Abstract
Background
In clinical practice, the vast array of potential drug combinations necessitates swift and accurate assessments of pharmacokinetic drug–drug interactions (DDIs), along with recommendations for adjustments. Current methodologies for clinical DDI evaluations primarily rely on basic extrapolations from clinical trial data. However, these methods are limited in accuracy owing to their lack of a comprehensive consideration of various critical factors, including the inhibitory potency, dosage, and type of the inhibitor, as well as the metabolic fraction and intestinal availability of the substrate.
Objective
This study aims to propose an efficient and accurate clinical pharmacokinetic-mediated DDI assessment tool, which comprehensively considers the effects of inhibitory potency and dosage of inhibitors, intestinal availability and fraction metabolized of substrates on DDI outcomes.
Methods
This study focuses on DDIs caused by cytochrome P450 3A4 enzyme inhibition, utilizing extensive clinical trial data to establish a methodology to calculate the metabolic fraction and intestinal availability for substrates, as well as the concentration and inhibitory potency for inhibitors (\({K}_{\text{i}}\) or \({k}_{\text{inact}}/{K}_{\text{I}}\)). These parameters were then used to predict the outcomes of DDIs involving 33 substrates and 20 inhibitors. We also defined the risk index for substrates and the potency index for inhibitors to establish a clinical DDI risk scale. The training set for parameter calculation consisted of 73 clinical trials. The validation set comprised 89 clinical DDI trials involving 53 drugs. which was used to evaluate the reliability of in vivo values of \({\text{K}}_{\text{i}}\) and \({k}_{\text{inact}}/{K}_{\text{I}}\), the accuracy of DDI predictions, and the false-negative rate of risk scale.
Results
First, the reliability of the in vivo \({K}_{\text{i}}\) and \({k}_{\text{inact}}/{K}_{\text{I}}\) values calculated in this study was assessed using a basic static model. Compared with values obtained from other methods, this study values showed a lower geometric mean fold error and root mean square error. Additionally, incorporating these values into the physiologically based pharmacokinetic-DDI model facilitated a good fitting of the C–t curves when the substrate’s metabolic enzymes are inhibited. Second, area under the curve ratio predictions of studied drugs were within a 1.5 × margin of error in 81% of cases compared with clinical observations in the validation set. Last, the clinical DDI risk scale developed in this study predicted the actual risks in the validation set with only a 5.6% incidence of serious false negatives.
Conclusions
This study offers a rapid and accurate approach for assessing the risk of pharmacokinetic-mediated DDIs in clinical practice, providing a foundation for rational combination drug use and dosage adjustments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
By fully utilizing the results from clinical drug trials, a newPK-mediated clinical drug–drug interaction risk assessment scale was established, which can accurately predict DDI outcomes. |
A series of parameters, including the in vivo metabolic fraction, intestinal availability, and the inhibition constants of inhibitors were calculated. These parameters, compared to results from in vitro experiments, can be more reliably applied in contexts such as PBPK models. |
1 Introduction
In clinical practice, over half the patients require the simultaneous use of two or more medications. Drug–drug interactions (DDIs) have always been an issue of special concern in clinical use because they can affect the exposure of the affected (victim) drug in the body, potentially leading to serious or even fatal adverse reactions [1, 2]. Reducing the toxic side effects caused by fluctuations in drug concentrations without delaying the treatment cycle is hindered by the lack of a highly efficient, feasible, accurate, and reliable method, aside from therapeutic drug monitoring tailored to specific medications. Although clinical DDI trials yield the most accurate risk results, the myriad of possible interaction combinations during the actual use of drugs make it impractical to assess all potential drug interactions in clinical trials.
Leveraging the results from existing clinical drug interaction studies to extrapolate DDIs has been recently proposed [3, 4] and has proven to be highly effective in clinical practice for predicting potential interactions [5]. This method utilizes midazolam as a standard drug and calculates the apparent inhibition ratio (IR) of the perpetrator drug based on clinical DDI trial results associated with midazolam. Subsequently, the contribution ratio (CR) of the affected drug has been determined in clinical DDI trials involving potent perpetrator drugs.
Using these two parameters, it is possible to extrapolate the exposure changes caused by other drug combinations using equation \(1/(1-\text{IR}\times \text{CR})\), allowing clinical trial data to rapidly and accurately guide the optimization of clinical prescriptions. However, this method still has shortcomings in the application of clinical DDI risk assessment because of three important issues. The first issue is the influence of intestinal metabolic sites on DDIs. This issue has been neglected. However, it is particularly pertinent for cytochrome P450 (CYP) 3A4, where the contribution of intestinal enzyme-mediated metabolism is significant and should not be overlooked [6]. Such an oversight could potentially compromise the precision of pharmacokinetic (PK) extrapolations. For substrates such as tacrolimus and sirolimus, the intestinal extraction exceeds 75% [7, 8]. Loue and Tod [5] utilized the methodology developed by Ohno et al. [3] and predicted DDIs when the aforementioned substrates were co-administered with CYP3A4 inhibitors. This prediction revealed significant discrepancies in both underestimation and overestimation. Clinical trials have demonstrated that an oral dose of 800 mg of posaconazole can lead to an 8.9-fold increase in the area under the curve (AUC) of sirolimus [9], whereas the method predicted only a 3.6-fold increase.
The second issue is that the IR of the perpetrator drug is calculated at a specific dosage, even though the effects of varying the perpetrator drug dosage on the substrate can differ significantly. For instance, a daily dose of 100 mg of fluconazole can cause a 2.1-fold increase in the AUC of triazolam, whereas a daily dose of 200 mg can elevate this value 4.4-fold [10]. In the context of complex clinical dosage adjustments, relying solely on the IR value does not fulfill the requirements for risk assessment.
The third issue is that the IR of the perpetrator drug and the CR of the victim drug lack concrete physiological significance and are not differentiated according to the type of inhibition exerted by the inhibitor. This limitation constrains the application scenarios for the parameter values. Hisaka et al. [4] examined the correlation between these parameters and the mechanistically meaningful parameters derived from in vitro experiments. This method could potentially replace in vitro experiments to obtain drug interaction parameters for use in physiologically based PK (PBPK) models to predict DDIs in special populations.
To address these issues, this study exploited the extensive available clinical trial data of drugs. Based on a mechanistic static model, the study fits parameters related to DDIs, including the metabolic fraction, intestinal permeability, inhibition constant of competitive inhibitors, inactivation efficiency of time-dependent inhibitors, and unbound concentration of the perpetrator drug at the enzyme-binding site. A framework was established to assess the clinical risks posed by drug combinations, enabling rapid and reliable quantitative assessment of DDIs during clinical use. Recommendations for their use are made based on these results.
2 Methods
2.1 Experimental Drugs
This study considered the CYP3A4 enzyme, which is frequently involved in DDIs, as a paradigm that included CYP3A4 substrates and inhibitors. By referencing the indicator drugs recommended on the US Food and Drug Administration (FDA) website [11] and reviewing previously published data [12,13,14,15], common CYP3A4 substrates and inhibitors were collected. Information was gathered regarding the main elimination pathways, metabolic profiles, types of CYP3A4 enzyme inhibition, and CYP3A4 selectivity of these drugs. The drugs were selected according to the following criteria. Substrates needed to be administered orally; with data available from clinical DDI trials involving two or more CYP3A4 inhibitors, resulting in an AUC ratio (AUCR) ≥1.15; primarily cleared by hepatic metabolism with assumed negligible renal clearance; with linear PK characteristics. Inhibitors needed to be administered orally; with clinical DDI trials performed with midazolam and other CYP3A4 substrates, yielding an AUCR ≥ 1.15; and with data of the type of CYP3A4 inhibition.
2.2 Development of the Mechanistic Static Model Method
Clinical DDI trials facilitate the acquisition of exposure change multiples, specifically the AUCR. The size of the AUCR is contingent on the binding affinity, potency, concentration, and extent of metabolic clearance of the substrate via CYP3A4 [16]. When the concentration of the substrate is below the Michaelis–Menten constant (Km), it is presumed that metabolic clearance by CYP3A4 is concentration independent. Furthermore, considering the presence of the CYP3A4 enzyme within the intestinal tract, the fraction of intestinal permeability affected by DDIs is considered into account [17].
