Abstract
RNA localization is a key mechanism in the regulation of protein expression. In neurons, this includes the axonal transport of select mRNAs based on the recognition of axonal localization motifs in these RNAs by RNA-binding proteins. Bioinformatic analyses of axonal RNAs suggest that selective inclusion of such localization motifs in mature mRNAs is one mechanism controlling the composition of the axonal transcriptome. The subsequent translation of axonal transcripts in response to specific stimuli provides precise spatiotemporal control of the axonal proteome. This axonal translation supports local phenomena including axon pathfinding, mitochondrial function, and synapse-specific plasticity. Axonal protein synthesis also provides transport machinery and signals for retrograde trafficking to the cell body to effect somatic changes including altering the transcriptional program. Here we review the remarkable progress made in recent years to identify and characterize these phenomena.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tight regulation of subcellular protein localization is required for establishing and maintaining the structural and functional polarity of neurons. A fundamental question is how genes expressed by a single nucleus can be regulated to achieve this specificity. One strategy that neurons employ to achieve spatial protein distribution is the transport of mRNAs to distal cellular sites where their subsequent translation can be regulated in response to appropriate signals. This local protein synthesis provides tight spatiotemporal control of the local proteome, supporting not only polarized function but also the capacity for rapid changes of the proteome in response to stimuli. While local neuronal translation has been long studied in dendrites, more recent efforts have focused on elucidating the roles and regulation of local translation in the axon. Here we focus on recent advances uncovering how particular mRNAs are transported to axons as well as the subsequent regulation of their translation in order to support axon outgrowth, mitochondrial function, synaptic plasticity, and signaling between axons and somata.
Localization of Transcripts to Axons
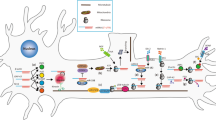
In cells, mRNA is packaged into large, multi-molecular granules along with proteins, such as RNA-binding proteins and molecular motors, which determine the localization of the mRNA within the cell. These granules can be transported into axons immediately or in response to specific events, such as injury or signaling by neuromodulators or growth factors [1•, 2, 3••, 4]. mRNAs dynamically associate with these granules based on selective binding between RNA sequence motifs and the RNA-binding proteins in the granules. For instance, the axonal transport of the transcript encoding β-actin (but not the one encoding the closely related γ-actin) is mediated by an interaction between a zip code motif in the β-actin RNA and the zipcode-binding protein ZBP1/IMP1 [5–7]. Individual proteins may be encoded by several different transcripts generated through alternative splicing and/or alternate polyadenylation site selection. These various transcripts may differ in whether they include the axonal localization motifs, thereby regulating which transcripts are transported into axons (Fig. 1a). This idea is consistent with the finding that axonal and somatic transcriptomes differ in their enrichment for particular transcript variants [8•]. Further, an individual axonal RNA is likely to have multiple motifs that collaborate to guide it to particular compartments within axons. For example, in the sea slug Aplysia californica, the transcript for the sensorin gene is directed to axonal processes by one motif in the 3′ UTR (untranslated region) and specifically to presynaptic terminals by a second motif in the 5′ UTR [9, 10]. The localization of an RNA to and within axons is thus mediated by selective association with appropriate granules based on the presence of particular motifs in that RNA.
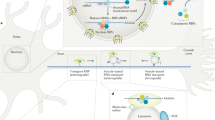
RNA transport and local translation play various roles in axons including: a Assuring that specific transcripts localize to the correct compartments; b Supplying growth cones with materials necessary for responding to guidance cues; c Supporting mitochondrial function; d Facilitating structural changes underlying synaptic plasticity; and e Providing proteins that are trafficked back from the axon to the soma during retrograde signaling. Listed is a selection of locally synthesized proteins whose involvement with each step has been characterized
The granules with which an individual RNA associates are influenced by competitive binding with both other RNAs and RNA-binding proteins. The β-actin zip code and similar motifs are expressed in many transcripts, such as that encoding GAP-43, which can compete with the β-actin mRNA for association with ZBP1. Experimentally manipulating the levels of the various zip code-containing transcripts or ZBP1 modulates which RNAs are transported into axons [11, 12•, 13]. Similarly, the RNA-binding protein TRF2-S (telomere repeat binding factor 2) can facilitate the axonal localization of its target mRNAs [14••]. This association and subsequent localization is antagonized by TRF2-S interaction with the RNA-binding protein Fragile X mental retardation protein (FMRP). Whether the relevant interaction between these two proteins occurs in the soma or axon is unclear as FMRP, though present in the cell bodies of all neurons, is found in the axons of select neuronal populations [15, 16]. The relative expression levels of both RNAs and RNA-binding proteins will thus influence the RNAs that are present in axons, thereby altering how axonal translation contributes to cellular function.
