Abstract
The chromosome numbers of eighteen species of Spiranthinae from Brazil and Mexico and two of Cranichidinae from Ecuador were analysed. Sixteen chromosome records are presented for the first time: Aulosepalum riodelayense (2n = 64), A. tenuiflorum (2n = 60), Cyclopogon luteoalbus (2n = 36), Dichromanthus aurantiacus (2n = 40), Eltroplectris calcarata (2n = 42), Pelexia funckiana (2n = 46), Sarcoglottis assurgens (2n = 46), S. richardiana (2n = 50), S. rosulata (2n = 33), S. sceptrodes (2n = 46), S. schaffneri (2n = 46), S. scintillans (2n = 46), Stenorrhynchos albidomaculatum (2n = 46), S. cf. speciosum (2n = 46), Ponthieva andicola (2n = 26) and P. pilosissima (2n = ±42). For Mesadenella cuspidata, a chromosomal number variation (2n = 38/42) was found. In addition, ideograms of five Brazilian species are presented. Chromosome data support phylogenetic relationships suggested by previous cytological and molecular studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Orchidaceae comprises approximately 25,000 species (Dressler 2005) distributed in 850 genera (Atwood 1986; Pridgeon et al. 1999; Chase et al. 2003). It has a cosmopolitan distribution, with the exception of the regions always covered with snow and extreme deserts, but is most abundant and diverse in humid tropical and subtropical forests. As for the classification, the most recent systems for Orchidaceae are those proposed by Dressler (1993) and Pridgeon et al. (1999, 2001, 2003, 2005, 2009, 2014), the latter based also on molecular data, proposing the division of the family into five subfamilies: Apostasioideae, Vanilloideae, Cypripedioideae, Orchidoideae and Epidendroideae. In the subfamily Orchidoideae, there are six tribes; one of them is the Cranichideae, with six subtribes: Cranichidinae, Galeottiellinae, Goodyerinae, Manniellinae, Pterostylidinae and Spiranthinae (Pridgeon et al. 2001, 2003).
Subtribe Cranichidinae has 16 genera and about 210 species, all of them endemic to the Neotropics (Cribb and Pridgeon 2003; Salazar et al. 2003), and is characterized by the non-resupinate flowers, comparatively short column and brittle pollinia (Dressler 1993). In turn, subtribe Spiranthinae comprises about 40 genera and 470 species almost exclusively restricted to the Neotropics, except for the cosmopolitan genus Spiranthes Rich. (Salazar 2003). Its members can be recognized by the tubular flowers, margins of the lip adhered to the sides of the column, forming a deep nectary, and the soft granulose pollinia (Dressler 1993).
There are few cytotaxonomic studies on Spiranthinae, and these are concentrated especially on the genus Spiranthes with x = 15, 22 and 2n = 30 to 74 (Vij and Vohra 1974; Sheviak 1982; Tanaka and Kamemoto 1984; Martínez 1985; Brandham 1999). For the Neotropical species, Martínez (1981, 1985), Felix and Guerra (2005), Daviña et al. (2009) and Grabiele et al. (2010, 2013) have made the most significant contributions. On Cranichidinae, chromosome counts are reported only for Ponthieva mandonii Rchb.f. (2n = 46; Martínez 1985) and six species of Prescottia R.Br. (one with n = 48 and the others with 2n = 48; Felix and Guerra 2005).
The aim of this study was to determine the chromosome numbers and other chromosomal features of eighteen species of Spiranthinae and two of Cranichidinae, as part of our ongoing systematic and evolutionary studies of these orchid groups (Guimarães 2014; Guimarães et al., unpublished data).
Materials and methods
Eighteen species of Spiranthinae from Brazil and Mexico, and two species of Cranichidinae from Ecuador were studied (Table 1). Samples were cultivated in the greenhouses of the “Núcleo de Pesquisa Orquidário do Estado/Instituto de Botânica” (NP-OE/IBt in São Paulo, Brazil) and of “Herbario AMO” (Mexico City, Mexico). Vouchers were deposited at the herbarium of the Institute of Botany (SP), Mexico’s National Herbarium (MEXU) and the herbarium of the Faculty of Superior Studies of the National Autonomous University of Mexico, campus Zaragoza (FEZA).