If the inhibition type of the perpetrator drug is competitive inhibition, the change in the AUCR of the victim drug exposure can be calculated as:
If the inhibition type of the perpetrator drug is time dependent, the change in AUCR of the victim drug exposure can be expressed as:
In Eqs. (1) and (2), \({\text{AUC}}^{\prime}\) represents the AUC of the victim drug after being affected by the perpetrator drug, \({F}_{g}\) represents intestinal availability, namely \({f}_{a}\times {f}_{g}\), \({f}_{m}\) represents the fraction of victim drug metabolized by enzymes (default refers to CYP3A4 enzyme), \({k}_{i,u}\) represents the inhibition constant for an unbound competitive inhibitor, \({k}_{\text{inact}}\) represents the maximal inactivation rate of time-dependent inhibitors, \({K}_{I,u}\) represents the unbound inactivator concentration at half \({k}_{\text{inact}}\), \({[I]}_{g,u}, {[I]}_{H,u}\) represent the unbound concentrations of the inhibitor in the liver and intestine, and \({k}_{\text{deg},g}, {k}_{\text{deg},H}\) represent the intestinal (\({\text{CYP}3\text{A}}_{\text{h}}\), 0.000321 min−1) and hepatic (\({\text{CYP}3\text{A}}_{\text{g}}\), 0.000481 min−1) degradation rates of CYP3A enzymes [18,19,20]. For a detailed derivation, please refer to the Electronic Supplementary Material (ESM).
It is evident from Eqs. (1) and (2) that the AUCR obtained from clinical DDI trials can be considered the result of the combined effects of four unknown parameters: intrinsic clearance (Fg), fraction metabolized (fm) of the substrate, the unbound concentration of the inhibitor ([I]u), and inhibition constant (Ki) or the ratio of inactivation rate constant to inhibition constant (kinact/KI). It is necessary to use a standard drug to establish the values of these parameters, as detailed in the process outlined in Fig. 1.
2.3 Standard Drug
This study requires the selection of a standard drug and setting initial values for the Fg and fm of this drug. In clinical trials, the sponsors typically use a single oral dose of midazolam to assess its inhibitory or inductive effects on CYP3A4. In vivo studies have confirmed that midazolam is a highly specific CYP3A4 substrate, undergoing clearance solely through CYP3A4 metabolism [21, 22]. The search terms “midazolam” and “CYP3A4” and “metabolite”, “midazolam” and “fraction gut” or “intestinal availability” were utilized in the PubMed search engine to query the published \({f}_{\text{m},\text{CYP}3\text{A}4}\) and \({F}_{g}\) values for midazolam. The search results are provided in Tables 1 and 2. The \({\text{f}}_{\text{m},\text{CYP}3\text{A}4}\) and \({F}_{g}\) values of the standard drug midazolam in this study were 0.92 and 0.55, respectively (the average values from the literature cited in Tables 1, 2).
2.4 Human Data Sources
The results of the clinical drug interaction trials in this study were derived from clinical trials registered on the ClinicalTrials.gov website (https://clinicaltrials.gov/). Data were obtained by searching for clinical pharmacology review reports on the FDA website and Cortellis Drug Discovery Intelligence database or through the published literature that reported trial outcomes. Data on the drug concentration–time curve incorporated in the study were digitized following recommended practices [23] using GetData Graph Digitizer version 2.26.0.20 (https://getdata-graph-digitizer.software.informer.com/download/). The extracted PK curves data from clinical trials at clinical doses were imported into Certara Phoenix WinNonlin 8.1 for compartmental modeling and non-compartmental analysis to calculate the values of PK parameters including AUC, ka, and apparent clearance. The oral bioavailability of the drugs was cited from the publicly available Human Oral Bioavailability Database (http://modem.ucsd.edu/adme/databases/databases_bioavailability.htm) [24, 25].
2.5 Determination of Unbound Inhibition Concentration for the Inhibitors
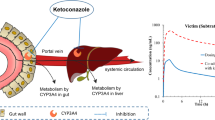
Because CYP3A4 is present in both the liver and small intestine, it is necessary to determine the unbound inhibitor concentration in these organs. As shown in Fig. 2, after absorption in the gastrointestinal tract, inhibitors are metabolized or transported through the small intestinal mucosa and enter the liver via the portal vein and hepatic artery.
Process of digestion and absorption of orally administered drugs. Drug bioavailability is collectively determined by the absorption fraction in the gastrointestinal tract, intestinal mucosal metabolism permeability fraction, and hepatic metabolism permeability fraction. The drug is initially absorbed from the intestinal lumen into epithelial cells. The amount of the absorbed drug is represented as \({f}_{a}\times {k}_{a}\times D\) f[27]. The drug then enters the hepatic portal vein through the intestinal mucosal layer. A portion of the drug may be transported back to the gut by transporters. Hepatic inlet drugs are supplied by both the portal vein and hepatic artery; the sum of these two parts is considered as the drug concentration within the liver. CYP cytochrome P450, D represents the oral dose, \({f}_{a}\) absorption fraction, \({f}_{h}\) represents the fraction of non-metabolic elimination in the liver, \({k}_{a}\) absorption rate constant
Although the free concentrations of inhibitors in the intestinal mucosa and around hepatic enzymes cannot be directly obtained from in vivo measurements, research by Ito et al. [26, 27] suggests that the free concentration of inhibitors in the intestinal mucosa and liver can be represented as the sum of the gastrointestinal absorbed fraction and systemic circulation concentration. Studies have compared the accuracy of drug interaction predictions based on static concentrations at the aforementioned sites, calculated using either \({C}_{\text{max}}\) (maximum systemic circulation concentration) or \({C}_{\text{ave}}\) (average systemic circulation concentration) derived from in vitro experiments [28,29,30]. Using the average concentration as the effective concentration of the inhibitor appears to provide a more conservative conclusion for risk assessment. This study based on the in vivo result, employed the free concentration at the portal vein inlet, denoted as \({\left[I\right]}_{g,u}\), to represent the free concentration of inhibitors in small intestinal mucosa, as determined in Equation (3). Similarly, the free concentration at the hepatic inlet was used to denote the free hepatic inhibitor concentration \({[I]}_{H,u}\), as shown in Eq. (4):
In the above equations, \({Q}_{h}\) represents the liver blood flow (96.6 L/h) [31, 32], \({Q}_{pv}\) represents the portal vein blood flow (73 L/h), which is approximately 75% of the hepatic blood flow, \({f}_{up}\) represents the free fraction of the test drug in plasma, \({f}_{ub}\) represents the free fraction of the test drug in the blood, \({k}_{a}\) represents the oral absorption rate of the test drug (h−1), BpR is the blood-to-plasma ratio, and τ is the dosing interval.
2.6 Calculation of In Vivo \({{\varvec{F}}}_{{\varvec{g}}}\) Value for the Substrates
Intestinal availability refers to the fraction of drugs that pass through the epithelial cells of the small intestinal mucosa and enter the portal vein. Currently, there are four methods to estimate the in vivo value of \({F}_{g}\). The first method is the comparison of the results of DDI studies following intravenous administration with the results obtained after oral administration [33]. The second method relies on grapefruit juice as a complete inhibitor of intestinal CYP3A4 enzymes. The DDI results of test drugs with grapefruit juice can effectively estimate the in vivo \({F}_{g}\) value [34]. The third method uses mathematical modeling approaches to derive the in vivo \({F}_{g}\) value from the body’s drug-time curves [35]. The fourth method infers the intestinal availability indirectly by obtaining information, such as bioavailability (F) and total clearance rate from the drug-time curves following both intravenous and oral administration of a drug [36].
In this study, the drugs included in the calculations were primarily metabolized and cleared by the liver, with negligible renal clearance. The total oral clearance rate, denoted as \({\text{CL}}_{\text{tot}}\), can be approximated as equivalent to the hepatic clearance rate, \({\text{CL}}_{\text{h}}\). Hence, intestinal availability, \({F}_{g}\), can be expressed as follows:
where F represents the oral bioavailability of the test drug, which can be obtained from the drug concentration–time curve of intravenous/oral administration and \({\text{CL}}_{\text{tot}}/F\) is obtained through a compartmental analysis of drug concentration–time curves in vivo. The in vivo \({F}_{g}\) values of the affected drugs can be calculated using Eq. (7).
2.7 Determination of Specific Parameters for In Vivo DDIs
Upon obtaining the results of the clinical drug interaction trials between the study inhibitors and midazolam, and the concentrations of the inhibitors, the in vivo \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values were calculated using Eqs. (1) and (2). Following the workflow depicted in Fig. 1, after determining the in vivo \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values of the inhibitors, data from a series of clinical drug interaction studies performed with inhibitors that are highly selective for CYP3A4 were used to estimate the in vivo \({f}_{m,\text{CYP}3\text{A}4}\) values for a range of substrates used in combination with these inhibitors.
2.8 Validation of Specific Parameters for In Vivo DDIs
To verify the accuracy and reliability of a series of in vivo DDI specificity parameters derived using the methodology outlined in this article for the assessment of interaction risks, cross-validation was performed using a validation set composed of clinical trials that did not incorporate the calculated parameters. Employing the basic static method described in the FDA DDI guidelines [37], we validated the discrepancies between the calculated specificity parameters for substrates and inhibitors for risk assessment and their observed values, intending to examine their reliability. Furthermore, this study incorporated the in vivo DDI specificity parameters calculated herein into Eqs. (1) and (2) to forecast the outcomes of clinical drug interactions, and compared these predictions with the actual measured values to assess predictive accuracy. The magnitude of the discrepancies was quantified using the geometric mean fold error (GMFE) and root mean square error (RMSE).
