, ndhC, ndhD, ndhF, ndhK, rpl32, and ycf4) in the plastome of R. teres. Part of the clpP and rps19 genes is duplicated in the LSC region, but one of both sequences is not functional (pseudogene).
Gene map of R. teres plastome. Genes drawn inside the circle are transcribed clockwise and genes drawn outside are expressed counterclockwise. Genes belonging to different functional groups are color coded. The darker gray in the inner circle corresponds to GC content, while the lighter gray corresponds to AT content. Dotted circle corresponds to 50% of AT/GC content. Two inverted repeat-regions IRA and IRB divide the rest of the circular DNA molecule into large (LSC) and small (SSC) single-copy regions
Mapping of SSRs, tandem repeats, and dispersed repeats
–We identified a total of 200 SSR loci in the plastome of R. teres of which most of them are mono- (150) and dipolymers (35). In contrast, tri- (4), tetra- (7), penta- (2), hexapolymers (2), and compounds (2) were identified in lower frequency. Most SSRs are composed of A/T bases, which means that 96.66% and 62.86% of all mapped here are mono- and dipolymers, respectively. The size, sequence, and location of the SSRs identified in the R. teres plastome are shown in Supplementary Table S2. From a total of 200 SSRs mapped, 111 are located in intergenic spaces (IGSs), 59 in coding sequences (CDSs), and 31 in introns. The SSRs located in CDSs encompass 26 genes, being the higher frequency identified in the ycf1 (9), ycf2 (7), rpoc2 (7), and accD (4) genes. Most of SSR loci located in introns occur in the genes, atpF (5), trnL-UAA (5), and petD (5).
A total of 23 tandem repeats (TRs) were identified in the plastome of R. teres, which were distributed in CDSs (13) and IGSs (10). The 13 TRs were found in CDSs of five genes, accD (6), ycf1 (4), rps18 (1), rps19 (1), and ycf2 (1). The consensus length of the TRs mapped here ranged from nine to 115 nucleotides and the copy number from two to six. The complete list of TRs mapped in the plastome of R. teres, including location, copy number, and consensus length, is shown in Supplementary Table S3.
Finally, we also mapped all directed and inverted repeats dispersed throughout the plastome sequence of R. teres with a minimum length of 30 bp and an identity of 90%. A total of 80 repeats were identified (Supplementary Table S4), which are divided into 66 directed and 14 inverted repeats. Regarding the directed repeats, 27 out of 66 are located in 27 different IGSs, but several of them are found in the trnI-GAU/rrn16S IGS (18). They were also identified in CDSs (34 out of 66), most of them situated in the accD (18), rps19 (7), and ycf1 (6) genes. The last five pairs are distributed in IGSs and CDSs. Concerning the inverted repeats, two pairs were found in IGSs, nine pairs are located in CDSs, and three pairs were detected in IGSs and CDSs.
Phylogeny of the suborder Cactineae based on plastid genes
–The phylogenetic position of R. teres within the suborder Cactineae was inferred from concatenated sequences of 58 plastid genes obtained from 22 species of the suborder Cactineae and S. oleracea as an outgroup species. The species included in the phylogenetic inference are listed in Supplementary Table S1. The maximum likelihood (ML) analysis produced a consensus tree with a log-likelihood (lnL) of -127.828.834 (Fig. 2).
Cactineae phylogenetic tree of 23 taxa (22 species of the suborder Cactineae and one outgroup species) based on 54 protein-coding plastid genes using the maximum likelihood (ML) method. Numbers (%) associated with branches are ML bootstrap support (BS) values. The branch length is proportional to the inferred divergence level. The scale bar indicates the number of inferred nucleic acid substitutions per site. The position of Rhipsalis teres is highlighted in red. Spinacia oleracea was used to root the tree
The tree topology shows an early divergence among Montiaceae and the other families, which formed a clade composed of Cactaceae, Portulacaceae, Talinaceae, Basellaceae, and Halophytaceae with high bootstrap support (BS). The three genera sampled within Montiaceae constituted a well-supported monophyletic clade (100% of ML-BS), being the genus Calandrinia closely related to Montia forming a sister group to Cistanthe. Concerning the clade composed of Cactaceae, Portulacaceae, Talinaceae, Basellaceae, and Halophytaceae, the family Halophytaceae diverged firstly from the others (97% of ML-BS), and subsequently, the family Basellaceae but with a low ML-BS (51%). Finally, the families Portulacaceae and Talinaceae formed a sister group to Cactaceae, with high branch support (100% of ML-BS). The relationship between Portulacaceae and Talinaceae was poorly supported (54% of ML-BL).
The genus Mammillaria (subfamily Cactoideae, family Cactaceae) is highly supported as a monophyletic clade, which is sister to the group formed by the genera Rhipsalis, Carnegiea, and Lophocereus. Carnegiea and Lophocereus formed a sister group to Rhipsalis, represented by the species R. teres sequenced here, with high branch support (100% of ML-BS). M. albiflora Backeb. (genus Mammillaria) diverged firstly (100% of ML-BS), and posteriorly, M. zephyranthoides Scheidw. (98% of ML-BS) from the other five species. The remaining species diverged into two clades (100% of ML-BS): one formed by M. pectinifera F.A.C. Weber and M. solisioides Backeb., and other formed by M. supertexta Mart. ex Pfeiff. which is sister to the group formed by M. crucigera Mart. and M. huitzilopochtli D.R. Hunt.
Structural features and gene content in Cactaceae plastomes
–Structural analyses of the rearrangements were carried out comparing the R. teres plastome and other plastomes of the family Cactaceae with S. oleracea, which bears the typical plastome structure found in most angiosperms. To facilitate the interpretation of rearrangements, we analyzed separately the LSC region from IR and SSC regions. Aiming to detail the structural dynamics of the IRs, we considered all the sequences including SSC, LSC-IRA border, LSC-IRB border, and IRs.
Comparing the LSC region of S. oleracea, which extend from the trnH-GUG gene to the rps19 gene, with plastomes of other species of the family Cactaceae by multiple alignment analyses, we were able to distinguish two distinct groups of plastomes: The first group includes the species R. teres, C. gigantea, and L. schottii (Supplementary Fig. S1), and the second one contains the species of the genus Mammilaria (Supplementary Fig. S2). These results corroborate with the phylogenetic relationships inferred among these species based on concatenated plastid genes (Fig. 2). R. teres plastome shares one translocation event with C. gigantea and L. schottii (LCB G; Supplementary Fig. S1) and one inversion of a plastome segment (LCB D; Supplementary Fig. S1). The C. gigantea and L. schottii plastomes also share another inversion (LCB E; Supplementary Fig. S1). On the other hand, the LCB E of the plastome of R. teres shows the same direction found in S. oleracea, but demonstrates the same inversion observed in C. gigantea and L. schottii. It indicates that the LCBs D and E inverted in the ancestor plastome of R. teres, C. gigantea, and L. schottii. Posteriorly, a new inversion event involving only the LCB E occurred in an ancestor lineage of R. teres changing the orientation of the genes present in this segment (LCBs D and E; Supplementary Fig. S1). Another unique structural feature of the plastome of R. teres is the presence of an additional inversion in the LSC region (LCB C; Supplementary Fig. S1) involving a large segment. To facilitate the understanding and visualization of the rearrangements that occurred in the plastome of R. teres, we delimitated the blocks of genes involved in the rearrangements using linear gene maps of the plastomes (Fig. 3).
Comparison of gene content and order between Rhipsalis teres, Carnegiea gigantea and Lophocereus schottii plastomes and the Spinacia oleracea plastome that presents the general structure of plastomes found in most angiosperms. The genes involved in primary inversions are highlighted by green squares, while a secondary inversion is highlighted by an orange square. Crossed and straight lines mean inversion and maintenance of the gene order, respectively. Gene losses are pointed out by red squares in the plastome of S. oleracea, which means most ndh genes. The remaining ndh genes in the plastomes of R. teres, C. gigantea, and L. schottii are indicated (* indicates a pseudogene). Black arrows indicate translocation and inversion events of a segment encompassing the clpP and rps12 genes in the plastomes of Cactaceae (subfamily Cactoideae) analyzed here (for more details, see Supplementary Fig. S4). Linear gene maps were drawn by using OGDRAW (Greiner et al. 2019). LSC, large single-copy regions; IRA/B, inverted repeat A/B region; SSC, small single-copy region
Moreover, a large inversion of a segment including the entire typical SSC region and part of the typical IR regions (more than half of the IRB and a small segment of the IRA) was identified in the plastomes of R. teres, C. gigantea and L. schottii (Fig. 3). The loss of several genes is also shared by R. teres, C. gigantea,, and L. schottii plastomes. It includes several genes such as trnV-UAC, rpl33, and most ndh genes. Indeed, from the 11 genes from plastid NDH complex commonly found in most angiosperms, only two are present as pseudogenes (ndhD and ndhB) and the remaining ones were lost from the plastomes of C. gigantea and L. schottii. In addition to the ndhD and ndhB pseudogenes, the R. teres plastome also contains degenerated and nonfunctional sequences of the ndhK, ndhC, and ndhF genes (Fig. 3).