Mitotic studies were performed in root tips pretreated with 8-hydroxyquinoline 0.2 mM (8-Hq) for 24 h at 4 °C (SP samples) and for 5 h at 18 °C (MEXU and FEZA samples), fixed in absolute ethanol:glacial acetic acid (3:1) for 3–24 h at room temperature (25 °C) and stored in a freezer at −20 °C. To prepare the slides, root tips were hydrolysed in 5 N HCl for 12–30 min at room temperature, frozen in liquid nitrogen to remove the coverslip, stained with 2 % Giemsa for the Brazilian specimens (Guerra 1983), hydrolysed in 1 N HCl for 12 min at 60 °C and stained according to the Feulgen technique for the remaining specimens (Mercado-Ruaro and Delgado-Salinas 1998). The meristems were soaked in a drop of 2 % aceto-orcein and then squashed.
For each species, chromosomes were counted in at least five cells, observed in a Carl Zeiss Jenaval (SP samples) and an Axioscop (MEXU, FEZA samples) optical microscopes and photographed using the Canon PowerShot S3 1S (SP samples) and AxioCam ERc5S (MEXU, FEZA samples) digital cameras.
The best mitotic metaphase spreads were selected from the SP samples for a general chromosome characterization. For five of them, we detailed the karyotypes and prepared the ideograms. Chromosomes were measured using Adobe Photoshop CS 5.1 software, determining the karyotype formula (KF), total chromosome length (TCL, calculated as the sum of all individual sizes of chromosomes), the average length of each chromosome (ALC) and the centromeric index (CI) for each chromosome pair (calculated as the ratio of the short arm and chromosome length). The chromosome types were classified according to the nomenclature of Levan et al. (1964).
Results
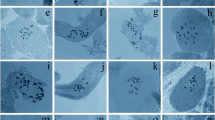
Within the subtribe Spiranthinae, Aulosepalum riodelayense (Burns-Bal.) Salazar presented 2n = 64; A. tenuiflorum (Greenm.) Garay, 2n = 60; Cyclopogon luteoalbus (A.Rich. & Galeotti) Schltr., 2n = 36; Dichromanthus aurantiacus (Lex.) Salazar & Soto Arenas, 2n = 40; Eltroplectris calcarata (Sw.) Garay & H.R.Sweet, 2n = 42; E. triloba (Lindl.) Pabst, 2n = 46; Pelexia funckiana (A.Rich. & Galeotti) Schltr., 2n = 46; Mesadenella cuspidata (Lindl.) Garay, 2n = 38 and 42 (variation possibility due to uncertainly or aneusomaty; see Discussion below); Sarcoglottis assurgens (Rchb.f.) Schltr., 2n = 46; Sarcoglottis cf. grandiflora (Lindl.) Klotzsch, 2n = 46; Sarcoglottis richardiana (Schltr.) Salazar & Soto Arenas, 2n = 50; Sarcoglottis rosulata (Lindl.) P.N.Don, 2n = 33; Sarcoglottis sceptrodes (Rchb.f.) Schltr., 2n = 46; Sarcoglottis schaffneri (Rchb.f.) Ames, 2n = 46; Sarcoglottis scintillans (E.W.Greenw.) Salazar & Soto Arenas, 2n = 46; Sauroglossum elatum Lindl., 2n = 46; Stenorrhynchos albidomaculatum Christenson, 2n = 46; and Stenorrhynchos cf. speciosum (Jacq.) Rich., 2n = 46 (Figs. 1–5, 11–26). All species studied, except A. riodelayense, A. tenuiflorum, C. luteoalbus, S. richardiana, S. scintillans, S. albidomaculatum and S. cf. speciosum, present a pair of acrocentric or submetacentric chromosomes, which are about twice the size of the remaining chromosomes, which are metacentric. Mesadenella cuspidata with 2n = 42 presents a pair of acrocentric chromosomes about four times as large as the remainder chromosomes (Table 2, Figs. 4, 8). The species A. riodelayense (Fig. 11) and A. tenuiflorum (Fig. 12) show a bimodal karyotype with six larger chromosomes and the others small.