Regulation of Axonal Translation
Extrinsic signaling on restricted subdomains of the axonal arbor can drive localized protein synthesis with high spatial and temporal resolution. As discussed below, this localized translation can occur with remarkable specificity occurring selectively in response to extracellular stimuli that occur on one side of a growth cone or only at activated synapses. This specificity is supported by the subcompartmental localization of signaling molecules and/or translational machinery based on selective association with components of the cytoskeleton. Binding of the growth cone chemoattractant netrin-1 to the axonal receptor DCC leads to clustering of translational machinery and an induction of axonal protein synthesis [17]. Interestingly, the actin cytoskeleton (located at the periphery of the growth cone where netrin is sensed) is required for the initiation of netrin-induced axonal translation via the initiation regulator eIF4E-BP1 (eukaryotic initiation factor 4E binding protein 1) [18]. In contrast, the microtubules (located at the core of the growth cone) are required for translational elongation via the elongation regulator eEF2 (eukaryotic elongation factor 2) and its kinase, eEF2K. The spatial restriction of axonal translation thus relies not only on the limited binding of extracellular cues, but also on the highly localized translational machinery.
The signaling mechanisms that control local axonal translation largely resemble those that occur in the cell body. The best studied of these is signaling through the PI3K (phosphatidylinositol 3-kinase) and mTOR (mammalian target of rapamycin) kinase cascades. As discussed above, netrin-1 binding activates PI3K, leading to phosphorylation and activation of mTOR and the subsequent phosphorylation of eIF4E-BP1 [18]. Similarly, NGF binding signals through PI3K and mTOR to induce axonal translation of the GTPase TC10 and the polarity complex protein Par3 [19•]. Whether the molecules that converge on mTOR signaling result in the translation of the same or separate sets of axonal mRNAs remains to be determined. However, activation of different cell surface receptors can elicit distinct translational programs within axons [20]. Furthermore, each of these ligands has disparate effects on axons apart from the induction of local translation. Thus, newly synthesized proteins will be incorporated into a different cellular environment depending on the other effects initiated by ligand binding.
Axonal Outgrowth
Local translation in axons has been well studied in the context of axon outgrowth during development or after injury (Fig. 1b). Axonal outgrowth can occur in two different modes: non-directional extension and growth directed by guidance cues. Non-directional extension does not appear to depend on local translation, since axons can elongate for 2–5 h in the presence of translation inhibitors [21–23]. Instead, local translation seems to be a strategy for responding to directional guidance cues [24, 25]. Interestingly, whether the response to a guidance cue involves local protein synthesis depends on the concentration of that cue, with lower concentrations generally inducing local translation [26]. Many guidance cues affect protein synthesis by increasing mTOR-mediated translation [27]. Since mTOR is activated in a concentration-dependent manner, the edge of the growth cone receiving the highest concentration of guidance cue has the greatest activation of protein synthesis, leading to asymmetric growth toward the source of the guidance cue [25]. Local translation affects growth by supplying new β-actin monomers, which display different post-translational modifications from soma-derived β-actin, allowing more efficient actin polymerization and thereby biasing cytoskeletal extension in one direction [28–30]. Local translation thus supports axon pathfinding by regulating the arrangement of the growth cone cytoskeleton.