2.9 Establishment of the PBPK model
The feasibility of using the drug interaction specificity parameters obtained from this study instead of in vitro experimental results for predicting DDIs using PBPK models was investigated. Physiologically based PK models were constructed for both the substrates and inhibitors using PK-Sim and MoBi (Open Systems Pharmacology Suite 10, available at www.open-systems-pharmacology.org), referencing model information from the OSP-PBPK-Model-Library (https://github.com/Open-Systems-Pharmacology/OSP-PBPK-Model-Library). The validated PBPK models were then used to simulate the PK profile changes of the substrate when combined with an inhibitor, which was subsequently compared with the observed clinical PK curves. The details of the model development are in the ESM.
2.10 Clinical DDI Risk Scale Evaluation
Currently, the risk of drug-induced interactions is categorized as strong, moderate, and weak, based on AUCR when co-administered with a probe drug. Such a basic qualitative classification is insufficient for risk assessment in complex clinical scenarios. In this study, by computing in vivo DDI specificity parameters, we introduced two novel metrics for evaluating the risk associated with CYP3A4 enzyme interactions: the risk index (RI) for substrates and the potency index (PI) for inhibitors, which serve as scales for risk measurement. The RI is defined as the maximal fold increase in the AUC of the substrate co-administered with a potent enzyme inhibitor (see Eq. 10). The PI is defined as the maximal theoretical fold increase in the AUC of the most sensitive enzyme substrate to the inhibitor at a given dose (see Eqs. 11 and 12).
For competitive inhibitors:
For time-dependent inhibitors:
3 Results
3.1 Inclusion of Experimental Drugs and Human Data
We included 33 marketed drugs that are primarily metabolized by CYP3A4 (Table 3) along with 20 commonly marketed drugs that inhibit CYP3A4 (Table 4). Clinical drug interaction studies for these drugs were identified by searching the FDA’s Drug Approvals and Databases (https://www.fda.gov/drugs) and querying the PubMed database with the keywords “[drug name]” and “drug interaction study” and “healthy volunteers”. A total of 53 in vivo drug interaction outcomes were included as a training set for the method to calculate DDI-specific parameters (Tables S1 and S2 of the ESM). Furthermore, 89 in vivo drug interaction outcomes were included as a validation set to verify the reliability of the predicted parameters, which can be found in Table S3 of the ESM.
3.2 Unbound Concentration for the Inhibitors and In Vivo Fg Values for the Substrates
According to Eqs. (3)–(6), the inhibitor concentration is related to the drug dosage, absorption constant, fraction absorbed, AUC, and free fraction, among other factors. Data on the drug concentration–time curve incorporated in the study were extracted using GetData Graph Digitizer version 2.26.0.20, and Certara Phoenix WinNonlin version 8.1 was utilized for non-compartmental analysis and compartment model fitting to obtain the AUC and ka values for specific inhibitor doses (Table S4 of the ESM). The unbound fraction was obtained by querying the Cortellis Drug Discovery Intelligence database. Based on Eqs. (3)–(6), the average concentration of the free inhibitor in the small intestinal mucosa and hepatic inlet at specific doses were calculated and are compiled in Table S5 of the ESM.
According to Eq. (7), the calculation of the \({\text{F}}_{\text{g}}\) for the substrates included in the study requires knowledge of the bioavailability and clearance rate of the substrate. We used the PK curves derived from the control group in clinical DDI trials where substrates are administered individually to perform compartmental model fitting, which yielded the \({\text{CL}}_{\text{tot}}/F\) of the substrate, approximating the hepatic clearance \({\text{CL}}_{\text{h}}/F\). The bioavailability was determined using the Human Oral Bioavailability Database summarized by Hou et al. [24, 25]. The calculated \({F}_{g}\) values for the substrates are listed in Table S6 of the ESM.
3.3 Calculation of In Vivo Apparent \({{\varvec{K}}}_{{\varvec{i}}}\) and \({{\varvec{k}}}_{\mathbf{i}\mathbf{n}\mathbf{a}\mathbf{c}\mathbf{t}}/{{\varvec{K}}}_{{\varvec{I}}}\) Values
To assess the potency of enzyme inhibitors, the current practice predominantly involves the use of in vitro systems, such as liver microsomes or human hepatocyte incubations. These systems test various concentration levels and standard substrates incubated with the enzyme, allowing the calculation of inhibition constants \({K}_{i}\) (for time-dependent inhibitors, the inactivation constant \({k}_{\text{inact}}/{K}_{I}\) is determined) [38, 39]. The results vary significantly depending on the incubation conditions. This study calculated the \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values based on human data obtained from the concomitant use of the inhibitor with midazolam, utilizing the model parameters in Table 5. The results are provided in Table S7 of the ESM.
According to the FDA DDI guidance, when a drug is determined to be an enzyme inhibitor in vitro, the severity of the consequences of the drug inhibiting the enzyme is assessed using a basic model [40] by calculating the R-value of the inhibitor. To compare whether the inhibitor parameters from different sources could reliably estimate the risk of the inhibitor in clinical use, the R values were calculated (Eqs. 13–16) using the \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) calculated in this study, the average of in vitro experiments obtained from the literature [18, 41,42,43] (see Table S8 of the ESM) and the IR value calculated as described by Ohno et al. [3] with the same dataset (see Table S1 of the ESM). Finally, these three sets of R values were compared with the clinical DDI results of the corresponding inhibitors in the validation set [44]. The results are shown in Fig. 3A, B. The data clearly show that the R values calculated using the \({K}_{i}\) and \({\text{k}}_{\text{inact}}/{\text{K}}_{\text{I}}\) values obtained in this study had smaller errors with the clinical AUCR in the validation set. The findings indicate that it better reflects the clinical DDI risk caused by the inhibitors. Moreover, Fig. 3C, D show that the \({K}_{i}\) and \({\text{k}}_{\text{inact}}/{\text{K}}_{\text{I}}\) values calculated in this study were consistently closer to the overall mean of the inhibitory potency parameters of the corresponding inhibitors obtained in the literature than those derived from the IR values.
A Comparison of basic model fitting results using \({K}_{i}\) or \({k}_{\text{inact}}/{K}_{I}\) values (Eqs. 13–16) obtained from this study, the method of Ohno et al., and in vitro experimental results from the literature with the clinically observed area under the curve ratio (AUCR) values of the validation set (Table S3 of the ESM). B Geometric mean fold error (GMFE) and root mean square error (RMSE) for basic model fitting using \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values from this study, the method of Ohno et al., and in vitro experimental results from the literature compared with clinically observed AUCR values of the validation set (Table S3 of the ESM). C Distribution of \({K}_{i}\) values of competitive inhibitors from this study, the method of Ohno et al., and in vitro experimental results from the literature, the diamonds represent a series of in vitro experimental Ki values from the literature listed in Table S8 of the ESM with the average of these in vitro experimental results shown in red lines and the standard error represented by black error bars; hexagons represent Ki values obtained using the method described by Ohno et al.; and pentagrams represent Ki values calculated in this study. D Distribution of \({k}_{\text{inact}}/{K}_{I}\) values of time-dependent inhibitors from this study, the method of Ohno et al., and in vitro experimental results from the literature, the diamonds represent a series of in vitro experimental \({k}_{\text{inact}}/{K}_{I}\) values from the literature listed in Table S8 of the ESM with the average of these in vitro experimental results shown in red lines and the standard error represented by black error bars; hexagons represent \({k}_{\text{inact}}/{K}_{I}\) values obtained using the method described by Ohno et al.; and pentagrams represent \({k}_{\text{inact}}/{K}_{I}\) values calculated in this study
For competitive inhibition:
For time-dependent inhibition:
The relationship between the IR and the boundary R values obtained by Ohno et al. [3] is as follows:
3.4 Calculation of In Vivo \({{\varvec{f}}}_{{\varvec{m}},\mathbf{C}\mathbf{Y}\mathbf{P}3\mathbf{A}4}\) Values
Accurately calculating the fraction of metabolized \({f}_{m}\) of a target drug is critical for assessing the infinite risk of drug interactions in clinical settings [5, 45]. Common methods for determining the contribution of a drug through a specific metabolic pathway include in vitro system incubation, back-calculation from human results, and human radiolabeled absorption, distribution, metabolism, and excretion studies.
This study employed inhibitors with strong CYP3A4 selectivity in conjunction with the clinical DDI trial results for the included substrates. The in vivo values of the target CYP3A4 substrates \({f}_{m,\text{CYP}3\text{A}4}\) were calculated using the \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values derived from the model parameters in Table 5, and the corresponding in vivo \({F}_{g}\) values calculated above. The results are provided in Table S9 of the ESM. We separately examined the risks of clinical DDI effects on substrates assessed using fm values alone, and using both \({F}_{g}\) and \({f}_{m}\) values. These results were compared to the corresponding substrate AUCR results for the validation set. As shown in Fig. 4, considering both \({\text{F}}_{\text{g}}\) and \({\text{f}}_{\text{m}}\) values of the substrate provides a better representation of the potential risk of clinical DDI and reduce the likelihood of false negatives.