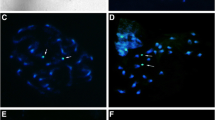
Chromosomes of species of Spiranthinae studied. 1 Eltroplectris calcarata (2n = 42). 2 Eltroplectris triloba (2n = 46). 3–4 Mesadenella cuspidata (2n = 38 and 2n = 42, respectively). 5 Sauroglossum elatum (2n = 46). Arrows indicate largest chromosomes in bimodal karyotypes; asterisks in 3 and 4 indicate the satellites. Bar = 1 µm
Chromosomes of species of Spiranthinae studied. 11 Aulosepalum riodelayense (2n = 64). 12 Aulosepalum tenuiflorum (2n = 60). 13 Cyclopogon luteoalbus (2n = 36). 14 Dichromanthus aurantiacus (2n = 40). 15 Pelexia funckiana (2n = 46). 16 Sarcoglottis assurgens (2n = 46). Arrows in 11–12, 14–16 indicate largest chromosomes in bimodal karyotypes; asterisks in 16 indicate the satellites. Bar = 5 µm (11–15); 10 µm (16)
The chromosome numbers for E. calcarata (Table 3, Figs. 1, 6), A. riodelayense (Fig. 11), A. tenuiflorum (Fig. 12), C. luteoalbus (Fig. 13), D. aurantiacus (Fig. 14), P. funckiana (Fig. 15), S. assurgens (Fig. 16), S. richardiana (Fig. 18), S. rosulata (Fig. 19), S. schaffneri (Fig. 20), S. sceptrodes (Fig. 21), S. scintillans (Fig. 22), S. albidomaculatum (Fig. 23) and S. cf. speciosum (Fig. 24) represent the first mitotic records for those species.
Chromosomes of species of Spiranthinae studied. 17 Sarcoglottis cf. grandiflora (2n = 46). 18 Sarcoglottis richardiana (2n = 50). 19 Sarcoglottis rosulata (2n = 33). 20 Sarcoglottis schaffneri (2n = 46). 21 Sarcoglottis sceptrodes (2n = 46). 22 Sarcoglottis scintillans (2n = 46). Arrows in 17, 19–21 indicate largest chromosomes in bimodal karyotypes; asterisks in 19 indicate the satellites. Bar = 5 µm (18–20); 10 µm (17, 21–22)
A secondary constriction (satellite) was observed on the largest chromosome pair of M. cuspidata (Table 2, Figs. 3, 4, 8, 9), S. assurgens (Fig. 16) and S. rosulata (Fig. 19).
The chromosome number (CN), total chromosome length (TCL) and karyotype formula (KF) of E. calcarata, E. triloba, M. cuspidata and S. elatum are given in Table 4.
Regarding the studied representatives of the subtribe Cranichidinae, Ponthieva andicola Rchb.f. has 2n = 26 (Fig. 25) and Ponthieva pilosissima (Senghas) Dodson, 2n = ±42 (Fig. 26). Both are first chromosome counts for these species. Ponthieva pilosissima presents 10 large chromosomes, 20 median chromosomes and ca. 12 small chromosomes in a trimodal karyotype. This difficulty of counting is due to the distinct degree of contraction of the different types of chromosomes, so that the small chromosomes may be earlier or later condensed and some of them have a secondary constriction. Thus, we were unable to determine the precise chromosome number of this species.
Discussion
Some of the counts obtained here differ from those found in literature. Martínez (1985) and Daviña et al. (2009) reported a 2n = 46 cytotype for a specimen of Mesadenella cuspidata collected in Argentina, while in this study we found a chromosomal number variation of 2n = 38/42 for a single specimen collected in Brazil. This difference in chromosomal number could be due to the difficulty of counting, to the possibility that these specimens could actually represent different species of Mesadenella, or merely to intra-polymorphism in this feature. Another possible explanation is that Martínez (1985) and Daviña et al. (2009) interpreted the secondary constrictions (satellites) of some chromosomes as distinct chromosomes, as was the case with the legume Oxyrhynchus (Palomino and Mercado 1983). Analysis of additional specimens of this species, preferably from different geographic origins, would help to clarify the issue.