Local Translation and Context-Dependent Growth Mechanisms
The ability of axons to extend new processes over large distances depends on the cellular context that these axons must navigate. For example, while axons throughout the body must grow during development, regrowth following injury in adult animals is restricted to peripheral axons. Interestingly, central axons can exhibit the capacity to regrow if provided an appropriate PNS substrate, and these regrowing CNS axons upregulate translational machinery in a manner very similar to that seen in peripheral axons [31•]. This difference in the localization of translational machinery may in part reflect the interactions with PNS versus CNS glia as regenerating peripheral axons have been proposed to receive ribosomes from surrounding glia [32, 33]. Additionally, peripheral axons exhibit a greater capacity for regeneration during development than in adult animals. This difference may reflect alterations in the axonal transcriptome, as embryonic axons are enriched for transcripts classically implicated in guidance and growth, while adult axons are enriched for transcripts associated with inflammation and immunity [34]. The differential ability of axons to regrow is thus reflected in differences in the levels of axonal translational machinery as well as the composition of the axonal transcriptome. Similar studies will be required to determine whether this pattern holds in other contexts, such as in axon outgrowth from central neurons generated via adult neurogenesis.
The Role of mRNA Degradation and Translational Inhibition in Axonal Guidance
Fine-tuning the local concentration of proteins requires balancing the signals that drive protein synthesis with those that repress this synthesis. For example, when a growth cone receives the guidance cue Sema3a, growth cone collapse is mediated by the translational suppression of MAP1B by FMRP [35, 36]. Similarly, the microRNA miR-132 promotes axonal extension by repressing the transcript for RASA1, a component of the Ras pathway that feeds into mTOR signaling [37]. These studies illustrate that the production of axonal proteins is also balanced by brakes that regulate their function.
In addition to inhibiting translation, protein synthesis levels can also be negatively regulated via mRNA degradation. Controlled degradation can serve to regulate the surface expression of guidance receptors to guide axons through complex, multi-step trajectories. Commissural axons crossing to the other side of the body must be sequentially attracted to and then repelled from the midline. During midline attraction, the transcript coding for the repulsive receptor Robo3.2 is translationally silent due to its transport within a repressive complex. Guidance cues at the midline de-repress the transcript, allowing for midline-sensitive Robo3.2 expression. However, many commissural axons remain close to the midline after crossing, so the levels of Robo3.2 protein must be kept in check. This is accomplished by a stop codon unique to the Robo3.2 splice form that induces nonsense-mediated decay after the first round of translation, ensuring that each transcript generates a limited number of Robo3.2 proteins [38••]. Nonsense-mediated decay is also used to limit the number of proteins produced in dendrites [39], suggesting that rapid turnover of mRNA after translation is a general mechanism for controlling local protein synthesis.
Mitochondrial Function
Several recent studies have indicated that axonal translation can support mitochondrial function in axons (Fig. 1c). In addition to serving as an energy source, neuronal mitochondria have diverse roles including storing calcium and modulating the membrane potential [40]. Many of these newly identified roles may be especially prevalent in axons, since injured neurons downregulate transcripts related to electron transport but upregulate other transcripts like those related to mitochondrial fusion and morphology [41]. Even proteins produced from transcripts with similar abundance in axons and somata may have different functions depending on their site of production. Axonally synthesized lamin B, which is distinct from somatically synthesized lamin B in its ability to associate with mitochondria, can be specifically downregulated without affecting somatic lamin B. This manipulation causes axonal degeneration, demonstrating that the two populations of lamin B play non-redundant roles [42]. The importance of axonal translation to mitochondrial function is highlighted by the observation that specifically decreasing the axonal expression of nuclear-encoded mitochondrial transcripts caused mice to display anxiety- and depression-like behaviors [43•]. These findings also emphasize that the pool of locally produced proteins may differ functionally from those proteins synthesized in the soma.