Relationship between substrate drug–drug interaction (DDI) sensitivity parameters and the clinical DDI area under the curve ratio (AUCR). A In vivo \({F}_{g}\) values and \({f}_{m}\) values of substrate determined using the study methodology were employed to assess the sensitivity of the substrate to DDIs: \(\text{AUCR}\approx 1/{(F}_{g}\times (1-{f}_{m})\). B In vivo \({f}_{m}\) values of substrates determined using the study methodology were used to assess substrate sensitivity to DDIs: \(\text{AUCR}\approx 1/(1-{f}_{m})\). The red color blocks represent false negatives, the gray color blocks represent false positives, and the green color blocks represent true positives/negatives
3.5 Validation of Clinical Drug Interaction Simulation Results
Upon obtaining a reliable inhibitor \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values, and substrate \({F}_{g}\) and \({f}_{m}\) values, common methodologies for predicting clinical DDI risks include static and PBPK models [46]. Initially, we applied Eqs. (1) and (2) to the aforementioned data to predict clinical DDI outcomes within the validation set. Concurrently, we utilized the method developed by Ohno et al. [3] to predict DDI outcomes in the validation set, comparing each with clinically observed values. As depicted in Fig. 5, the predictions made using our method showed that 81% of the results varied within 1.5-fold of the clinically observed values, which is superior to the outcomes achieved using the method of Ohno et al. [3] (Fig. 5A, a). In scenarios involving high-risk clinical DDIs with AUCR > 2.5 (Fig. 5B, b) and cases where Fg < 0.5 (Fig. 5C, c), our method demonstrated superior accuracy. Under all circumstances, the overall prediction error of our method was less than that of the method of Ohno et al. [3] (Fig. 5D, d).
Comparison of in vivo drug–drug interaction simulation outcomes with values observed in clinical trials. This figure illustrates data of a comparative analysis between the results of in vivo drug–drug interaction simulations and actual measured values from clinical trials. The simulations were conducted using both \({K}_{i}\),\({k}_{\text{inact}}/{K}_{I}\), and \({f}_{m}\) values obtained from this study, and inhibition ratio (IR) and contribution ratio (CR) values derived from the method of Ohna et al. The comparison involves 89 clinical drug–drug interaction measured values from a validation set. The vertical axis represents the simulated area under the curve (AUCR) values and the horizontal axis displays the AUCR values observed in clinical trials. A Results of simulations using the method of Ohna et al. compared with clinical observed values. a Results of simulations using the method from this study compared with clinical observed values. B Subset of the validation set with AUCR > 2.5, comparing simulations based on the method of Ohna et al. with clinically observed values. b Subset of the validation set with AUCR > 2.5, comparing simulations from this study against clinical observed values. C Subset of the validation set with intrinsic clearance (Fg) < 0.5, comparing simulations based on the method of Ohna et al with clinically observed values. c Subset of the validation set with Fg < 0.5, comparing simulations from this study with clinical observed values. D Comparison between the geometric mean fold error (GMFE) of the results simulated in this study and those by Ohna et al. relative to clinical observed values. d Comparison of the root mean square error (RMSE) between the simulation results of this study and those by Ohna et al. relative to clinical observed values
To describe the impact of temporal variation in the in vivo concentrations of inhibitors and substrates on the PK profile of the substrate due to DDIs, PBPK models are commonly utilized to simulate interaction combinations for which clinical trials have not been performed. Recent studies have analyzed new drug applications submitted to the FDA [47, 48]. These studies have revealed that 56–67% of these applications employ PBPK modeling to assess the risk of DDIs. These applications predominantly use Simcyp™ (https://www.certara.com/ software/simcyp-pbpk/), GastroPlus® (https://www.simulations-plus.com/software/gastroplus/), and PK-SIM® (https://www.open-systems-pharmacology.org/) software for PBPK modeling. Although built-in models and parameters for common inhibitors are available in these software packages, the inhibitory potency parameters are still largely derived from in vitro experimental results.
In this study, we established PBPK models for the investigational drugs using PK-SIM and MoBi. After optimization using human PK curves, we used the inhibitory potency parameters obtained from our research to simulate drug interactions in the validation set. Tables S10–S27 of the ESM list the clinical trial references used for training and test sets in the self-built models. Table S28 of the ESM lists the models from the OSP-PBPK-Model-Library cited in this study. Tables S29–S48 of the ESM list the parameter values and sources of all models, as well as parameters related to enzymes and transporters. See Figs. 3.10–3.35 for the goodness of fit of the model and sensitivity analysis results. The results demonstrated that utilizing the inhibitory parameters \({K}_{i}\) and \({k}_{\text{inact}}/{K}_{I}\) values derived from our study to fit the substrate concentration–time curves affected by the inhibitor yielded good agreement with the observed data, as illustrated in Fig. 6.
Integration of inhibitor parameters calculated using the study methodology in a physiologically based pharmacokinetic model to predict drug interactions. a Fluconazole 100 mg twice daily (BID) for 2 days and zolpidem 5 mg at day 2, clinical results from [98]. b Ketoconazole 200 mg BID for 3 days and alprazolam 1 mg once daily (QD) at day 3, clinical results from [97]. c Fluvoxamine 100 mg QD for 6 days and zolpidem 5 mg at day 6, clinical results from [99]. d Itraconazole 200 mg BID for 4 days and tacrolimus 3 mg at day 4, clinical results from [100]. E Voriconazole 400 mg BID at day 2, 200 mg BID for 3 days and tacrolimus 3 mg at day 5, clinical results from [101]. f Cimetidine 400 mg bid for 2 weeks and triazolam 0.5 mg at day 14, clinical results from [102]. g Diltiazem 60 mg three time daily (TID) for 2 days and triazolam 0.25 mg at day 2, clinical results from [103]. h Ritonavir 200 mg bid for 7 days and tadalafil 20 mg at day 3, clinical results from [104]. i Aprepitant 125 mg QD and bosutinib 500 mg, clinical results from [105], j Erythromycin 500 mg tid for 2 days and simvastatin 40 mg at day 2, clinical results from [106]. k Posaconazole 400 mg BID for 7 days and tacrolimus 0.05 mg/kg at 7 days, clinical results from [107]. l Clarithromycin 500 mg BID for 2 days and triazolam 0.125 mg at day 2, clinical results from [108]. m Saquinavir 1200 mg TID for 8 days and sildenafil 100 mg at 8 days, clinical results from [109]. n Azithromycin 500 mg QD for 3 days and sildenafil 100 mg at day 3, clinical results from [110]. o Roxithromycin 300 mg QD for 5 days and lovastatin 80 mg at day 5, clinical results from [111]. p Ranitidine 300 mg TID for 5 days and nifedipine 20mg at day 5, clinical results from [112]. q Nefazodone 100 mg BID for 9 days and alprazolam 1 mg bid for 6 days, clinical results from [113]. r Verapamil 80 mg tid for 2 days and simvastatin 40 mg at day 2, clinical results from [106]. The solid line represents the physiologically based pharmacokinetic fitted concentration–time curves of the victims affected by the perpetrators; the solid dots represent the measured values of victims concentration–time curves in clinical drug interaction trials; and the shading represents the arithmetic standard deviation of physiologically based pharmacokinetic simulations for the population
3.6 Application of Clinical Drug Interaction Risk Scales
Currently, the clinical drug interaction risk quantification system established based on the method by Ohno et al. is widely used [49], sorting drugs according to their substrate CR and inhibitor IR value. However, the CR value does not differentiate between the contributions of intestinal and hepatic enzymes. At the same time, the IR value does not consider significant changes in the inhibitory potency caused by different doses of the inhibitor. The RI for substrates and PI for inhibitors established in this study address these issues. We placed the clinically measured values from the validation set into the AUCR risk intervals obtained after sorting drugs based on their RI and PI values (Fig. 7a) to investigate whether our risk scale can reliably reflect the clinical DDI risk levels and compared them with the AUCR risk intervals obtained using the method of Ohno et al. [3] (Fig. 7b). Using the RI and PI values as quantitative measures reduced the probability of false-negative assessments of clinical DDI risk from 15.73% to 5.6%. From the figure, it is evident that the IR-CR system led to an uneven distribution of risk intervals for AUCR. In the low-to medium-risk intervals, where the AUCR is < 3, alterations in IR and CR values by 0.5 have minimal impacts on the outcomes. However, in the higher risk intervals where AUCR is 3, a change of 0.5 in IR and CR values resulted in more than a twofold difference in outcomes, which is unsuitable for the preliminary assessment of DDI risk.