Within the genus Mammilaria, Solórzano et al. (2019) identified three distinct plastome structures among the seven species. The rearrangements found in the LSC region include inversions and translocations of large and small segments (Supplementary Fig. S2; Solórzano et al. 2019). Concerning the rearrangements in the LSC, all the three structures are very similar, except for two additional inversions exclusively identified in only one of the structures (Supplementary Fig. S2; Solórzano et al. 2019). The genes encompassed by the rearrangements are shown in Supplementary Fig. S3. Most of these rearrangements differ from the rearrangements harbored by the plastomes of R. teres, C. gigantea, and L. schottii. Nevertheless, some features are present in all species of the family Cactaceae analyzed here. One of these features is the translocation of the segment involving the clpP gene and the first exon of the rps12 gene (highlighted by arrows in Fig. 3 and Supplementary Fig. S3). This segment is typically located between the rpl20 and psbB genes in S. oleracea, whereas in plastomes of the family Cactaceae it is inverted and found between the trnS-GCU and trnG-UCC genes (Supplementary Fig. S4). Only the segment located between the trnS-GCU and trnG-UCC genes bears a functional copy of the clpP gene without introns, while the other segment contains a nonfunctional fragment of this gene. Except for the R. teres plastome, the segments found in both locations bear a copy of the first exon of the rps12 gene (Supplementary Fig. S4).
Moreover, all plastomes of the family Cactaceae analyzed here bear an inversion between the ndhB and ycf1 genes (green square 3; Fig. 3 and green square 4; Supplementary Fig. S3). This inversion involves part of the IR and all SSC regions from an ancestor plastome, which is analogous to S. oleracea plastome. Despite this conserved large inversion, expansion and contraction events of the IRs in plastomes of the family Cactaceae did not follow the same pattern and consequently affected the sizes and presence of IRs. The structural dynamics of the IRs within Cactaceae are shown in Fig. 4, which bring detailed information related to the presence or absence of the IRs, gene content of the IRs, presence or absence of the SSC between the IRs, and the LSC/IR borders. The size of the IRs in the plastome of R. teres is approximately 8.5 kb, which indicates the occurrence of large contraction events at LSC-IRB and SSC-IRA junctions during the evolution of this species if we compare it with the IR structure found in S. oleracea (Fig. 4).
View of gene content and order of IRs, SSC, and part of the LSC bordering the IRs. The genes located in each region (LSC, IR, and SSC) or a plastome without quadripartite structure are color-coded as indicated by the legend. The red square highlights the block of genes involved in the inversion shared by all plastomes of Cactaceae (subfamily Cactoideae) analyzed here. LSC, large single-copy regions; IRA/B, inverted repeat A/B region; SSC, small single-copy region
The dimensions of LSC, SSC, and IR regions in Cactaceae plastomes are highly variable and especially determined by events of contraction/expansion of IRs (Table 2). Furthermore, some differences in the pattern of gene degeneration and loss among the species also contributed to the variability in the plastome dimensions. All Cactaceae plastomes analyzed here contain the conserved set of four rRNA and 29 tRNA genes including R. teres plastome (Table 1). The functional loss of trnV-UAC gene seems to be a feature of the subfamily Cactoideae since it has degenerated in all species analyzed here. Among the protein-coding genes, only 60 genes are conserved in Cactoideae, whereas the 19 remaining were lost or degenerated in all species or at least in some of them. The pattern of gene degeneration and loss across the phylogeny inferred here is shown in Fig. 5. Besides the loss of the trnV-UAC gene, all species share the loss or degeneration of most ndh genes (9 out of 11 genes). Only the ndhG and ndhJ genes are probably functional in the genera Mammillaria and R. teres, respectively. Lastly, none functional copy of the rpl23 and rpl33 genes was identified in all plastomes of the subfamily Cactoideae. On the other hand, the rpl36, rps16, rps18, ycf1, ycf2, and ycf4 genes are presumably functional in some species, whereas in others are pseudogenes.
The pattern of gene degeneration or loss in species of the subfamily Cactoideae (Cactaceae) plotted across the phylogeny presented in Fig. 2. a Pattern of degeneration or losses within the ndh suite. b Pattern of degeneration and or losses in other plastid genes. The pseudogenes highlighted by (*) are annotated as functional in the database. However, we considered them pseudogenes due to the high reduction of length (sequence size) of the putative coding sequence when compared with other functional sequences
Gene divergence analysis in plastid protein-coding genes within Cactaceae
–From the 66 protein-coding genes identified in plastomes of Cactaceae, our gene divergence analysis indicates that most of them (53 out of 66 genes) are evolving with a low to medium substitution rate (branch length < 0.05 in all species). From the 53 genes, 38 exhibited mean branch lengths below 0.002. Contrarily, the remaining 13 out of 66 genes showed branch lengths > 0.05 in one or more species of the family, being six of them (accD, ycf1, psbM, clpP, infA, and rpl22) the most divergent ones (Supplementary Fig. S5). They demonstrate the extensive variation of branch lengths among the species (Fig. 6), being the highest variation of the values among the species found in the psbM gene. The psbM gene shows a very low substitution rate in R. teres, C. gigantea, and L. schottii, whereas in Mammillaria species its mutation rate ranged from 0.18 (M. albiflora) to 0.49 (M. crucigera). Nevertheless, the highest divergence in the subfamily Cactoideae (Cactaceae) was observed in the accD gene, which exhibited branch lengths ranging from 0.32 (M. crucigera) to 0.51 (M. zephyranthoides). Similarly, the clpP gene also shows branch lengths varying from 0.11 (C. gigantea) to 0.25 (M. huitzilopochtli). Concerning the ycf1 gene, absent in M. zephyranthoides (Fig. 5), exhibited higher substitution rates among Mammillaria species (0.26) in comparison with R. teres (0.13), L. gigantea (0.12), and L. schotti (0,18). A similar pattern was observed for the ycf2 gene (also absent in M. zephyranthoides), which is more divergent in Mammillaria species in comparison with the others. On the other hand, the rpl22 gene shows a lower substitution rate in Mammillaria species in comparison with the others being the highest divergence identified in R. teres. Lastly, the infA gene is also grouped among the highly divergent genes containing branch lengths from 0.05 (R. teres) to 0.12 (M. supertexta).
The sequence of the accD gene in the plastomes of Cactaceae comprises open reading frames (ORFs) of a wide range of lengths among the species analyzed here, potentially encoding proteins from 490 (L. schotti) to 1,347 (M. huitzilopochtli) amino acids. However, most of these ORFs have several ATG found in frame next to the N-terminus region that may work as a start codon. Sliding window analysis involving the entire region encompassing the putative accD gene and surrounding genes shows a high level of nucleotide diversity in almost all sequence of the accD gene (Fig. 7a). This analysis was based on the alignment among the closely related species R. teres, C. giganteae, and L. schottii. The species of the genus Mammillaria were excluded for the reason that rearrangements found in this region hinder adequate alignments. The hotspots of nucleotide polymorphisms are concentrated on the region around the N-terminus of the gene and its 5′ untranslated region (5′-UTR) next to the trnM-CAU gene. Only the C-terminus of the sequence of accD gene is conserved (Pi above 0.2) as well as surrounding genes (Fig. 7a). The alignment involving the ORFs that putatively encodes the accD gene from all cactus species analyzed here shows that the conserved C-terminus also occurs in the species of the genus Mammillaria. Putative start codons were mapped by alignment analyses of the region around the N-terminus, which resulted in proteins containing different sizes as follows: 490 (L. schottii), 831 (M. pectinifera), 910 (M. zephyranthoides), 994 (M. albiflora), 1,066 (C. gigantea), 1,104 (M. solisioides), 1,114 (R. teres), 1,129 (M. supertexta), 1,205 (M. crucigera), and 1,211 (M. huitzilopochtli) amino acids.