Synapse Formation and Signaling
In comparison to local translation during axon outgrowth, relatively little is known about how axonal translation impacts synapses. Several lines of evidence from both vertebrate and, especially, invertebrate systems suggest a role for local protein synthesis in supporting structural changes underlying presynaptic plasticity (Fig. 1d). Long-term facilitation (LTF) requires presynaptic protein synthesis in both crayfish and Aplysia [44–46]. These studies have implicated that many of the signaling pathways found to regulate protein synthesis in other cellular contexts, including the TOR (target of rapamycin), MAP kinase, calcineurin, and PI3K pathways [47, 48]. Interestingly, FMRP has a presynaptic role in Aplysia in the regulation of synaptic depression induced by FMRFamide, but not in serotonin-induced LTF, indicating the presence of distinct regulatory mechanisms underlying different forms of plasticity [49]. Local translation also likely contributes to several forms of presynaptic plasticity in the vertebrate nervous system. In nerve-muscle preparations from Xenopus, BDNF (brain-derived neurotrophic factor) induces a presynaptic translation-dependent synaptic potentiation [50]. Presynaptic translation has also been suggested to support long-term depression (LTD) and long-term potentiation (LTP) in rodent corticostriatal and hippocampal mossy fiber axons [51, 52]. Local synthesis of proteins directly in the presynaptic compartment thus contributes to specific forms of synaptic plasticity in both invertebrate and vertebrate nervous systems.
Bidirectional Synaptic Signaling Regulates Presynaptic Translation
Critical to elucidating the functional roles of presynaptic protein synthesis is identifying the transcripts that are localized to and translated in the presynaptic compartment. Here, significantly less progress has been made than in illuminating the general regulatory principles. Best characterized of the presynaptic transcripts is the transcript encoding the neuropeptide sensorin in Aplysia neurons. Sensorin RNA localizes to synapses shortly after synapse formation and blocking translation of this RNA inhibits the formation of new synapses [46, 53, 54]. Serotonin-induced LTF, which markedly increases the size and complexity of synapses, induces local protein synthesis of sensorin in isolated neurites [9]. Importantly, this local translation only occurs if the postsynaptic cell is stimulated, as it requires calcium-dependent postsynaptic release of netrin, which binds to the presynaptic DCC receptor [1•]. The detailed study of the regulation of local sensorin production thus reveals the importance of bidirectional communication across the synapse in the regulation of presynaptic protein synthesis.
Regulation of Synaptic Vesicles
Several presynaptically synthesized proteins have also been identified in vertebrate neurons, with findings to date converging on a role for presynaptic translation in regulating the structure and dynamics of the vesicle pool. Locally translated β-catenin mRNA rapidly accumulates at nascent synaptic sites [55•]. This β-catenin helps to restrain the vesicle pool at these synapses. Loss of β-catenin, either through cell-wide knockout or by specifically targeting axonal translation using siRNAs, leads to increased synaptic release during lower stimulation intensities and an inability to sustain release during prolonged stimulation [55•, 56]. Local translation of β-catenin may thus serve to stabilize the vesicle pool and constrain the release probability as synapses mature [57].
Investigation of additional presynaptic transcripts suggests that separate pools of mRNAs may be used under different conditions in response to distinct signaling events. Inhibiting all translation briefly (for up to several hours) at nascent synapses leads to a decrease in the recycling pool of synaptic vesicles without a change in the size of the readily releasable pool [58]. This inhibition can lead to a complete destabilization and loss of nascent synapses. In contrast, mature synapses do not require ongoing local translation and are not affected by this manipulation. Local translation of the calmodulin-dependent kinase CaMKIIα is required to maintain these new synapses. The stabilization of newly formed synapses thus appears to, at least in some conditions, require ongoing local synthesis of CaMKIIα. However, while acute block of local translation decreases the recycling vesicle pool, longer term blockade specifically of cap-dependent protein synthesis leads to an increase in the size of the recycling vesicle pool [59]. This effect at least in part involves modulation of cdk5 signaling via local translation of the cdk5 activator p35. Both CaMKII and cdk5 can phosphorylate β-catenin [60, 61], as well as other proteins important for the regulation of the synaptic vesicle pools. The opposing roles of locally translated CaMKII and cdk5 in the regulation of vesicles suggest that distinct signals may employ translational programs that include one or the other. Although the endogenous signals that regulate presynaptic translation in the vertebrate nervous system have not been well characterized, it is of note that netrin regulates axonal β-catenin translation during outgrowth in both hippocampal and thalamocortical axons [62, 63]. The cues that regulate local presynaptic translation may thus be broadly conserved in both Aplysia and mammals. Further, β-catenin translation is regulated by FMRP, which is found at presynaptic sites in select axonal populations throughout the nervous system [15, 16, 64]. Interestingly, FMRP is mutated in the autism-related disorder Fragile X syndrome, while β-catenin is mutated in both autism and intellectual disability [65, 66]. Thus, alterations in presynaptic protein synthesis may contribute to the symptoms seen in autism patients. Together, these findings suggest a model in which local protein synthesis controls presynaptic β-catenin levels as well as the post-translational regulation of its activity in order to modulate the size and dynamics of the synaptic vesicle pool in a synapse-specific manner.