The variation of the chromosome number in M. cuspidata in the present study may be due to the difficulty of counting or to the occurrence of aneusomaty. Davide et al. (2007) defined polysomaty and aneusomaty as the intraindividual variations in chromosome number, related to the occurrence of polyploidy and aneuploidy, respectively. In the case of aneusomaty, it would represent the first observation of this condition in Spiranthinae, although it has been documented in orchids of the rupicolous epidendroid genus Hoffmannseggella of Laeliinae (Yamagishi-Costa and Forni-Martins 2009, 2013). Few cases of aneusomaty in plants are described in literature, but this phenomenon is common in families such as Orobanchaceae (Greilhuber and Weber 1975), Boraginaceae (Bigazzi and Selvi 2003) and Fabaceae (Rodrigues et al. 2009).
Martínez (1985) also noted that the species of Spiranthinae with 2n = 46 often show a bimodal karyotype, with one pair of large chromosomes and the remainder ones much smaller. The present study confirms this bimodal karyotype (with some exceptions), as has been found in other species of the subtribe by Felix and Guerra (2005). This karyotype bimodality supports the inclusion of the genera Eltroplectris and Mesadenella in the Stenorrhynchos clade sensu Salazar et al. (2003). On the other hand, Sarcoglottis assurgens, S. cf. grandiflora, S. sceptrodes, S. schaffneri and S. scintillans share the same chromosome number (2n = 46) of other species of Sarcoglottis and Pelexia previously analysed (Martínez 1985; Dematteis and Daviña 1999; Felix and Guerra 2005; Daviña et al. 2009), which reinforces the proposal that Sarcoglottis really belongs in the Pelexia clade sensu Salazar et al. (2003). The meaning of the discordant chromosome numbers of Sarcoglottis richardiana (2n = 50) and S. rosulata (2n = 33) remains unclear and must be carefully studied by analysis of additional specimens of both species.
According to Martínez (1985), the basic number x = 23 for the Spiranthinae, as well as the bimodal chromosome condition, seems to be the distinguishing characteristic of the subtribe, which Felix and Guerra (2005) suggest as a significant feature to separate it from subtribe Goodyerinae, which belongs to a different lineage within the tribe Cranichideae (Salazar et al. 2003). The origin for this high basic number, compared to Goodyerinae (with x = 10, 14?, Brandham 1999; Felix and Guerra 2005), is not clear but could be due to polyploidy, although the single pair of large chromosomes does not support this hypothesis, unless the polyploids have chromosomes derived from different species, i.e. the origin would be fully allopolyploid as supposed by Martínez (1985).
The evolutionary significance of a single pair (or three pairs in Aulosepalum ssp.) of larger chromosomes in a bimodal karyotype also remains obscure, especially in cases where this condition is present in species with different chromosome numbers, e.g. Eltroplectris sp. with 2n = 26 (Martínez 1985; Daviña et al. 2009) and Sacoila sp. with 2n = 46 (Cocucci 1956; Martínez 1985; Felix and Guerra 2005; Daviña et al. 2009; Grabiele et al. 2010). Furthermore, this kind of karyotype appears to be quite rare in other groups of Orchidaceae. Felix and Guerra (2000) analysed 44 species of cymbidioid orchids and found bimodality in only two of them. Apparently, a bimodal karyotype is a specialized genomic architecture that could be selected for its functionality (Stebbins 1971; Kenton et al. 1990).
Ponthieva pilosissima, with 2n = ±42, is a typical case of trimodal karyotype. This condition was observed only in Hyacinthaceae (Forrest and Jong 2004), Ruscaceae (Yamashita and Tamura 2004; Meng et al. 2005) and Orchidaceae (this study). All these families belong to the order Asparagales, and this type of karyotype could be a homoplasy (parallel/convergent character) among those groups.