Sliding window analyses. a Plastome region around the accD gene and its neighboring genes among related species R. teres, C. gigantea, and L. schottii. b and c Plastome regions involved in the translocation and inversion of a segment encompassing the clpP gene and the first exon of the rps12 gene (see more details in Supplementary Fig. S4). The genes are color-coded following the plastome map of R. teres in Fig. 1. Genes highlighted with (*) are degenerated in one or more cacti species. Pi, nucleotide diversity. Window length, 20 bp. Step size, 5 bp
To understand the evolution of the clpP gene and the first exon of the rps12 gene in the family Cactaceae, both genes located in the duplicated and inverted segment identified here (Supplementary Figs. S3 and S4; Fig. 3), we also performed a sliding windows analysis comprising both duplicated segments and surrounding genes (Fig. 7b and 7c). The region containing the clpP gene (Fig. 7b) shows higher peaks of nucleotide diversity around the clpP in comparison with the copy found in the other region (Fig. 7c). The first one (Fig. 7b) is functional given that the other segment contains a degenerated copy of the clpP gene (Supplementary Fig. S4). Differently, the first exon of the rps12 gene in the duplicated segment (Fig. 7c) shows a higher conservation rate in comparison with the sequence found in Fig. 7b. The alignment of both duplicated sequences enwrapping the rps12 gene shows that in the plastomes of R. teres and M. zephyranthoides the duplicated isoform (Fig. 7b) degenerated into a pseudogene (Supplementary Fig. S6). The sequence found in the plastome of R. teres is only a fragment that does not match with the sequences found in other species of Cactaceae (Supplementary Fig. S4); therefore, we did not consider it in the plastome annotation of R. teres. Expect for R. teres and M. zephyranthoides, all the other species seem to bear two functional copies of the rps12 gene. Phylogenetic relationships covering the duplicated sequences indicate that both sequences of rps12 are highly conserved among species of the genus Mammillaria, C. gigantea, and L. schotti (Supplementary Fig. S7).
Prediction of RNA editing sites in plastid genes of the family Cactaceae
–The predictive analysis of the RNA editing sites in plastid protein-coding genes of the family Cactaceae was performed using the program PREP, which revealed a total of 39 sites distributed in 15 genes. All putative RNA editing sites occurred in the first (33%) or second (67%) codon positions and all editions changed a cytidine (C) to a uridine (U). Most putative RNA editing sites (59%) changed the encoded amino acid from polar to apolar (23 out of 39). Only three editions induced amino acid changes from apolar to polar. The other 13 editions do not change the amino acid polarity (eight apolar–apolar changes and five polar–polar changes). A list of the RNA editing sites, targeted genes, and amino acid changes identified in the family Cactaceae is shown in Supplementary Table S5.
Only 13 out of 39 sites are shared by all cactus species, the other 26 sites potentially represent polymorphic RNA editing sites within Cactaceae. To identify the pattern of events of gain or loss of RNA editing sites in Cactaceae, we plotted the polymorphic RNA editing sites against the phylogeny inferred here (Fig. 8). Nine RNA editing sites occur specifically among the taxa sampled, being rpoC1(157) and rpoC2(606) in R. teres, rpl2(77) and rpoC1(530) in L. schottii, atpA(354) in M. supertexta, and rpl2(159), rpoB(172), rpoB(519), and rpoB(809) in M. zephyranthoides. Another seven RNA editing sites appear distinctively in closely related species: three in all species of Mammillaria [petB (163), rpoC2 (756), and rps2 (20)], three shared by C. gigantea and L. schottii [atpF (14), psbF (26), and rpoC2 (770)], and one present in the clade including R. teres, C. gigantea, and L. schottii [rpoA (28)]. Five sites are widely distributed in almost all species sampled here, except in one or few related taxa, which dismiss the editions by the occurrence of mutations C-to-T in the plastid DNA: atpA (305), matK (115), rpl20 (87), rpoB (184), and rps2 (83). The site rpoC2 (658) occurs in R. teres and the group of three related species within Mammillaria. Lastly, four RNA editing sites were identified in amino acid positions that are not conserved in these proteins within Cactaceae: atpA (354), rpoA (294), rpoC1 (45), e rpoC2 (809).
Distribution of putative RNA editing sites across the subfamily Cactoideae (Cactaceae) phylogeny based on concatenated plastid genes as shown in Fig. 2. The codons highlighted in red have an editing site and the codons in blue codify non-conserved amino acids
4 Discussion
Cactineae phylogeny and relationships within Cactaceae
–The suborder Cactineae (heterotypic synonym: Portulacineae) comprises eight families: Anacampserotaceae, Basellaceae, Cactaceae, Didiereaceae, Halophytaceae, Montiaceae, Portulacaceae, and Talinaceae (Ocampo and Columbus 2010). Here, we presented a phylogenetic analysis based on concatenated plastid genes of suborder Cactineae including taxa from six of these families. The families Anacampserotaceae and Didiereaceae were not sampled here because no complete plastome is available in the organelle database (Genebank). A consensus among several phylogenetic approaches is the position of Montiaceae as the first divergent family (including the phylogeny showed here), which is sister to all other families of the suborder (Ocampo and Columbus 2010; Moore et al. 2018; Walker et al. 2018; Wang et al. 2019). Nevertheless, several incongruences remain to be resolved concerning relationships among the families within the suborder Cactineae. The family Basellaceae is a classic example reported as sister to Didiereaceae in a Cactineae phylogeny based on plastid sequences (Ocampo and Columbus 2010), whereas it is sister to Halophytaceae if nuclear sequences are used for the inference (Moore et al. 2018).
The position of the family Cactaceae has been contradictory within the order Caryophyllales in various inferences. Cactaceae is sister to a clade composed of Portulacaceae and Anacampserotaceae in phylogenetic analyses using transcriptomic (Walker et al. 2018; Wang et al. 2019) and genomic data (Moore et al. 2018). On the other hand, a phylogeny based on plastid sequences revealed Cactaceae as sister to only Portulacaceae, being Cactaceae and Portulacaceae sister to Anacampserotaceae (Ocampo and Columbus 2010). In our Cactineae phylogeny, Cactaceae is sister to a clade composed of Portulacaceae and Talinaceae. However, the absence of complete plastome sequences from the family Anacampserotaceae impedes us to infer relationships among Cactaceae and other families. Similarly, the absence of data from Didiereaceae hinders us to accurately infer about the relationships of the family Basellaceae. It is noteworthy to note that most branches are well supported, except for the bifurcation of the family Basellaceae, the clade including Portulacaceae, Talinaceae, and Cactaceae (51% of BS), and the bifurcation indicating Portulacaceae sister to Talinaceae (54% of BS). Therefore, the increase in plastome sequences of different taxa mainly of the families Anacampserotaceae and Didiereaceae will allow us to infer with accuracy the phylogeny of the suborder Cactineae.
Concerning the family Cactaceae, we sampled here for the phylogenetic inference of nine species containing the complete plastome available in the organelle database and R. teres that we sequenced and reported here. The species belong to four genera: Carnegiea (1), Lophocereus (1), Mammillaria (7), and Rhipsalis (1). All these genera belong to the subfamily Cactoideae: the genus Mammillaria classified into the tribe Cacteae, which is sister to the Core Cactoideae including the other three genera. Within the Core Cactoideae, the genera Carnegiea and Lophocereus belong to the Core Cactoideae I, whereas Rhipsalis belongs to the Core Cactoideae II (Guerrero et al. 2019). The relationships of the plastid phylogenetic inferences that we reported for the family Cactaceae are following these classifications. Furthermore, plastome structural analyses and comparison among the species revealed similar patterns of classification: Carnegiea (Sanderson et al. 2015) and Lophocereus plastomes are similar and closely related to Rhipsalis plastome. Similarly, the three different plastome structures of Mammillaria (Solórzano et al. 2019) are closely related to each other and formed a clade distinct from Carnegiea, Lophocereus, and Rhipsalis.
Evolutionary trends in plastomes of the subfamily Cactoideae (Cactaceae): rearrangements, gene losses, highly divergent genes, and polymorphism of RNA editing sites
–Overall, plastomes of land plants contain a conserved set of genes and a typical quadripartite structure composed of two single-copy regions (LSC and SSC) separated by two inverted repeats (IRs) (Wicke et al. 2011; Rogalski et al. 2015; Daniell et al. 2016). Normally, IR borders have a small variation in most angiosperms, but some plant lineages show diversified IRs containing expansion and/or contraction events affecting significantly gene order and content or even absence of IRs (Zhu et al. 2016; Lopes et al. 2018b; Shrestha et al. 2019). Unusual gene order has been also reported in the single-copy regions, especially in the LSC, because of complex rearrangements undergone by ancestral plastomes (Lopes et al. 2019; Shrestha et al. 2019; Solórzano et al. 2019).