Retrograde Signaling
Retrograde Transport of Transcription Factors
In addition to providing proteins for use locally in the axon, axonal translation can also serve as the source for retrograde signals transmitted from the axon to the soma (Fig. 1e). For example, local protein synthesis of transcription factors in the distal axon can lead to somatic transcriptional responses during development and in response to injury. Axonal exposure to NGF in developing neurons can initiate local synthesis of the transcription factor CREB (cAMP response element-binding protein), which is then trafficked to the soma [67]. The subsequent specific somatic transcriptional responses to this CREB signaling are critical for neuronal survival early in development. NGF may also impact CREB function through the local translation of the inositol metabolism enzyme IMPA1 (myo-inositol monophosphatase-1). Disrupting Impa1 translation in axons alters both nuclear CREB activation and axonal integrity [2]. In a sciatic nerve and dorsal root ganglia axonal injury model, trafficking of axonally translated STAT3 via the importin/dynein complex influences neuronal survival [68]. Likewise, axonal synthesis of the transcription factor SMAD1/5/8 in the trigeminal ganglia mediates the somatic transcriptional response to BDNF and BMP4 (bone morphogenetic protein 4) with effects on neuronal specification [69]. Similarly, axonal synthesis of Luman/CREB3, an intra-axonal ER transmembrane protein that can be cleaved to produce an active transcription factor, helps regulate axonal regeneration after injury [70•]. Together, these studies demonstrate that axonal translation can impact gene expression profiles via retrograde transport of axonally synthesized transcription factors.
Retrograde Signaling in Response to Injury
Retrograde signaling in response to axonal injury communicates information from distal cues to the soma and enacts transcriptional change to aid in neuronal survival and regeneration. After axotomy, there is a rapid and transient increase in intracellular calcium concentration, which propagates from the site of injury back to the soma and acts as a priming signal for the transcriptional injury response [71–73]. The increase in calcium concentration activates PKCμ (protein kinase Cμ), which subsequently induces nuclear export of HDAC5 (histone deacetylase 5) into the axon [73]. HDAC5 export enhances histone acetylation to activate pro-regenerative genetic cascades necessary for axon regeneration. Injury induces axonal translation of filamin A, which binds to HDAC5 to facilitate tubulin deacetylation and localization to the growth cone while promoting regeneration [74•]. This response is likely cell type specific, as it was not seen in an optic nerve model [72]. These studies demonstrate that, in addition to the trafficking of transcription factors, retrograde transport of locally translated products plays a role in the epigenetic response to injury.
Axonal protein synthesis also plays an important role in the nuclear localization of a signal-dependent retrograde signaling complex. In this system, importin-α, importin-β1, and dynein bind signals and transcription factors at the site of axonal injury and transport them on microtubules to the soma to effect a specific transcriptional response [75]. This retrograde transport is supported by injury-induced axonal synthesis of several components of this complex including importin-β1, RanBP1 (RAN-binding protein 1), and the intermediate filament vimentin [76–78]. Importin-β1 is encoded by two transcripts that vary in the length of their 3′ UTR. Specifically targeting the axon-specific long variant eliminates axonal importin-β1 synthesis without disrupting the somatic synthesis [76]. This loss of axonally translated importin-β1 both reduced and delayed the somatic transcriptional response to injury in vivo resulting in delayed functional recovery. RanBP1 stimulates dissociation of RanGTP from newly synthesized importin-β1, which can then form a complex with importin-α and dynein [77]. The injury-induced Ca++ wave increases activity of calpain, which cleaves the newly synthesized vimentin to facilitate importin-dependent retrograde transport of the active, phosphorylated form of the kinase ERK [78, 79]. Interestingly, investigation in a pseudorabies virus infection model in superior cervical ganglia cells identified several other retrograde transport-associated proteins synthesized locally in the axon: peripherin (another intermediate filament), annexin A2 (a calcium-regulated phospholipid-binding protein mediating interactions between vesicles and the actin cytoskeleton), and Pafah1b1 (a dynein regulator) are locally translated and required for efficient viral trafficking from axons to somata [80]. These findings thus highlight the importance of the axonal translation of transport machinery for soma-directed trafficking in both physiological and pathophysiological conditions.