Application of clinical drug–drug interaction risk scales. A Comparison of drug–drug interaction risk stratification using a risk index (RI) and a potency index (PI) from this study with clinically observed values. B: Comparison of drug risk stratification using inhibition ratio (IR) and contribution ratio (CR) from Ohna et al. with clinically observed values. The vertical axis denotes the magnitude of the inhibitor’s PI or IR values. The horizontal axis indicates the magnitude of the substrate’s RI or CR values. Color mapping reflects the magnitude of area under the curve ratio (AUCR) values associated with the predicted RI and PI values. Dots symbolize the coordinate positions in the graph for 89 pairs of substrate and inhibitor combinations from the validation set, with the color mapping of the dots representing the clinical observed AUCR values. All axes are logarithmic; the RI is derived from Eq. 10; the PI is obtained from Eqs. 11 and 12; IR is calculated as IR = (1/AUCR+1)/CR, as proposed by Ohno et al., where CR is computed as CR = (1/AUCR+1)/IR per Ohno et al. and IR is calculated using equation IR = (1/AUCR + 1)/CR based on the training set data of this study, derived from the method proposed by Ohno et al., CR is determined using equation CR = (1/AUCR + 1)/IR based on the training set data of this study, also from the method by Ohno et al.
In summary, this study included predicted AUCR values for 660 pairs of clinical drug interactions at specific inhibitor dosages, as illustrated in Fig. 8. In clinical use, these drugs allow for a clear pre-determination of safe co-administration with certain drugs based on RI or PI values, identifying combinations that may lead to high-risk DDIs. Moreover, for two potentially co-administered drugs, the fold increase in the AUC of one drug due to co-administration can be rapidly calculated, enabling precise dose adjustment recommendations. Similarly, assuming the inhibitor has linear PK characteristics, the free concentration of the inhibitor at the corresponding dose calculated in this study can be converted to concentrations at other doses. The fold increase in substrate AUC caused by adjusted doses of the inhibitor could be rapidly calculated. To facilitate the application of this method in clinical practice, we developed a simplified program based on Microsoft Excel 2021, as detailed in the mini-program in the ESM.
Matrix plot of the predicted fold increase in substrates area under the curve (AUC) due to inhibitors at specific dosages. The z-axis denotes the magnitude of the predicted AUC ratio (AUCR) values. The left axis arranges the substrates according to their risk index values. The right axis organizes the inhibitors by their potency index values at specific dosages. bid twice daily, pred predicted, tid three times daily
4 Discussion and Conclusions
Pharmacokinetic-mediated DDIs are among the primary causes of clinically relevant adverse drug reactions [50]. Studies indicate that DDIs in the USA result in an annual range of emergency department visits from 2600 to 220,000, impacting between 1.9 and 5 million hospitalized patients [51, 52]. These events, preventable and avoidable [53], often occur because of a lack of accessible tools and assessment databases for clinical pharmacists to promptly mitigate and provide adjustment recommendations [54]. The concomitant use and overlay of post-marketing drugs are complex and variable, making it impossible to fully assess and investigate DDIs in preclinical evaluations and clinical trials. The results of clinical drug interaction trials should be more fully utilized to serve as vital reference material for clinical pharmacists to assess the myriad of DDIs in clinical practice. This study took advantage of abundant clinical drug interaction trial data to calculate critical DDI-specific parameters, such as \({K}_{i}\),\({k}_{\text{inact}}/{K}_{I}\),\({f}_{m}\), and \({F}_{g}\), for effective and accurate extrapolation of interaction results, and to establish a quantitative clinical DDI risk assessment framework. Compared with previously published studies, this study achieved an accurate prediction of the DDI outcome of the inhibitors with the substrates at any dose.
Based on the reliable calculation of the in vivo DDI specific parameters, we have developed a Clinical Drug Interaction Risk Scale and the mini-program for clinical applications. If the RI value is < 2, the probability of adverse reactions due to PK DDIs in patients with special conditions, such as non-hepatic or renal dysfunction, is low. In such cases, intensive monitoring may not be necessary, and the primary focus should be on ensuring therapeutic effectiveness when co-medication. Conversely, if the RI value exceeds 2, it indicates that the drug is a more sensitive substrate for an enzyme. When co-administered with a high dose of enzyme inhibitor, the drug’s exposure in vivo increases significantly. Therefore, prescriptions should be carefully selected and managed when co-administration is necessary. The PI is determined using the intended clinical dose concentrations. If the inhibitor’s PI value is > 2, a quantitative assessment process must be initiated, in which the fold increase in the drug’s AUC when co-administered is calculated, and the reduction of the inhibitor concentration for an acceptable increase in substrate AUCR is determined. These actions specify the dose adjustment range for the inhibitor when co-administration with the drug. For example, ibrutinib, an orally active inhibitor targeting Bruton’s tyrosine kinase, undergoes significant first-pass and metabolic clearance, which are both mediated by CYP3A4 [55, 56]. Based on clinical trial results, which showed a 23-fold increase in AUC when co-administered with a daily dose of 400 mg of ketoconazole, its RI value was calculated as 22.7, classifying it as a high-risk victim drug. However, no clinical DDI trial data are currently available for this drug in combination with other CYP3A4 inhibitors. A clinical case reported in 2016 [57] described a 68-year-old patient with relapsed mantle cell lymphoma and hypertension who experienced severe diarrhea and dizziness, which led to unconsciousness and hospitalization 1 week after taking 560 mg of ibrutinib along with verapamil plus trandolapril 180 mg/2 mg. According to the risk assessment framework of this study, the PI value for a daily dose of verapamil of 180 mg was 4. Further quantitative predictions indicated that verapamil increased the AUC of ibrutinib three-fold, resulting in an exposure that exceeded the safe therapeutic window of ibrutinib. Employing the methodology of this study, if the daily dose of verapamil was reduced to 120 mg, the AUC of ibrutinib was predicted to increase by 2.27-fold. Therefore, for patients undergoing treatment with ibrutinib who require verapamil, the daily dose of verapamil should not exceed 120 mg. For drugs with wider therapeutic windows, an increase in exposure to inhibitors that does not exceed the therapeutic window is considered safe. The RI value for lovastatin is 17.68, and a daily verapamil dose of 240 mg can increase its AUC 3.5-fold. However, because of its broad therapeutic window, the combination of verapamil and lovastatin is not associated with adverse reactions [58, 59].
The methodology of this study involved the retrograde calculation of specific parameters for victim and perpetrator drugs based on human trial results. This is predicated on six assumptions. First, midazolam, a standard substrate for CYP3A4, is metabolized solely through CYP3A4-mediated clearance, with an \({\text{f}}_{\text{m CYP}3\text{A}4}\) of 0.92 and \({F}_{g}\) of 0.55. Second, the renal clearance of the included substrates is considered negligible. Third, the in vivo concentration of substrates is much lower than Km, thus the dosing does not affect the \({f}_{m\text{ CYP}3\text{A}4}\) value. Fourth, only the impact of CYP3A4 on interactions is considered, with CYP3A4 distributed in both intestinal epithelial cells and hepatocytes. Fifth, the free concentration of the inhibitor at the enzyme-binding site in intestinal epithelial cells is represented by the average concentration at the hepatic portal vein, and at the hepatocyte-binding site by the average concentration at the liver inlet. Sixth, for time-dependent inhibition, it is assumed that the free concentration of the inhibitor is much less than its KI. With these assumptions, the inhibitory constant of the inhibitor against CYP3A4 is calculated based on the concomitant administration results with midazolam. Subsequently, the human trial results of the combination of selective CYP3A4 inhibitors with other substrates were used to calculate the \({f}_{m\text{ CYP}3\text{A}4}\) of these substrates.
It is noteworthy that the prediction of DDIs in special populations requires particular attention. More reliable results could be obtained through verification using PBPK models, with a thorough understanding of drug clearance pathways, interactions with endogenous substances, and the physiological parameters of special populations. However, this approach is time consuming and labor intensive in clinical practice. When assessing the risk of drug interactions on the exposure of victim drugs in the real world, the method should be simple and accessible and yield reliable results to provide sufficient confidence when adjusting drug dosages. The assessment system in this study offers a certain level of simplicity compared with simulations using PBPK models and a degree of accuracy over the currently popular static models, making it a useful reference for clinicians and patients in adjusting medication prescriptions. However, this study still has several unresolved issues: for enzyme inhibitors, there are no inhibitors that selectively target only one enzyme. The common CYP3A4 inhibitors included in this study, along with their metabolites, exhibited varying degrees of induction or inhibition of other enzymes, such as CYP2D6 and CYP2C9. Although these interactions can be accurately predicted, the role of CYP3A4 in these interactions may have been overestimated. For victim drugs, after the metabolic clearance pathways are inhibited, renal clearance of the parent drug may compensate for the decrease in clearance rate. This study overlooked this aspect of clearance, which could potentially lead to an overestimation of risks. Moreover, victim drugs may exert inhibitory or inductive effects on metabolic enzymes, resulting in bidirectional interactions that affect the fit of the outcomes.
References
Diasio RB. Sorivudine and 5-fluorouracil; a clinically significant drug–drug interaction due to inhibition of dihydropyrimidine dehydrogenase. Br J Clin Pharmacol. 1998;46(1):1–4.