Another unusual evolutionary event observed in plastomes of some plant lineages is a massive loss of genes, which is an extremely rare phenomenon in photosynthetic plants (Wicke et al. 2011). Some of these unusual features seem to evolve as isolated cases in one or few species belonging to a plant group containing conserved plastomes such as the family Arecaceae (Barret et al. 2016; Lopes et al. 2018a, 2019). On the other hand, several plant lineages containing unusual plastomes genuinely bear a set of uncommon features, which include several rearrangements, expanded and/or contracted IRs, pseudogenes, and losses of genes and introns. Among these lineages, we can highlight several angiosperm families Geraniaceae, Campanulaceae, Passifloraceae, and Fabaceae (Cai et al. 2008; Haberle et al. 2008; Guisinger et al. 2011; Shrestha et al. 2019). Similarly, previous reports related to C. gigantea and Mammillaria plastomes (Sanderson et al. 2015; Solórzano et al. 2019) revealed a massive loss of genes, various pseudogenes, and complex rearrangements, which place the family Cactaceae among the plant lineages with unusual plastome evolution.
To comprehend in more detail the evolution of plastomes within Cactaceae, we collected all complete plastomes available in the organelle database and added to them the plastome of R. teres completely sequenced here. If we consider structural aspects of plastomes in Cactaceae demonstrated so far, our data suggest that the rearrangements found in the single-copy regions occurred earlier and are more conserved than contraction/expansion events characterized in the IRs. These rearrangements follow a phylogenetic tendency within the subfamily Cactoideae by showing distinct patterns of rearrangements between the tribe Cacteae (represented here by Mammillaria) and the Core Cactoideae (represented here by R. teres, C. gigantea, and L. schottii). Nevertheless, two rearrangements are present in all plastomes of these species, which may be candidates for using as phylogenetic markers at the level of subfamily or even family. One of them is the translocation of a segment including the clpP and rps12 (first exon) genes, which resulted in the duplication of the segment. Curiously, the functional copy of the clpP gene seems to be the new one that emerged after the translocation event. The clpP is one of the most divergent genes identified in the plastomes of the family Cactaceae sampled here (Fig. 6) as well as the loss of two introns commonly found in plastomes of most angiosperms. Diversely, in most species analyzed both sequences of the rps12 gene seem to be functional, conserving most amino acids in their putative translated sequence. Experimental approaches will be necessary to investigate whether both sequences are correctly transcribed, spliced, and translated (Hildebrand et al. 1988). Unlike the other species, R. teres shows an advanced degeneration of the duplicated isoform of the rps12 gene, which resulted in the loss of this isoform and the maintenance of only one functional gene. The second structural feature conserved among the cacti analyzed here is the inversion of a large segment comprising most of IRB, all SSC, and a small part of the IRA, considering the quadripartite structure of a typical plastome (Supplementary Fig. S8). Presumably, this complex inversion changed dramatically the typical plastome structure by generating largely directed tandem repeats involving two remaining segments from both IRs. Subsequently, the posterior deletion of one of the tandem repeats by homologous recombination originated the putative structure of the ancestral plastome (Supplementary Fig. S8). If we consider this hypothetic ancestral plastome, the diversity of IR boundaries (or complete loss of IRs) could be produced by specific and recent events of IR contraction and/or expansion.
The plastomes of the subfamily Cactoideae undergone a massive loss of genes, which accounts for 12 genes, among pseudogenes and completely lost ones. Most of them belong to the ndh suite, which encodes subunits of the plastid NAD(P)H dehydrogenase-like (NDH) complex. The NDH complex acts in ATP generation by participating in the cyclic electron flow (Strand et al. 2019). Mutant plants lacking this complex did not show a visible phenotype under favorable growth conditions, but growth and development were affected by environmental stress conditions (Horváth et al. 2000). Several lineages across land plants contain pseudogenes or lost from one to all genes encoding subunits of the NDH complex from plastome (Martín and Sabater 2010; Blazier et al. 2011; Lin et al. 2015, 2017; Ni et al. 2017; Strand et al. 2019). No evidence of functional transfer to the nucleus has been reported for any ndh subunit in the cactus C. gigantea (Sanderson et al. 2015) and even other species lacking plastid ndh genes (Li et al. 2015, 2017; Ruhlman et al. 2015; Strand et al. 2019). It is still unclear why some plant lineages evolved under relaxed selective pressure for retention or loss of ndh genes and what is the relationship among the absence of the NDH complex and a particular lifestyle or ecological niche (Strand et al. 2019).
Additionally, several plastid ribosomal protein genes were lost or are pseudogenes within the subfamily Cactoideae. The rpl23 and rpl33 are pseudogenes or were lost in all species of this subfamily sequenced to date. Both genes encode ribosomal proteins of the large subunit 50S (Rogalski et al. 2008b; Fleischmann et al. 2011). In spinach (family Amaranthaceae, order Caryophyllales), the prokaryotic-type ribosomal protein L23 (encoded by rpl23 gene) was replaced by a eukaryotic-type L23 version encoded by the nucleus and imported into the plastids (Bubunenko et al. 1994; Schmitz-Linneweber et al. 2001). Similarly, the rpl23 is a pseudogene in other families of the order Caryophyllales such as Polygonaceae (Logacheva et al. 2008), and Caryophyllaceae (Sloan et al. 2014; Raman and Park 2015). Given its essential role in plastid translation (Fleischmann et al. 2011), it is highly likely that a eukaryotic-type L23 also replaces the function of the plastid rpl23 gene in Cactaceae. On the other hand, the rpl33 gene is not essential for plastid translation but is required under stress conditions (Rogalski et al. 2008b). Its function during plastid translation is highly required in the initial phase of seedling growth in which a massive protein biosynthesis is necessary for the formation of protein complexes in the thylakoid membranes. If we consider that cacti grow slowly in their natural environments, a relaxed pressure to maintain the rpl33 gene functional may be expected (Rogalski et al. 2008b; Ehrnthaler et al. 2014). To investigate a possible gene transfer to the nucleus of the rpl33 gene, we checked the nucleotide collection of Cactaceae database using the rpl33 sequence of Portulaca oleracea L.. Interestingly, we found some partial CDS of five cactus species containing high query cover (83–98%) and identity (92–98%). Curiously, three out of them (Blossfeldia liliputana Werderm., Maihuenia poeppigii (Otto ex Pfeiff.) F.A.C.Weber, and Weingartia kargliana Rausch), are cacti found in high altitudes of the Andean region (Anderson 2001). The other two species, Pereskiopsis diguetii (F.A.C.Weber) Britton & Rose and Pereskia sacharosa Griseb., belong to the early diverging lineages within Cactaceae that bear the plesiomorphic morphological states of cacti (Anderson 2001; Guerrero et al. 2019). Curiously, the loss or pseudogenization of the rpl33 gene in angiosperm is an extremely rare event; it occurred only in some legume species (Guo et al. 2007; Tangphatsornruang et al. 2010). The other three genes (rps16, rps18, and rpl36) encoding ribosomal proteins are pseudogenes in most species of the subfamily Cactoideae (Cactaceae). The rps16 and rps18 genes encode essential ribosomal proteins of the small subunit (30 S) and are essential for cellular viability (Rogalski et al. 2006; Fleischmann et al. 2011). Considering their essential function on plastid ribosome, it is reasonable to suggest a possible transfer of these genes to the nucleus as observed for some ribosomal protein genes in several angiosperm lineages. Although the rpl36 gene is conserved in angiosperm plastomes, it is not essential for plastid translation, but its absence revealed severe morphological aberrations, almost incapacity of autotrophic growth, and no seed production (Fleischmann et al. 2011). Rationally, most cactus plants grow slowly in natural conditions, which theoretically require a less demand for protein biosynthesis. Thus, we cannot discard in these plants a possible adaptation of the ribosome to slow growth.
Also, we detected the loss of the trnV-UAC gene in all plastomes of the subfamily Cactoideae analyzed here. Since it is a very important and rare evolutionary feature, it was overlooked in the previous reports related to plastome sequences of Cactaceae (Sanderson et al. 2015; Solórzano et al. 2019). According to conventional wobble rules of plastomes of higher plants, two tRNAs for the amino acid valine, tRNA-Val(UAC) and tRNA-Val(GAC), are encoded by the trnV-UAC and trnV-GAC genes, respectively (Bock 2007; Alkatib et al. 2012b). However, only the tRNA-Val(GAC) is encoded by the plastomes of the subfamily Cactoideae (Cactaceae) analyzed here. Interestingly, a study based on reverse genetics in plastids concerning the contributions of wobbling and superwobbling to plastid translation showed that the trnV-UAC gene is essential for cellular viability, whilst the trnV-GAC gene is dispensable (Alkatib et al. 2012b). Consequently, the lack of the tRNA-Val(UAC) would impair plastid translation (Rogalski et al. 2008a; Alkatib et al. 2012a, b). Thus, the tRNA data obtained from the species of the subfamily Cactoideae analyzed here indicate a specific mechanism, which supplies the plastids with tRNA-Val(UAC) from the cytosol. In plants, tRNA import from the cytosol to mitochondria has been reported for all tRNAs that are not encoded by the mitochondrial genome (Dietrich et al. 1996; Alfonzo and Söll 2009; Salinas-Giegé et al. 2015; Murcha et al. 2016). Although the tRNA-Val(UAC) is essential for plastid translation and cell viability (Alkatib et al. 2012b), no evidence of tRNA import into plastids was demonstrated to date for photosynthetic plants (Legen et al. 2007; Rogalski et al. 2008a; Alkatib et al. 2012a, b; Agrawal et al. 2020).