This study confirms some previous observations (Martínez 1985; Daviña et al. 2009) on chromosome numbers in Spiranthinae, but also contributes new chromosome counts, expanding the cytogenetic knowledge of the subtribe, and also of the family Orchidaceae. Further analyses are needed to clarify the different chromosome numbers found for M. cuspidata and to understand the trimodal karyotype reported for P. pilosissima.
References
Atwood JT (1986) The size of the Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 9:171–186
Bigazzi M, Selvi F (2003) Chromosome variation in Anatolian species of Nonea Medik. (Boraginaceae), with special reference to endemics and N. persica. Caryologia 56:509–519
Brandham P (1999) Cytogenetics. In: Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) Genera Orchidacearum, vol 1, General Introduction, Apostasioideae, Cypripedioideae. Oxford University Press, Oxford, pp 67–80
Chase MW, Cameron KM, Barrett RS, Freudenstein JV (2003) DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KW, Kell SP, Barrett RL, Cribb PJ (eds) Orchid Conservation. Natural History Publications, Kota Kinabalu, pp 69–89
Cocucci AE (1956) Cromosomas gaméticos de Stenorrhynchos australis Lindl. y Tradescantia radiata Clarke. Trab Mus Bot (Cordoba) 2:781–783
Cribb PJ, Pridgeon AM (2003) Subtribe Cranichidinae. In: Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) Genera Orchidacearum, vol 3, Orchidoideae (part 2) Vanilloideae. Oxford University Press, Oxford, pp 23–24
Davide LC, Techio VH, Nunes JD, Pereira AV (2007) Variação cromossômica numérica em Pennisetum. Ci Agrotecnol 31:398–405
Daviña JR, Grabiele M, Cerutti JC, Hojsgaard DH, Almada RD, Insaurralde IS, Honfi AI (2009) Chromosomes studies in Orchidaceae from Argentina. Genet Mol Biol 32:811–821
Dematteis M, Daviña JR (1999) Chromosome studies on some orchids from South America. Selbyana 20:235–238
Dressler RL (1993) Phylogeny and classification of the orchid family. Dioscorides Press, Portland
Dressler RL (2005) How many orchid species? Selbyana 26:155–158
Felix LP, Guerra M (2000) Cytogenetics and cytotaxonomy of some Brazilian species of Cymbidioid orchids. Genet Mol Biol 23:957–978
Felix LP, Guerra M (2005) Basic chromosome numbers of terrestrial orchids. Plant Syst Evol 254:131–148
Forrest LL, Jong K (2004) Karyotype asymmetry in Galtonia and Pseudogaltonia (Hyacinthaceae). Edinburgh J Bot 60:569–579
Grabiele M, Honfi AI, Cerutti JC, Fernandez V, Franco D, Daviña JR (2010) Cytogenetic analysis in the terrestrial orchid Sacoila argentina (Griseb.) Garay from Paraguay. Bot Stud 51:337–342
Grabiele M, Cerutti JC, Hojsgaard DH, Almada RD, Daviña JR, Honfi AI (2013) Comparative cytogenetics in Cyclopogon (Orchidaceae). Biologia (Bratislava) 68:48–54
Greilhuber J, Weber A (1975) Aneusomaty in Orobanche gracilis. Plant Syst Evol 124:67–77
Guerra M (1983) O uso de Giemsa na citogenética vegetal: comparação entre a coloração simples e o bandeamento. Cienc Cult 35:190–193
Guimarães LRS (2014) Filogenia e citotaxonomia do clado Stenorrhynchos (Spiranthinae, Cranichideae, Orchidoideae, Orchidaceae). PhD Thesis, Institute of Botany
Kenton A, Dickie JB, Langton DH, Bennett MD (1990) Nuclear DNA amount and karyotype symmetry in Cypella and Hesperoxiphion (Tigridieae; Iridaceae). Evol Trends Pl 4:59–69
Levan A, Fredga K, Sandberg AA (1964) Nomenclature for centromeric position of chromosomes. Hereditas 52:201–220
Martínez AJ (1981) Notas citotaxonomicas sobre el género Cyclopogon Presl. Parodiana 1:139–148
Martínez AJ (1985) The chromosomes of orchids VIII. Spiranthinae and Cranichidinae. Kew Bull 40:139–147