Backman JT, Kyrklund C, Neuvonen M, Neuvonen PJ. Gemfibrozil greatly increases plasma concentrations of cerivastatin. Clin Pharmacol Ther. 2002;72(6):685–91.
Ohno Y, Hisaka A, Suzuki H. General framework for the quantitative prediction of CYP3A4-mediated oral drug interactions based on the AUC increase by coadministration of standard drugs. Clin Pharmacokinet. 2007;46(8):681–96.
Hisaka A, Ohno Y, Yamamoto T, Suzuki H. Prediction of pharmacokinetic drug–drug interaction caused by changes in cytochrome P450 activity using in vivo information. Pharmacol Ther. 2010;125(2):230–48.
Loue C, Tod M. Reliability and extension of quantitative prediction of CYP3A4-mediated drug interactions based on clinical data. AAPS J. 2014;16(6):1309–20.
Galetin A, Gertz M, Houston JB. Contribution of intestinal cytochrome P450-mediated metabolism to drug–drug inhibition and induction interactions. Drug Metab Pharmacokinet. 2010;25(1):28–47.
Floren LC, Bekersky I, Benet LZ, Mekki Q, Dressler D, Lee JW, et al. Tacrolimus oral bioavailability doubles with coadministration of ketoconazole. Clin Pharmacol Ther. 1997;62(1):41–9.
Mary FP, Louis YL, Lim HK, Kecheng L, Aram O, Mei-Yi Z, et al. Identification of a novel route of extraction of sirolimus in human small intestine: roles of metabolism and secretion. J Pharmacol Exp Ther. 2002;301(1):174.
Moton A, Ma L, Krishna G, Martinho M, Seiberling M, McLeod J. Effects of oral posaconazole on the pharmacokinetics of sirolimus. Curr Med Res Opin. 2009;25(3):701–7.
Varhe A, Olkkola KT, Neuvonen PJ. Effect of fluconazole dose on the extent of fluconazole–triazolam interaction. Br J Clin Pharmacol. 1996;42(4):465–70.
US FDA. Drug development and drug interactions: table of substrates, inhibitors and inducers. 2020. https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers. Accessed 24 Aug 2022.
Fuhr U, Hsin C-H, Li X, Jabrane W, Sörgel F. Assessment of pharmacokinetic drug–drug interactions in humans: in vivo probe substrates for drug metabolism and drug transport revisited. Annu Rev Pharmacol Toxicol. 2019;59(1):507–36.
Nagar S, Argikar UA, Tweedie DJ. Enzyme kinetics in drug metabolism: fundamentals and applications. Methods Mol Biol. 2014;1113:1–6.
Kenworthy KE, Bloomer JC, Clarke SE, Houston JB. CYP3A4 drug interactions: correlation of 10 in vitro probe substrates. Br J Clin Pharmacol. 1999;48(5):716–27.
Liu YT, Hao HP, Liu CX, Wang GJ, Xie HG. Drugs as CYP3A probes, inducers, and inhibitors. Drug Metab. 2007;39(4):699–721.
Hisaka A, Ohno Y, Yamamoto T, Suzuki H. Theoretical considerations on quantitative prediction of drug–drug interactions. Drug Metab Pharmacokinet. 2010;25(1):48–61.
Thelen K, Dressman JB. Cytochrome P450-mediated metabolism in the human gut wall. J Pharm Pharmacol. 2009;61(5):541–58.
Obach RS, Walsky RL, Venkatakrishnan K. Mechanism-based inactivation of human cytochrome p450 enzymes and the prediction of drug–drug interactions. Drug Metab Dispos. 2007;35(2):246–55.
Rowland Yeo K, Walsky RL, Jamei M, Rostami-Hodjegan A, Tucker GT. Prediction of time-dependent CYP3A4 drug–drug interactions by physiologically based pharmacokinetic modelling: impact of inactivation parameters and enzyme turnover. Eur J Pharm Sci. 2011;43(3):160–73.
Chan CYS, Roberts O, Rajoli RKR, Liptrott NJ, Siccardi M, Almond L, et al. Derivation of CYP3A4 and CYP2B6 degradation rate constants in primary human hepatocytes: a siRNA-silencing-based approach. Drug Metab Pharmacokinet. 2018;33(4):179–87.
Paine MF, Davis CL, Shen DD, Marsh CL, Raisys VA, Thummel KE. Can oral midazolam predict oral cyclosporine disposition? Eur J Pharm Sci. 2000;12(1):51–62.
Kim RB, Wandel C, Leake B, Cvetkovic M, Fromm MF, Dempsey PJ, et al. Interrelationship between substrates and inhibitors of human CYP3A and P-glycoprotein. Pharm Res. 1999;16(3):408–14.
Wojtyniak JG, Britz H, Selzer D, Schwab M, Lehr T. Data digitizing: accurate and precise data extraction for quantitative systems pharmacology and physiologically-based pharmacokinetic modeling. CPT Pharmacometr Syst Pharmacol. 2020;9(6):322–31.
Hou T, Wang J, Zhang W, Xu X. ADME evaluation in drug discovery. 6. Can oral bioavailability in humans be effectively predicted by simple molecular property-based rules? J Chem Inf Model. 2007;47(2):460–3.
Falcón-Cano G, Molina C, Cabrera-Pérez M. ADME Prediction with KNIME: development and validation of a publicly available workflow for the prediction of human Oral Bioavailability. J Chem Inf Model. 2020;60(6):2660–7.
Ito K, Iwatsubo T, Kanamitsu S, Ueda K, Suzuki H, Sugiyama Y. Prediction of pharmacokinetic alterations caused by drug–drug interactions: metabolic interaction in the liver. Pharmacol Rev. 1998;50(3):387–412.
Ito K, Iwatsubo T, Kanamitsu S, Nakajima Y, Sugiyama Y. Quantitative prediction of in vivo drug clearance and drug interactions from in vitro data on metabolism, together with binding and transport. Annu Rev Pharmacol Toxicol. 1998;38:461–99.
Tseng E, Eng H, Lin J, Cerny MA, Tess DA, Goosen TC, et al. Static and dynamic projections of drug–drug interactions caused by cytochrome P450 3A time-dependent inhibitors measured in human liver microsomes and Hhepatocytes. Drug Metab Dispos. 2021;49(10):947–60.
Fahmi OA, Hurst S, Plowchalk D, Cook J, Guo F, Youdim K, et al. Comparison of different algorithms for predicting clinical drug–drug interactions, based on the use of CYP3A4 in vitro data: predictions of compounds as precipitants of interaction. Drug Metab Dispos. 2009;37(8):1658–66.
Filppula AM, Parvizi R, Mateus A, Baranczewski P, Artursson P. Improved predictions of time-dependent drug–drug interactions by determination of cytosolic drug concentrations. Sci Rep. 2019;9(1):5850.
Rostami-Hodjegan A, Tucker G. ‘In silico’ simulations to assess the ‘in vivo’ consequences of ‘in vitro’ metabolic drug–drug interactions. Drug Discov Today Technol. 2004;1(4):441–8.
Yang J, Jamei M, Yeo KR, Rostami-Hodjegan A, Tucker GT. Misuse of the well-stirred model of hepatic drug clearance. Drug Metab Dispos. 2007;35(3):501.
Varma MV, Obach RS, Rotter C, Miller HR, Chang G, Steyn SJ, et al. Physicochemical space for optimum oral bioavailability: contribution of human intestinal absorption and first-pass elimination. J Med Chem. 2010;53(3):1098–108.
Gertz M, Davis JD, Harrison A, Houston JB, Galetin A. Grapefruit juice–drug interaction studies as a method to assess the extent of intestinal availability: utility and limitations. Curr Drug Metab. 2008;9(8):785–95.
Yau E, Petersson C, Dolgos H, Peters SA. A comparative evaluation of models to predict human intestinal metabolism from nonclinical data. Biopharm Drug Dispos. 2017;38(3):163–86.
Kato M, Chiba K, Hisaka A, Ishigami M, Kayama M, Mizuno N, et al. The intestinal first-pass metabolism of substrates of CYP3A4 and P-glycoprotein-quantitative analysis based on information from the literature. Drug Metab Pharmacokinet. 2003;18(6):365–72.
US FDA. In vitro drug interaction studies: cytochrome P450 enzyme- and transporter-mediated drug interactions. Guidance for industry. 2020. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/in-vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions. Accessed 7 May 2020.
Hewitt NJ, Gómez Lechón MJ, Houston JB, Hallifax D, Brown HS, Maurel P, et al. Primary hepatocytes: current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab Rev. 2007;39(1):159–234.
Chen Y, Liu L, Monshouwer M, Fretland AJ. Determination of time-dependent inactivation of CYP3A4 in cryopreserved human hepatocytes and assessment of human drug–drug interactions. Drug Metab Dispos. 2011;39(11):2085.