Moreover, our analyses focusing on plastid protein-coding genes pointed out highly divergent genes and several polymorphic RNA editing sites in Cactaceae. Some genes such as accD, ycf1, psbM, clpP, and rpl22 showed branch length values above 0.2 in one or more species. The accD gene is the highest divergent gene in Cactaceae according to our data. The plastome of R. teres, contains mainly in the N-terminus several tandem repeats. This gene encodes the β-carboxyl transferase subunit of the heteromeric acetyl-CoA carboxylase (ACCase), which is responsible for the catalysis of the first committed step of the fatty acids biosynthesis in plastids (Rogalski and Carrer 2011; Salie and Thelen 2016). This gene is essential for cell viability (Kode et al. 2005). The accD gene was considered a pseudogene in Mammilaria and Carnegiea plastomes (Sanderson et al. 2015; Solórzano et al. 2019). However, all accD genes analyzed here have conserved C-terminus sequences, which is located in the carboxyl transferase domain (Lee et al. 2004). Although the accD sequence is highly divergent in length among the cactus species, the conservation of carboxyl transferase domain argues in favor of the functionality of accD in the subfamily Cactoideae (Cactaceae) as observed in other plant lineages such as Passiflora (Rabah et al. 2019; Shrestha et al. 2019; Pacheco et al. 2020a, b, c).
RNA editing found in plant organelles is a post-transcriptional mechanism, which frequently changes cytidines (C) to uridines (U) or more rarely U to C (Bock 2000; Takenaka et al. 2013; Ichinose and Sugita 2016). Overall, the changes occur in the first or the second position of the codons and can alter the amino acid and/or create start and stop codons (Takenaka et al. 2013; Ichinose and Sugita 2016). In our analysis, we identified 26 polymorphic RNA editing sites among the species sampled here of the subfamily Cactoideae. The distribution pattern of these sites across the Cactoideae phylogeny suggests that 16 sites may represent recent acquisitions of specific RNA editing sites. Such results indicate a very dynamic and fast-evolving RNA editing mechanism acting within the subfamily Cactoideae. Among the recent gains of RNA editing sites, nine of them occur in a different species: the rpoC1 (157) and rpoC2 (606) sites in R. teres; the rpl2 (77) and rpoC1 (530) sites in L. schottii; the rpl2 (159), rpoB (172), rpoB (519), and rpoB (809) sites in M. zephyranthoides; and the atpA (354) site in M. supertexta. The other four sites possibly evolved in an ancestral plastome of closely related species, which are the atpF (14), and rpoC2 (770) sites shared by C. gigantea and L. schottii, and the matK (115) site shared by a clade within Mammillaria. Lastly, three sites rpoA (294), rpoC1 (45), and rpoC2 (809) may represent acquisitions that evolved independently across the subfamily Cactoideae. Some of the polymorphic RNA editing sites found in the subfamily Cactoideae seem to be lost recently via mutations that fixed a T in the plastid DNA at the RNA editing site. These losses are the atpA (305) (M. albiflora), matK (218) (L. schotti and C. gigantea), rpl20 (87) (M. zephyranthoides), rpoB (184) (Mammillaria), and rps2 (83) (M. albiflora). Finally, the limitation of taxa in our Cactoideae phylogeny impedes us to infer the evolutionary pattern of the RNA editing sites, petB (163), rpoA (28), rpoC2 (756), rpoC2 (809), and rps2 (20). We presented here the first mapping analysis of RNA editing sites in the subfamily Cactoideae and family Cactaceae. If we compare the RNA editing sites found here with other species of Caryophyllales such as S. oleracea, a significantly different pattern is detected for the species of Cactaceae. In the plastome of S. oleracea, as also observed in most angiosperms, the ndh genes accumulate most RNA editing sites (Tsudzuki et al. 2001), while the ndh suite has undergone significant degeneration in the subfamily Cactoideae (Sanderson et al. 2015; Solórzano et al. 2019).
Importance of plastid markers for genetic studies within Cactaceae
The family Cactaceae is among the most threatened taxonomic groups mainly due to high anthropogenic pressures on their habitats and devaluation of its ecological and economic importance. Some plastome regions containing high mutation rate, such as the SSR loci, represent useful molecular markers due to their high levels of intraspecific nucleotide polymorphism combined with the nonrecombinant nature and uniparental inheritance of plastids in most land plants. Such plastid sequences have been explored in a wide range of genetic studies in natural populations (Provan et al. 2001; Rogalski et al. 2015). Here, we identify 200 SSR loci in the R. teres plastome, most of them in noncoding sequences, which are useful for assessing the genetic diversity of R. teres natural populations and other species of the genus. Additionally, the accD gene represents a promising plastid marker for genetic studies given that it is a highly divergent gene and contains several tandem repeats in its sequence. Moreover, clpP and accD gene sequences may be useful to complement datasets to be utilized for accurate phylogenies at lower taxa in Cactaceae.
The cactus lineage diverged from closely related species approximately 30–35 million years ago (Mya), which is associated with the emergence of pronounced morphological, anatomical, and metabolic adaptive changes. If we consider the evolutionary modifications undergone by species of the subfamily Cactoideae (Cactaceae), it is possible to connect several unusual features of plastomes with environmental adaptation and speciation since many multiprotein complexes in plastids have chimeric composition, which is composed of subunits encoded by the nucleus and plastomes, and a coevolution of nuclear and plastid genomes may play a significant role in speciation (Greiner and Bock 2013). Here, we reported the complete plastome of R. teres, an epiphytic cactus species belonging to the Core Cactoideae. Although some rearrangements mapped here are specific for one or few related species, two rearrangements configure structural features found in all analyzed cactus plastomes. The first one is a translocation of the duplicated segment including the clpP gene and the first exon of rps12 gene, and the other is a large inversion of the segment including the set of genes from ndhB to ycf1. The second rearrangement probably determined a reorganization of the IR boundaries creating two large tandem repeats. Posteriorly, deletion of one repeat by homologous recombination originated an ancestor plastome that served as a basic structure for all early contraction and/or expansion events that gave rise to the diversified IR boundaries found among these cactus plastomes. Moreover, we mapped all SSRs and other repetitive sequences in R. teres plastome, which represent useful sources of molecular markers to be applied in genetic studies in natural populations. Other common features among these cactus plastomes are several losses and pseudogenization of important genes, including most ndh genes, rpl23, rpl33, and trnV-UAC genes. Additionally, highly divergent genes and many polymorphic RNA editing sites were also characterized. According to our analyses, the loss or pseudogenization of rpl33 (C. gigantea, L. schottii; M. albiflora, M. crucigera, M. huitzilopochtli, M. pectinifera, M. solisioides, and M. supertexta) and rpl36 (M. albiflora, M. crucigera, M. huitzilopochtli, M. pectinifera, M. solisioides, and M. supertexta) genes may suggest a relaxed plastid translation in these species, which is presumably allowed by the slow growth of cactus species. Moreover, the lack of the trnV-UAC gene indicates that a mechanism of tRNA import from cytosol was developed by the species of the subfamily Cactoideae given that plastid translation cannot work without this tRNA according to wobbling and superwobbling rules. Finally, our data show that plastomes of Cactaceae undergone unique features during the evolution, which can be directly related to environmental adaptation and speciation.