Retrograde Signaling in the Alzheimer’s Brain
Although most of the research in axonal translation in the context of retrograde signaling has focused on the PNS, studies of Alzheimer’s disease (AD) provide compelling insights into the role this phenomenon plays in the CNS. Axonal transport is well known to be disrupted in AD and other neurodegenerative diseases [81, 82], leading to often-severe pathologies in the brain. AD is the leading cause of age-related dementia and is largely manifested through axonal pathogenesis including tau hyperphosphorylation and exposure to amyloid plaques [83, 84]. These plaques arise when pathogenic cleavage of APP (amyloid precursor protein) leads to formation of both Aβ plaques and oligomers. Aβ oligomers can bind to receptors, including mGluR5, to initiate signal cascades that alter neuronal physiology [85]. Axonal exposure to pathogenic Aβ oligomers interferes with BDNF retrograde signaling, and is thought to be sufficient to induce neurodegeneration without somatic exposure [84, 86]. Axonal exposure to these pathogenic oligomers also induces axonal recruitment of mRNAs including the transcript encoding ATF4 (activating transcription factor 4) [3••]. ATF4 is locally synthesized and retrogradely trafficked to the soma, initiating neuronal expression of the ER stress-associated transcription factor CHOP and subsequent cell death, indicating a contribution of intra-axonal transcription to neurodegeneration in AD patients. Other mRNA transcripts with increased localization in the Aβ-exposed axons included APP, ApoE (apolipoprotein E), clusterin, and fermentin family homolog 2; all of these proteins have characterized roles in AD pathology [87–90]. These data point to a previously unexplored path by which pathology spreads from the distal axon via retrograde trafficking of axonally synthesized proteins and also add to the relatively small but growing body of evidence for axonal translation in the mature CNS.
Conclusions
Neurons use local protein synthesis to establish and maintain their exquisite structural and functional polarity. Significant progress has been made in the last several decades in revealing the importance of local axonal translation to the structure and function of axons. Recent technical advances in subcellular transcriptome analysis have allowed the identification of many axonal transcripts and suggest patterns in the composition of the axonal transcriptome. The number of transcripts identified by these approaches indicates that axonal translation supports many functions that have yet to be characterized. It should be noted that these approaches will be biased toward the most highly expressed axonal transcripts. However, low levels of axonal expression for a particular transcript does not negate the possibility of an important functional role for that transcript, as exemplified by the requirement for axonal synthesis of TC10 despite vanishingly low axonal levels of the TC10 mRNA [19•]. Additionally, it remains to be determined how association with specific RNA granules segregates the axonal transcriptome into distinct functional pools, each of which can respond to distinct cell signaling events. Further, neurons are remarkably diverse in their morphology and function. The extent to which the axonal transcriptome and its regulated translation differ among these varied neuronal types remains to be determined. Finally, while an increasing number of experiments has been conducted in live organisms, most studies of axonal translation have been performed in cell culture. It will be of great interest to further elucidate how axonal translation is regulated by and contributes to the biology of living organisms.
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
• Kim S, Martin KC. Neuron-wide RNA transport combines with netrin-mediated local translation to spatially regulate the synaptic proteome. eLife. 2015;4:e04158. By investigating how presynaptic translation can be limited specifically to activated synapses despite broad distribution of presynaptic RNAs, this study identified that postsynaptic activity induces calcium-dependent release of netrin. This netrin binds to its presynaptic receptor to induce local translation only at activated synapses.