Vieira MLT, Kirby B, Ragueneau-Majlessi I, Galetin A, Chien JYL, Einolf HJ, et al. Evaluation of various static in vitro-in vivo extrapolation models for risk assessment of the CYP3A inhibition potential of an investigational drug. Clin Pharmacol Ther. 2014;95(2):189–98.
Zhou S, Yung Chan S, Cher Goh B, Chan E, Duan W, Huang M, et al. Mechanism-based inhibition of cytochrome P450 3A4 by therapeutic drugs. Clin Pharmacokinet. 2005;44(3):279–304.
Ito K, Brown HS, Houston JB. Database analyses for the prediction of in vivo drug–drug interactions from in vitro data. Br J Clin Pharmacol. 2004;57(4):473–86.
Tseng E, Eng H, Lin J, Cerny MA, Tess DA, Goosen TC, et al. Static and dynamic projections of drug–drug interactions caused by cytochrome P450 3A time-dependent inhibitors measured in human liver microsomes and hepatocytes. Drug Metab Dispos. 2021;49(10):947.
Bachmann KA. Inhibition constants, inhibitor concentrations and the prediction of inhibitory drug drug interactions: pitfalls, progress and promise. Curr Drug Metab. 2006;7(1):1–14.
Hua L, Chiang CW, Cong W, Li J, Wang X, Cheng L, et al. The cancer drug fraction of metabolism database. CPT Pharmacometr Syst Pharmacol. 2019;8(7):511–9.
Peng Y, Cheng Z, Xie F. Evaluation of pharmacokinetic drug–drug interactions: a review of the mechanisms, in vitro and in silico approaches. Metabolites. 2021;11(2):75.
Zhang X, Yang Y, Grimstein M, Fan J, Grillo JA, Huang S-M, et al. Application of PBPK modeling and simulation for regulatory decision making and its impact on US prescribing information: an update on the 2018–2019 submissions to the US FDA’s Office of Clinical Pharmacology. J Clin Pharmacol. 2020;60(S1):S160–78.
Grimstein M, Yang Y, Zhang X, Grillo J, Huang SM, Zineh I, et al. Physiologically based pharmacokinetic modeling in regulatory science: an update from the US Food and Drug Administration’s Office of Clinical Pharmacology. J Pharm Sci. 2019;108(1):21–5.
Hisaka A, Kusama M, Ohno Y, Sugiyama Y, Suzuki H. A proposal for a pharmacokinetic interaction significance classification system (PISCS) based on predicted drug exposure changes and its potential application to alert classifications in product labelling. Clin Pharmacokinet. 2009;48(10):653–66.
Palleria C, Di Paolo A, Giofrè C, Caglioti C, Leuzzi G, Siniscalchi A, et al. Pharmacokinetic drug–drug interaction and their implication in clinical management. J Res Med Sci. 2013;18(7):601–10.
Magro L, Moretti U, Leone R. Epidemiology and characteristics of adverse drug reactions caused by drug–drug interactions. Expert Opin Drug Saf. 2012;11(1):83–94.
CDC. National Center for Health Statistics. FastStats: hospital utilization. 2018. https://www.cdc.gov/nchs/fastats/hospital.htm. Accessed 18 May 2024.
Scheife RT, Hines LE, Boyce RD, Chung SP, Momper JD, Sommer CD, et al. Consensus recommendations for systematic evaluation of drug–drug interaction evidence for clinical decision support. Drug Saf. 2015;38(2):197–206.
Moreau F, Simon N, Walther J, Dambrine M, Kosmalski G, Genay S, et al. Does DDI-predictor help pharmacists to detect drug–drug interactions and resolve medication issues more effectively? Metabolites. 2021;11(3):173.
Scheers E, Leclercq L, de Jong J, Bode N, Bockx M, Laenen A, et al. Absorption, metabolism, and excretion of oral 14C radiolabeled ibrutinib: an open-label, phase I, single-dose study in healthy men. Drug Metab Dispos. 2015;43(2):289–97.
de Jong J, Skee D, Murphy J, Sukbuntherng J, Hellemans P, Smit J, et al. Effect of CYP3A perpetrators on ibrutinib exposure in healthy participants. Pharmacol Res Perspect. 2015;3(4): e00156.
Lambert Kuhn E, Levêque D, Lioure B, Gourieux B, Bilbault P. Adverse event potentially due to an interaction between ibrutinib and verapamil: a case report. J Clin Pharm Ther. 2016;41(1):104–5.
Wiggins BS, Saseen JJ, Page RL 2nd, Reed BN, Sneed K, Kostis JB, et al. Recommendations for management of clinically significant drug–drug interactions with statins and select agents used in patients with cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2016;134(21):e468–95.
Rätz Bravo AE, Tchambaz L, Krähenbühl-Melcher A, Hess L, Schlienger RG, Krähenbühl S. Prevalence of potentially severe drug–drug interactions in ambulatory patients with dyslipidaemia receiving HMG-CoA reductase inhibitor therapy. Drug Saf. 2005;28(3):263–75.
Lu C, Berg C, Prakash SR, Lee FW, Balani SK. Prediction of pharmacokinetic drug–drug interactions using human hepatocyte suspension in plasma and cytochrome P450 phenotypic data. III. In vitro-in vivo correlation with fluconazole. Drug Metab Dispos. 2008;36(7):1261.
Desbans C, Hilgendorf C, Lutz M, Bachellier P, Zacharias T, Weber JC, et al. Prediction of fraction metabolized via CYP3A in humans utilizing cryopreserved human hepatocytes from a set of 12 single donors. Xenobiotica. 2014;44(1):17–27.
Nguyen HQ, Kimoto E, Callegari E, Obach RS. Mechanistic modeling to predict midazolam metabolite exposure from in vitro data. Drug Metab Dispos. 2016;44(5):781.
Obach RS, Walsky RL, Venkatakrishnan K, Gaman EA, Houston JB, Tremaine LM. The utility of in vitro cytochrome P450 inhibition data in the prediction of drug–drug interactions. J Pharmacol Exp Ther. 2006;316(1):336.
Brown HS, Ito K, Galetin A, Houston JB. Prediction of in vivo drug–drug interactions from in vitro data: impact of incorporating parallel pathways of drug elimination and inhibitor absorption rate constant. Br J Clin Pharmacol. 2005;60(5):508–18.
Chen M, Nafziger AN, Bertino JS. Drug-metabolizing enzyme inhibition by ketoconazole does not reduce interindividual variability of CYP3A activity as measured by oral midazolam. Drug Metab Dispos. 2006;34(12):2079.
Ohno Y, Hisaka A, Suzuki HJ. General framework for the quantitative prediction of CYP3A4-mediated oral drug interactions based on the AUC increase by coadministration of standard drugs. Clin Pharmacokinet. 2007;46:681–96.
Gertz M, Harrison A, Houston JB, Galetin A. Prediction of human intestinal first-pass metabolism of 25 CYP3A substrates from in vitro clearance and permeability data. Drug Metab Dispos. 2010;38(7):1147.
Karlsson FH, Bouchene S, Hilgendorf C, Dolgos H, Peters SA. Utility of in vitro systems and preclinical data for the prediction of human intestinal first-pass metabolism during drug discovery and preclinical development. Drug Metab Dispos. 2013;41(12):2033–46.
Hatley O. Mechanistic prediction of intestinal first-pass metabolism using in vitro data in preclinical species and in man [Student thesis: Phd]: Manchester Pharmacy School 2014.
Bohets H, Lavrijsen K, Hendrickx J, van Houdt J, van Genechten V, Verboven P, et al. Identification of the cytochrome P450 enzymes involved in the metabolism of cisapride: in vitro studies of potential co-medication interactions. Br J Pharmacol. 2000;129(8):1655–67.
Rehmel J, Ferguson-Sells L, Morse BL, Li B, Dickinson GL. Physiologically based pharmacokinetic modeling of tadalafil to inform pediatric dose selection in children with pulmonary arterial hypertension. CPT Pharmacometr Syst Pharmacol. 2022;11(2):173–84.
Warrington JS, Shader RI, von Moltke LL, Greenblatt DJ. In vitro biotransformation of sildenafil (Viagra): identification of human cytochromes and potential drug interactions. Drug Metab Dispos. 2000;28(4):392–7.
Shiraga T, Niwa T, Teramura Y, Kagayama A, Tsutsui M, Ohno Y, et al. Oxidative metabolism of tacrolimus and its metabolite by human cytochrome P450 3A subfamily. Drug Metab Pharmacokinet. 1999;14(4):277–85.
Picard N, Djebli N, Sauvage FL, Marquet P. Metabolism of sirolimus in the presence or absence of cyclosporine by genotyped human liver microsomes and recombinant cytochromes P450 3A4 and 3A5. Drug Metab Dispos. 2007;35(3):350–5.