References
Agrawal S, Karcher D, Ruf S, Bock R (2020) The functions of chloroplast Glutamyl-tRNA in translation and tetrapyrrole biosynthesis. Plant Physiol 183:263–276. https://doi.org/10.1104/pp.20.00009
Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R (2010) Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell 22:2838–2855. https://doi.org/10.1105/tpc.110.073908
Alfonzo JD (2009) Söll D (2009) Mitochondrial tRNA import – the challenge to understand has just begun. Biol Chem 390:717–722. https://doi.org/10.1515/BC.2009.101
Alkatib S, Fleischmann T, Scharff LB, Bock R (2012a) Evolutionary constraints on the plastid tRNA set decoding methionine and isoleucine. Nucleic Acids Res 40:6713–6724. https://doi.org/10.1093/nar/gks350
Alkatib S, Scharff LB, Rogalski M, Fleischmann TT, Matthes A, Seeger S, Schöttler MA, Ruf S, Bock R (2012b) The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet 8:e1003076. https://doi.org/10.1371/journal.pgen.1003076
Anderson EF (2001) The cactus family. Timber Press, Portland, Oregon
Arakaki M, Christin PA, Nyffeler R, Lendel A, Eggli U, Ogburn RM, Spriggs E, Moore MJ, Edwards EJ (2011) Contemporaneous and recent radiations of the world’s major succulent plant lineages. Proc Natl Acad Sci USA 108:8379–8384. https://doi.org/10.1073/pnas.1100628108
Barrett CF, Baker WJ, Comer JR, Conran JG, Lahmeyer SC, Leebens-Mack JH, Li J, Lim GS, Mayfield-Jones DR, Perez L, Medina J, Pires JC, Santos C, Stevenson DW, Zomlefer WB, Davis JI (2016) Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol 209:855–870. https://doi.org/10.1111/nph.13617
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. https://doi.org/10.1093/nar/27.2.573
Blazier JC, Guisinger MM, Jansen RK (2011) Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol 76:263–272. https://doi.org/10.1007/s11103-011-9753-5
Bock R (2000) Sense from nonsense: how the genetic information of chloroplasts is altered by RNA editing. Biochimie 82:549–557. https://doi.org/10.1016/s0300-9084(00)00610-6
Bock R (2007) Structure, function, and inheritance of plastid genomes. In: Bock R (ed) Cell and molecular biology of plastids. Springer, Berlin, pp 29–63
Bock R (2017) Witnessing genome evolution: experimental reconstruction of endosymbiotic and horizontal gene transfer. Annu Rev Genet 51:1–22. https://doi.org/10.1146/annurev-genet-120215-035329
Bubunenko MG, Schmidt J, Subramanian AR (1994) Protein substitution in chloroplast ribosome evolution. A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J Mol Biol 240:28–41. https://doi.org/10.1006/jmbi.1994.1415
Cai Z, Guisinger M, Kim HG, Ruck E, Blazier JC, McMurtry V, Kuehl JV, Boore J, Jansen RK (2008) Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol 67:696–704. https://doi.org/10.1007/s00239-008-9180-7
Daniell H, Lin CS, Yu M, Chang WJ (2016) Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol 17:134. https://doi.org/10.1186/s13059-016-1004-2
Darling AC, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. https://doi.org/10.1101/gr.2289704
Díaz M, Barba de la Rosa AP, Héliès-Toussaint C, Guéraud F, Nègre-Salvayre A (2017) Opuntia spp.: characterization and benefits in chronic diseases. Oxid Med Cell Longev 2017:8634249. https://doi.org/10.1155/2017/8634249
Dietrich A, Small I, Cosset A, Weil JH, Maréchal-Drouard L (1996) Editing and import: strategies for providing plant mitochondria with a complete set of functional transfer RNAs. Biochimie 78:518–529. https://doi.org/10.1016/0300-9084(96)84758-4
Drescher A, Ruf S, Calsa T, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22:97–104. https://doi.org/10.1046/j.1365-313x.2000.00722.x
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Ehrnthaler M, Scharff LB, Fleischmann TT, Hasse C, Ruf S, Bock R (2014) Synthetic lethality in the tobacco plastid ribosome and its rescue at elevated growth temperatures. Plan Cell 26:765–776. https://doi.org/10.1105/tpc.114.123240
Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schöttler MA, Bock R (2011) Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23:3137–3155. https://doi.org/10.1105/tpc.111.088906
Goettsch B, Hilton-Taylor C, Cruz-Piñón G et al (2015) High proportion of cactus species threatened with extinction. Nat Plants 1:15142. https://doi.org/10.1038/nplants.2015.142
Greiner S, Bock R (2013) Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35:354–365. https://doi.org/10.1002/bies.201200137
Greiner S, Lehwark P, Bock R (2019) OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res 47:W59–W64. https://doi.org/10.1093/nar/gkz238
Griffiths H, Males J (2017) Succulent plants. Curr Biol 27:R890–R896. https://doi.org/10.1016/j.cub.2017.03.021
Guerrero PC, Majure LC, Cornejo-Romero A, Hernández-Hernández T (2019) Phylogenetic relationships and evolutionary trends in the cactus family. J Hered 110:4–21. https://doi.org/10.1093/jhered/esy064
Guisinger MM, Kuehl JV, Boore JL, Jansen RK (2011) Extreme reconfiguration of plastid genomes in the angiosperm family geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol 28:583–600. https://doi.org/10.1093/molbev/msq229
Guo X, Castillo-Ramírez S, González V, Bustos P, Fernández-Vázquez JL, Santamaría RI, Arellano J, Cevallos MA, Dávila G (2007) Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8:228. https://doi.org/10.1186/1471-2164-8-228
Haberle RC, Fourcade HM, Boore JL, Jansen RK (2008) Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol 66:350–361. https://doi.org/10.1007/s00239-008-9086-4
Hernández-Hernández T, Brown JW, Schlumpberger BO, Eguiarte LE, Magallón S (2014) Beyond aridification: multiple explanations for the elevated diversification of cacti in the New World Succulent Biome. New Phytol 202:1382–1397. https://doi.org/10.1111/nph.12752
Hildebrand M, Hallick RB, Passavant CW, Bourque DP (1988) Trans-splicing in chloroplasts: the rps 12 loci of Nicotiana tabacum. Proc Natl Acad Sci USA 85:372–376. https://doi.org/10.1073/pnas.85.2.372
Horváth EM, Peter SO, Joët T, Rumeau D, Cournac L, Horváth GV, Kavanagh TA, Schäfer C, Peltier G, Medgyesy P (2000) Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol 123:1337–1350. https://doi.org/10.1104/pp.123.4.1337
Ichinose M, Sugita M (2016) RNA editing and its molecular mechanism in plant organelles. Genes 8:5. https://doi.org/10.3390/genes8010005
Inglese P, Mondragon C, Nefzaoui A, Sáenz C (2017) Crop ecology, cultivation and uses of cactus pear. Food and Agriculture Organization of the United Nations (FAO) and International Center for Agricultural Research in the Dry Areas (ICARDA)
Kikuchi S, Asakura Y, Imai M, Nakahira Y, Kotani Y, Hashiguchi Y, Nakai Y, Takafuji K, Bédard J, Hirabayashi-Ishioka Y, Mori H, Shiina T, Nakai M (2018) A Ycf2-FtsHi heteromeric AAA-ATPase complex is required for chloroplast protein import. Plant Cell 30:2677–2703. https://doi.org/10.1105/tpc.18.00357
Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339:571–574. https://doi.org/10.1126/science.1229262
Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44:237–244. https://doi.org/10.1111/j.1365-313X.2005.02533.x
Krech K, Ruf S, Masduki FF, Thiele W, Bednarczyk D, Albus CA, Tiller N, Hasse C, Schöttler MA, Bock R (2012) The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol 159:579–591. https://doi.org/10.1104/pp.112.196642
Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R (2001) REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res 29:4633–4642. https://doi.org/10.1093/nar/29.22.4633
Lee SS, Jeong WJ, Bae JM, Bang JW, Liu JR, Harn CH (2004) Characterization of the plastid-encoded carboxyltransferase subunit (accD) gene of potato. Mol Cells 17:422–429
Legen J, Wanner G, Herrmann RG, Small I, Schmitz-Linneweber C (2007) Plastid tRNA Genes trnC-GCA and trnN-GUU are essential for plant cell development. Plant J 51:751–762. https://doi.org/10.1111/j.1365-313X.2007.03177.x