Meng Y, Nie ZL, Yang YP, Gu ZJ (2005) Karyomorphology of Maianthemum sensu lato (Polygonatae, Ruscaceae). J Plant Res 118:155–162
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: phaseolinae). Am J Bot 85:1–9
Palomino G, Mercado P (1983) Cytological studies in the genus Oxyrhynchus (Leguminosae). In: Brandham PE, Bennett MD (eds) Kew Chromosome Conference II. George Allen and Unwin, London
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (1999) Genera Orchidacearum, vol 1: general introduction, Apostasioideae, Cypripedioideae. Oxford University Press, Oxford
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (2001) Genera Orchidacearum, vol 2: Orchidoideae (part 1). Oxford University Press, Oxford
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (2003) Genera Orchidacearum, vol 3: Orchidoideae (part 2), Vanilloideae. Oxford University Press, Oxford
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (2005) Genera Orchidacearum, vol 4: Epidendroideae (part one). Oxford University Press, Oxford
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (2009) Genera Orchidacearum, vol 5: Epidendroideae (part two). Oxford University Press, Oxford
Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) (2014) Genera Orchidacearum, vol 6: Epidendroideae (part three). Oxford University Press, Oxford
Rodrigues RS, Corrêa AM, Forni-Martins E, Tozzi AMGA (2009) Números cromossômicos em espécies de Acosmium Schott e Leptolobium Vogel (Leguminosae, Papilionoideae). Acta Bot Bras 23:902–906
Salazar GA (2003) Subtribe Spiranthinae. In: Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN (eds) Genera Orchidacearum, vol 3, Orchidoideae (part 2) Vanilloideae. Oxford University Press, Oxford, pp 164–278
Salazar GA, Chase MW, Soto Arenas MA, Ingrouille M (2003) Phylogenetics of Cranichideae with emphasis on Spiranthinae (Orchidaceae, Orchidoideae): evidence from plastid and nuclear DNA sequences. Am J Bot 90:777–795
Sheviak CJ (1982) Biosystematic study of the Spiranthes cernua complex. Bull NY State Mus 48:1–73
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, London
Tanaka R, Kamemoto H (1984) Chromosomes in orchids: counting and numbers. In: Arditti J (ed) Orchid biology: reviews and perspectives III. Cornell University Press, Ithaca, pp 323–410
Vij SP, Vohra N (1974) Cytomorphological studies in the genus Spiranthes Rich. Cytologia 39:139–143
Yamagishi-Costa J, Forni-Martins ER (2009) Hybridization and polyploidy: cytogenetic indications for Hoffmannseggella (Orchidaceae) species evolution. Int J Bot 5:93–99
Yamagishi-Costa J, Forni-Martins ER (2013) Chromosomal diversity in two synchronopatric species of Hoffmannseggella (Orchidaceae). Nordic J Bot 31:505–509
Yamashita J, Tamura MN (2004) Phylogenetic analyses and chromosome evolution in Convallarieae (Ruscaceae sensu lato), with some taxonomic treatments. J Plant Res 117:363–370
Acknowledgments
The authors are grateful to Fernanda Lo Schiavo and Leandro C.S. Ventura (NP-OE/IBt) for assistance in the laboratory, and Eric Hágsater (AMO Herbarium) and Gabriela Cruz-Lustre (UNAM) for providing plant material for this study. This study was supported by a scholarship of the São Paulo Research Foundation (FAPESP) [Grant Numbers 2010/16353-1, 2012/19778-9] to LRSG and by a productivity grant of the Brazilian Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [grant number 304506/2013-3] to FB.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guimarães, L.R.S., Mercado-Ruaro, P., Corrêa, A.M. et al. Chromosome studies in Spiranthinae and Cranichidinae (Orchidaceae). Braz. J. Bot 38, 333–342 (2015). https://doi.org/10.1007/s40415-014-0130-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-014-0130-x