Wang RW, Kari PH, Lu AY, Thomas PE, Guengerich FP, Vyas KP. Biotransformation of lovastatin. IV. Identification of cytochrome P450 3A proteins as the major enzymes responsible for the oxidative metabolism of lovastatin in rat and human liver microsomes. Arch Biochem Biophys. 1991;290(2):355–61.
Neuvonen PJ, Kantola T, Kivistö KT. Simvastatin but not pravastatin is very susceptible to interaction with the CYP3A4 inhibitor itraconazole. Clin Pharmacol Ther. 1998;63(3):332–41.
Houston JB, Galetin A. Methods for predicting in vivo pharmacokinetics using data from in vitro assays. Curr Drug Metab. 2008;9(9):940–51.
Galetin A, Burt H, Gibbons L, Houston JB. Prediction of time-dependent CYP3A4 drug–drug interactions: impact of enzyme degradation, parallel elimination pathways, and intestinal inhibition. Drug Metab Dispos. 2006;34(1):166–75.
Pan X, Yamazaki S, Neuhoff S, Zhang M, Pilla RV. Unraveling pleiotropic effects of rifampicin by using physiologically based pharmacokinetic modeling: assessing the induction magnitude of P-glycoprotein-cytochrome P450 3A4 dual substrates. CPT Pharmacometr Syst Pharmacol. 2021;10(12):1485–96.
US FDA. Multi-discipline review application number: 213246Orig1s000. 2020. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/213246Orig1s000MultidisciplineR.pdf. Accessed 7 Aug 2020.
US FDA. Multi-discipline review application number: 213591Orig1s000. 2020. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/213591Orig1s000Approv.pdf. Accessed 15 Mar 2023.
US FDA. Multi-discipline review application number: 212608Orig1s000. 2019. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/212608Orig1s000Approv.pdf. Accessed 10 Aug 2023.
US FDA. Clinincal pharmacology and biopharmaceutics review application number: 207997Orig1s000, 207997Orig2s000. 2016. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2017/207997Orig1Orig2s000lbl.pdf. Accessed 10 Aug 2023.
Dowty ME, Lin J, Ryder TF, Wang W, Walker GS, Vaz A, et al. The pharmacokinetics, metabolism, and clearance mechanisms of tofacitinib, a janus kinase inhibitor, in humans. Drug Metab Dispos. 2014;42(4):759–73.
US FDA. Multi-discipline review application number: 212725Orig1s000 212726Orig1s000. 2019. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/212725Orig1s000,%20212726Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 213217Orig1s000. 2019. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/213217Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 211192Orig1s000. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/211192Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 210496Orig1s000. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210496Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 211349Orig1s000. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/211349Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 211155Orig1s000, 211155Orig2s000. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/211155Orig1Orig2s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 210951Orig1s000. 2018. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210951orig1s000multidiscipliner.pdf. Accessed 10 Aug 2023.
US FDA. Multi-discipline review application number: 210259Orig1s000. 2017. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2017/210259Orig1s000MultidisciplineR.pdf. Accessed 10 Aug 2023.
US FDA. Clinical pharmacology and biopharmaceutics review(s) application number: 208462Orig1s000. 2016. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/208462Orig1s000MedR.pdf. Accessed 10 Aug 2023.
US FDA. Clinical pharmacology and biopharmaceutics review(s) application number: 206192Orig1s000. 2014. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2015/206192orig1s000clinpharmr.pdf. Accessed 12 Sept 2022.
US FDA. Clinical pharmacology and biopharmaceutics review(s) application number: 206162Orig1s000. 2014. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/206162Orig1s000PharmR.pdf. Accessed 15 Aug 2022.
Adiwidjaja J, Boddy AV, McLachlan AJ. Potential for pharmacokinetic interactions between Schisandra sphenanthera and bosutinib, but not imatinib: in vitro metabolism study combined with a physiologically-based pharmacokinetic modelling approach. Br J Clin Pharmacol. 2020;86(10):2080–94.
Greenblatt DJ, Wright CE, von Moltke LL, Harmatz JS, Ehrenberg BL, Harrel LM, et al. Ketoconazole inhibition of triazolam and alprazolam clearance: differential kinetic and dynamic consequences. Clin Pharmacol Ther. 1998;64(3):237–47.
Greenblatt DJ, von Moltke LL, Harmatz JS, Mertzanis P, Graf JA, Durol AL, et al. Kinetic and dynamic interaction study of zolpidem with ketoconazole, itraconazole, and fluconazole. Clin Pharmacol Ther. 1998;64(6):661–71.
Vlase L, Popa A, Neag M, Muntean D, Achim M, Leucuţa SE. Effect of fluvoxamine on the pharmokinetics of zolpidem: a two-treatment period study in healthy volunteers. Clin Exp Pharmacol Physiol. 2012;39(1):9–12.
Vanhove T, Annaert P, Knops N, de Loor H, de Hoon J, Kuypers DRJ. In vivo CYP3A4 activity does not predict the magnitude of interaction between itraconazole and tacrolimus from an extended release formulation. Basic Clin Pharmacol Toxicol. 2019;124(1):50–5.
Imamura CK, Furihata K, Okamoto S, Tanigawara Y. Impact of cytochrome P450 2C19 polymorphisms on the pharmacokinetics of tacrolimus when coadministered with voriconazole. J Clin Pharmacol. 2016;56(4):408–13.
Pourbaix S, Desager JP, Hulhoven R, Smith RB, Harvengt C. Pharmacokinetic consequences of long term coadministration of cimetidine and triazolobenzodiazepines, alprazolam and triazolam, in healthy subjects. Int J Clin Pharmacol Ther Toxicol. 1985;23(8):447–51.
Varhe A, Olkkola KT, Neuvonen PJ. Diltiazem enhances the effects of triazolam by inhibiting its metabolism. Clin Pharmacol Ther. 1996;59(4):369–75.
US FDA. Clinical pharmacology and biopharmaceutics reviews application number: 21-368. 2003. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2003/21-368_Cialis_Medr_P1.pdf. Accessed 24 Aug 2021.
Ono C, Hsyu PH, Abbas R, Loi CM, Yamazaki S. Application of physiologically based pharmacokinetic modeling to the understanding of bosutinib pharmacokinetics: prediction of drug–drug and drug–disease interactions. Drug Metab Dispos. 2017;45(4):390–8.
Kantola T, Kivistö KT, Neuvonen PJ. Erythromycin and verapamil considerably increase serum simvastatin and simvastatin acid concentrations. Clin Pharmacol Ther. 1998;64(2):177–82.
Sansone-Parsons A, Krishna G, Martinho M, Kantesaria B, Gelone S, Mant TG. Effect of oral posaconazole on the pharmacokinetics of cyclosporine and tacrolimus. Pharmacotherapy. 2007;27(6):825–34.
Greenblatt DJ, von Moltke LL, Harmatz JS, Counihan M, Graf JA, Durol AL, et al. Inhibition of triazolam clearance by macrolide antimicrobial agents: in vitro correlates and dynamic consequences. Clin Pharmacol Ther. 1998;64(3):278–85.
Muirhead GJ, Wulff MB, Fielding A, Kleinermans D, Buss N. Pharmacokinetic interactions between sildenafil and saquinavir/ritonavir. Br J Clin Pharmacol. 2000;50(2):99–107.
Muirhead GJ, Faulkner S, Harness JA, Taubel J. The effects of steady-state erythromycin and azithromycin on the pharmacokinetics of sildenafil in healthy volunteers. Br J Clin Pharmacol. 2002;53(Suppl. 1):37s–43s.
Bucher M, Mair G, Kees F. Effect of roxithromycin on the pharmacokinetics of lovastatin in volunteers. Eur J Clin Pharmacol. 2002;57(11):787–91.
Smith SR, Kendall MJ, Lobo J, Beerahee A, Jack DB, Wilkins MR. Ranitidine and cimetidine; drug interactions with single dose and steady-state nifedipine administration. Br J Clin Pharmacol. 1987;23(3):311–5.
Greene D, Salazar DE, Dockens RC, Kroboth PD, Barbhaiya RHJJocp. Coadministration of nefazodone and benzodiazepines: III. A pharmacokinetic interaction study with alprazolam. J Clin Psychopharmacol. 1995;15(6):399–408.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Funding
No funding was received for the preparation of this article.
Conflicts of interest/competing interests
Tong Yuan, Fulin Bi, Kuan Hu, Yuqi Zhu, Yan Lin, and Jin Yang have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The datasets generated during and/or analyzed during the current study are available from supplemental data or the corresponding author on reasonable request.
Code availability
The code generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
TY and YL wrote the manuscript; TY and JY designed the research; FB, TY, and YZ performed the research; TY analyzed the data; all authors critically reviewed several drafts and approved the final manuscript.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yuan, T., Bi, F., Hu, K. et al. Clinical Trial Data-Driven Risk Assessment of Drug–Drug Interactions: A Rapid and Accurate Decision-Making Tool. Clin Pharmacokinet 63, 1147–1165 (2024). https://doi.org/10.1007/s40262-024-01404-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-024-01404-0