Lin CS, Chen JJ, Huang YT, Chan MT, Daniell H, Chang WJ, Hsu CT, Liao DC, Wu FH, Lin SY, Liao CF, Deyholos MK, Wong GKS, Albert VA, Chou ML, Chen CY, Shih MC (2015) The location and translocation of ndh genes of chloroplast origin in the Orchidaceae family. Sci Rep 5:9040. https://doi.org/10.1038/srep09040
Lin CS, Chen JJW, Chiu CC et al (2017) Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J 90:994–1006. https://doi.org/10.1111/tpj.13525
Logacheva MD, Samigullin TH, Dhingra A, Penin AA (2008) Comparative chloroplast genomics and phylogenetics of Fagopyrum esculentum ssp Ancestrale - a wild ancestor of cultivated buckwheat. BMC Plant Biol 8:59. https://doi.org/10.1186/1471-2229-8-59
Lopes AS, Pacheco TG, Nimz T, Vieira LN, Guerra MP, Nodari RO, de Souza EM, Pedrosa FO, Rogalski M (2018a) The complete plastome of macaw palm [Acrocomia aculeata (Jacq.) Lodd. ex Mart.] and extensive molecular analyses of the evolution of plastid genes in Arecaceae. Planta 247:1011–1030. https://doi.org/10.1007/s00425-018-2841-x
Lopes AS, Pacheco TG, Santos KGD, Vieira LN, Guerra MP, Nodari RO, de Souza EM, Pedrosa FO, Rogalski M (2018b) The Linum usitatissimum L. plastome reveals atypical structural evolution, new editing sites, and the phylogenetic position of Linaceae within Malpighiales. Plant Cell Rep 37:307–328. https://doi.org/10.1007/s00299-017-2231-z
Lopes AS, Pacheco TG, Silva ON, Cruz LM, Balsanelli E, Souza EM, Pedrosa FO, Rogalski M (2019) The plastomes of Astrocaryum aculeatum G. Mey. and A. murumuru Mart. show a flip-flop recombination between two short inverted repeats. Planta 250:1229–1246. https://doi.org/10.1007/s00425-019-03217-z
Lopes AS, Pacheco TG, Vieira LN, Guerra MP, Nodari RO, de Souza EM, Pedrosa FO, Rogalski M (2018c) The Crambe abyssinica plastome: Brassicaceae phylogenomic analysis, evolution of RNA editing sites, hotspot and microsatellite characterization of the tribe Brassiceae. Gene 671:36–49. https://doi.org/10.1016/j.gene.2018.05.088
Lowe TM, Chan PP (2016) tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. https://doi.org/10.1093/nar/gkw413
Maciel VBV, Yoshida CMP, Goycoolea FM (2019) Agronomic cultivation, chemical composition, functional activities and applications of Pereskia species—a mini review. Curr Med Chem 26:4573–4584. https://doi.org/10.2174/0929867325666180926151615
Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015) A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207:437–453. https://doi.org/10.1111/nph.13264
Majure LC, Puente R, Griffith MP, Judd WS, Soltis PS, Soltis DE (2012) Phylogeny of Opuntia s.s. (Cactaceae): clade delineation, geographic origins, and reticulate evolution. Am J Bot 99:847–864. https://doi.org/10.3732/ajb.1100375
Majure LC, Baker MA, Cloud-Hughes M, Salywon A, Neubig KM (2019) Phylogenomics in Cactaceae: a case study using the chollas sensu lato (Cylindropuntieae, Opuntioideae) reveals a common pattern out of the Chihuahuan and Sonoran deserts. Am J Bot 106:1327–1345. https://doi.org/10.1002/ajb2.1364
Martín M, Sabater B (2010) Plastid ndh genes in plant evolution. Plant Physiol Biochem 48:636–645. https://doi.org/10.1016/j.plaphy.2010.04.009
Moore AJ, Vos JM, Hancock LP, Goolsby E, Edwards EJ (2018) Targeted enrichment of large gene families for phylogenetic inference: phylogeny and molecular evolution of photosynthesis genes in the Portullugo Clade (Caryophyllales). Syst Biol 67:367–383. https://doi.org/10.1093/sysbio/syx078
Mower JP (2009) The PREP suite: predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res 37:W253–W259. https://doi.org/10.1093/nar/gkp337
Murcha MW, Kubiszewski-Jakubiak S, Teixeira PF, Gügel IL, Kmiec B, Narsai R, Ivanova A, Megel C, Schock A, Kraus S, Berkowitz O, Glaser E, Philippar K, Maréchal-Drouard L, Soll J, Whelan J (2016) Plant-specific preprotein and amino acid transporter proteins are required for tRNA import into mitochondria. Plant Physiol 172:2471–2490. https://doi.org/10.1104/pp.16.01519
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
Ni Z, Ye Y, Bai T, Xu M, Xu LA (2017) Complete chloroplast genome of Pinus massoniana (Pinaceae): gene rearrangements, loss of ndh Genes, and short inverted repeats contraction. Expansion Mol 22:1528. https://doi.org/10.3390/molecules22091528
Nyffeler R (2002) Phylogenetic relationships in the cactus family (Cactaceae) based on evidence from trnK/matK and trnL-trnF sequences. Am J Bot 89:312–326. https://doi.org/10.3732/ajb.89.2.312
Ocampo G, Columbus JT (2010) Molecular phylogenetics of suborder Cactineae (Caryophyllales), including insights into photosynthetic diversification and historical biogeography. Am J Bot 97:1827–1847. https://doi.org/10.3732/ajb.1000227
Ogburn RM, Edwards EJ (2010) The ecological water-use strategies of succulent plants. Adv Bot Res 55:179–255
Pacheco TG, Lopes AS, Oliveira JD, Otoni WC, Balsanelli E, Pedrosa FO, Souza EM, Rogalski M (2020a) The complete plastome of Passiflora cirrhiflora A. Juss.: structural features, RNA editing sites, hotspots of nucleotide diversity and molecular markers within the subgenus Deidamioides. Braz J Bot 43:839–853. https://doi.org/10.1007/s40415-020-00655-y
Pacheco TG, Lopes AS, Viana GD, Silva ON, Silva GM, Vieira LN, Guerra MP, Nodari RO, Souza EM, Pedrosa FO, Otoni WC, Rogalski M (2019) Genetic, evolutionary and phylogenetic aspects of the plastome of annatto (Bixa orellana L.), the Amazonian commercial species of natural dyes. Planta 249:563–582. https://doi.org/10.1007/s00425-018-3023-6
Pacheco TG, Lopes AS, Welter JF, Yotoko KSC, Otoni WC, Vieira LN, Guerra MP, Nodari RO, Balsanelli E, Pedrosa FO, Souza EM, Rogalski M (2020b) Plastome sequences of the subgenus Passiflora reveal highly divergent genes and specific evolutionary features. Plant Mol Biol 104:21–37. https://doi.org/10.1007/s11103-020-01020-z
Pacheco TG, da Morais SG, Lopes AS, Oliveira JD, Rogalski JM, Balsanelli E, Souza EM, Pedrosa FO, Rogalski M (2020) Phylogenetic and evolutionary features of the plastome of Tropaeolum pentaphyllum Lam (Tropaeolaceae). Planta 252:17. https://doi.org/10.1007/s00425-020-03427-w
Provan J, Powell W, Hollingsworth PM (2001) Chloroplast microsatellites: new tools for studies in plant ecology and evolution. Trends Ecol Evol 16:142–147. https://doi.org/10.1016/s0169-5347(00)02097-8
Rabah SO, Shrestha B, Hajrah NH, Sabir MJ, Alharby HF, Sabir MJ, Alhebshi AM, Sabir JSM, Gilbert LE, Ruhlman TA, Jansen RK (2019) Passifora plastome sequencing reveals widespread genomic rearrangements. J Syst Evol 57:1–14. https://doi.org/10.1111/jse.12425
Raman G, Park S (2015) Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS One. https://doi.org/10.1371/journal.pone.0141329
Ramírez-Rodríguez Y, Martínez-Huélamo M, Pedraza-Chaverri J, Ramírez V, Martínez-Tagüeña N, Trujillo J (2020) Ethnobotanical, nutritional and medicinal properties of Mexican drylands Cactaceae fruits: recent findings and research opportunities. Food Chem 312:126073. https://doi.org/10.1016/j.foodchem.2019.126073
Ritz CM, Martins L, Mecklenburg R, Goremykin V, Hellwig FH (2007) The molecular phylogeny of Rebutia (Cactaceae) and its allies demonstrates the influence of paleogeography on the evolution of South American mountain cacti. Am J Bot 94:1321–1332. https://doi.org/10.3732/ajb.94.8.1321
Ritz CM, Reiker J, Charles G, Hoxeyc P, Huntc D, Lowryc M, Stuppyd W, Taylore N (2012) Molecular phylogeny and character evolution in terete-stemmed Andean opuntias (Cactaceae-Opuntioideae). Mol Phylogenet Evol 65:668–681. https://doi.org/10.1016/j.ympev.2012.07.027
Rogalski M, Nascimento VL, Fraga HP, Guerra MP (2015) Plastid genomics in horticultural species: importance and applications for plant population genetics, evolution, and biotechnology. Front Plant Sci 6:586. https://doi.org/10.3389/fpls.2015.00586
Rogalski M, Karcher D, Bock R (2008) Superwobbling facilitates translation with reduced tRNA sets. Nat Struct Mol Biol 15:192–198. https://doi.org/10.1038/nsmb.1370
Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34:4537–4545. https://doi.org/10.1093/nar/gkl634
Rogalski M, Schöttler MA, Thiele W, Schulze WX, Bock R (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20:2221–2237. https://doi.org/10.1105/tpc.108.060392
Roy PS, Rao GJ, Jena S, Samal R, Patnaik A, Patnaik SSC, Jambhulkar NN, Sharma S, Mohapatra T (2016) Nuclear and chloroplast DNA variation provides insights into population structure and multiple origin of native aromatic rices of Odisha, India. PLoS One 11:e0162268. https://doi.org/10.1371/journal.pone.0162268
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Ruhlman TA, Chang WJ, Chen JJ, Huang YT, Chan MT, Zhang J, Liao DC, Blazier JC, Jin X, Shih MC, Jansen RK, Lin CS (2015) NDH expression marks major transitions in plant evolution and reveals coordinate intracellular gene loss. BMC Plant Biol 15:100. https://doi.org/10.1186/s12870-015-0484-7
Salie MJ, Thelen JJ (2016) Regulation and structure of the heteromeric acetyl-CoA carboxylase. Biochim Biophys Acta 1861:1207–1213. https://doi.org/10.1016/j.bbalip.2016.04.004
Salinas-Giegé T, Giegé R, Giegé P (2015) tRNA biology in mitochondria. Int J Mol Sci 16:4518–4559. https://doi.org/10.3390/ijms16034518
Sanderson MJ, Copetti D, Búrquez A, Bustamante E, Charboneau JLM, Eguiarte LE, Kumar S, Lee HO, Lee J, McMahon M, Steele K, Wing R, Yang TJ, Zwickl D, Wojciechowski MF (2015) Exceptional reduction of the plastid genome of saguaro cactus (Carnegiea gigantea): loss of the ndh gene suite and inverted repeat. Am J Bot 102:1115–1127. https://doi.org/10.3732/ajb.1500184
Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R (2001) The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol Biol 45:307–315. https://doi.org/10.1023/a:1006478403810
Shrestha B, Weng ML, Theriot EC, Gilbert LE, Ruhlman TA, Krosnick SE, Jansen RK (2019) Highly accelerated rates of genomic rearrangements and nucleotide substitutions in plastid genomes of Passiflora subgenus Decaloba. Mol Phylogenet Evol 138:53–64. https://doi.org/10.1016/j.ympev.2019.05.030
Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR (2014) A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol Phylogenet Evol 72:82–89. https://doi.org/10.1016/j.ympev.2013.12.004
Solórzano S, Chincoya DA, Sanchez-Flores A, Estrada K, Díaz-Velásquez CE, González-Rodríguez A, Vaca-Paniagua F, Dávila P, Arias S (2019) De novo assembly discovered novel structures in genome of plastids and revealed divergent inverted repeats in Mammillaria (Cactaceae, Caryophyllales). Plants 8:392. https://doi.org/10.3390/plants8100392
Stefenon VM, Klabunde G, Lemos RPM, Rogalski M, Nodari RO (2019) Phylogeography of plastid DNA sequences suggests post-glacial southward demographic expansion and the existence of several glacial refugia for Araucaria angustifolia. Sci Rep 9:2752. https://doi.org/10.1038/s41598-019-39308-w
Strand DD, D’Andrea L, Bock R (2019) The plastid NAD(P)H dehydrogenase-like complex: structure, function and evolutionary dynamics. Biochem J 476:2743–2756. https://doi.org/10.1042/BCJ20190365
Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A (2013) RNA editing in plants and its evolution. Annu Rev Genet 47:335–352. https://doi.org/10.1146/annurev-genet-111212-133519
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tangphatsornruang S, Sangsrakru D, Chanprasert J, Uthaipaisanwong P, Yoocha T, Jomchai N, Tragoonrung S (2010) The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: structural organization and phylogenetic relationships. DNA Res 17:11–22. https://doi.org/10.1093/dnares/dsp025
Thiel T, Michalek W, Varshney RK, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422. https://doi.org/10.1007/s00122-002-1031-0
Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7:1105–1120. https://doi.org/10.1093/mp/ssu022
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S (2017) GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res 45:W6–W11. https://doi.org/10.1093/nar/gkx391
Tsai CC, Chou CH, Wang HV, Ko YZ, Chiang TY, Chiang YC (2015) Biogeography of the Phalaenopsis amabilis species complex inferred from nuclear and plastid DNAs. BMC Plant Biol 15:202. https://doi.org/10.1186/s12870-015-0560-z
Tsudzuki T, Wakasugi T, Sugiura M (2001) Comparative analysis of RNA editing sites in higher plant chloroplasts. J Mol Evol 53:327–332. https://doi.org/10.1007/s002390010222
Vieira LN, Faoro H, Fraga HPF, Rogalski M, de Souza EM, de Oliveira PF, Nodari RO, Guerra MP (2014) An improved protocol for intact chloroplasts and cpDNA isolation in conifers. PLoS ONE 9:e84792. https://doi.org/10.1371/journal.pone.0084792
Vieira LN, Rogalski R, Faoro H, Fraga HPF, dos Anjos KG, Picchi GFA, Nodari RO, Pedrosa FO, de Souza EM, Guerra MP (2016) The plastome sequence of the endemic Amazonian conifer, Retrophyllum piresii (Silba) C.N.Page, reveals different recombination events and plastome isoforms. Tree Genet Genomes 12:1–11. https://doi.org/10.1007/s11295-016-0968-0
Walker JF, Yang Y, Feng T, Timoneda A, Mikenas J, Hutchison V, Edwards C, Wang N, Ahluwalia S, Olivieri J, Walker-Hale N, Majure LC, Puente R, Kadereit G, Lauterbach M, Eggli U, Flores-Olvera H, Ochoterena H, Brockington SF, Moore MJ, Smith SA (2018) From cacti to carnivores: Improved phylotranscriptomic sampling and hierarchical homology inference provide further insight into the evolution of Caryophyllales. Am J Bot 105:446–462. https://doi.org/10.1002/ajb2.1069
Wang N, Yang Y, Moore MJ, Brockington SF, Walker JF, Brown JW, Liang B, Feng T, Edwards C, Mikenas J, Olivieri J, Hutchison V, Timoneda A, Stoughton T, Puente R, Majure LC, Eggli U, Smith SA (2019) Evolution of Portulacineae marked by gene tree conflict and gene family expansion associated with adaptation to harsh environments. Mol Biol Evol 36:112–126. https://doi.org/10.1093/molbev/msy200
Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76:273–297. https://doi.org/10.1007/s11103-011-9762-4
Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. https://doi.org/10.1093/bioinformatics/bth352
Zappi D, Taylor N, Larocca J, Calvente A (2011) Domínios Fitogeográficos. In: Silva SR, Zappi D, Taylor N, Machado M (eds) Plano de ação nacional para conservação das cactáceas. Série Espécies Ameaçadas n 24. ICMBIO, Brasília
Zhu A, Guo W, Gupta S, Fan W, Mower JP (2016) Evolutionary dynamics of the plastid inverted repeat: the effects of expansion, contraction, and loss on substitution rates. New Phytol 209:1747–1756. https://doi.org/10.1111/nph.13743
Acknowledgements
This research was supported by the National Council for Scientific and Technological Development, Brazil (CNPq—Grants 459698/2014-1, 310654/2018-1 and 436407/2018-3). We are grateful to INCT-FBN and for the scholarships granted by the CNPq to ASL, TGP, KLGM, JDO, EB, EMS, FOP, and MR. Research fellowships granted by Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) to GMS and MCS are also gratefully acknowledged. We are also grateful to the Núcleo de Análise de Biomoléculas (NuBiomol) of the Universidade Federal de Viçosa for providing the software CLC Genomics.
Author information
Authors and Affiliations
Contributions
MR, EMS, and FOP conceived and designed the research. GMS, ASL, TGP, KLGM, MCS, JDO, VAB, EB, and MR conducted experiments and analyzed the data. ASL, TGP, EMS, FOP, and MR contributed with reagents and materials. GMS, ASL, and MR wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Morais da Silva, G., de Santana Lopes, A., Gomes Pacheco, T. et al. Genetic and evolutionary analyses of plastomes of the subfamily Cactoideae (Cactaceae) indicate relaxed protein biosynthesis and tRNA import from cytosol. Braz. J. Bot 44, 97–116 (2021). https://doi.org/10.1007/s40415-020-00689-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-020-00689-2