Andreassi C, Zimmermann C, Mitter R, Fusco S, De Vita S, Devita S, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301.
•• Baleriola J, Walker CA, Jean YY, Crary JF, Troy CM, Nagy PL, et al. Axonally synthesized ATF4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158(5):1159–72. Alzheimer’s pathology appears to spread over long distances between brain regions that are connected with each other. This paper reveals that axon exposure to pathogenic Aβ oligomers induces the axonal transport and subsequent axonal translation of transcripts that can induce neurodegeneration.
Merianda TT, Coleman J, Kim HH, Kumar Sahoo P, Gomes C, Brito-Vargas P, et al. Axonal amphoterin mRNA is regulated by translational control and enhances axon outgrowth. J Neurosci. 2015;35(14):5693–706.
Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127(2):441–51.
Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17(4):2158–65.
Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J Neurosci. 1998;18(1):251–65.
• Minis A, Dahary D, Manor O, Leshkowitz D, Pilpel Y, Yaron A. Subcellular transcriptomics—dissection of the mRNA composition in the axonal compartment of sensory neurons. Dev Neurobiol. 2014;74(3):365–81. One of the most exhaustive surveys of an axonal transcriptome to date. Bioinformatic analysis revealed short sequence motifs that are enriched in axonal transcripts, which are candidate cis elements responsible for axonal localization.
Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, et al. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324(5934):1536–40.
Meer EJ, Wang DO, Kim S, Barr I, Guo F, Martin KC. Identification of a cis-acting element that localizes mRNA to synapses. Proc Natl Acad Sci. 2012;109(12):4639–44.
Donnelly CJ, Willis DE, Xu M, Tep C, Jiang C, Yoo S, et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30(22):4665–77.
• Donnelly CJ, Park M, Spillane M, Yoo S, Pacheco A, Gomes C, et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J Neurosci. 2013;33(8):3311–22. This work shows that RNAs with similar structures can compete for association with RNA granules that mediate axonal transport. As a result, modulating the relative axonal levels of β-actin and GAP-43 mRNAs can influence the size and shape of the axonal arbor.
Fallini C, Rouanet JP, Donlin-Asp PG, Guo P, Zhang H, Singer RH, et al. Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons. Dev Neurobiol. 2014;74(3):319–32.
•• Zhang P, Abdelmohsen K, Liu Y, Tominaga-Yamanaka K, Yoon J-H, Ioannis G, et al. Novel RNA- and FMRP-binding protein TRF2-S regulates axonal mRNA transport and presynaptic plasticity. Nat Commun. 2015;6:8888. This study identified TRF2-S as a novel RNA binding protein that directs the axonal transport of its target mRNAs to support axonal elongation and presynaptic transmitter release. This transport role is antagonized by competitive interactions with another RNA binding protein, FMRP. Competition between mRNA and FMRP for binding to TRF2-S is thus an important mechanism for regulating axonal RNA transport.
Akins MR, LeBlanc HF, Stackpole EE, Chyung E, Fallon JR. Systematic mapping of fragile X granules in the mouse brain reveals a potential role for presynaptic FMRP in sensorimotor functions. J Comp Neurol. 2012;520(16):3687–706.
Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J Neurosci. 2009;29(5):1514–24.
Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141(4):632–44.
Piper M, Lee AC, van Horck FPG, McNeilly H, Lu TB, Harris WA, et al. Differential requirement of F-actin and microtubule cytoskeleton in cue-induced local protein synthesis in axonal growth cones. Neural Develop. 2015;10:3.
• Gracias NG, Shirkey-Son NJ, Hengst U. Local translation of TC10 is required for membrane expansion during axon outgrowth. Nat Commun. 2014;5:3506. Axon growth requires a rapid and directed expansion of the plasma membrane. These studies indicate that local translation of TC10 regulates an exocyst-mediated increase in plasma membrane phospholipids.
Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178(6):965–80.
Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32(6):1013–26.
Leung K-M, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–56.
Höpker VH, Shewan D, Tessier-Lavigne M, Poo M, Holt C. Growth-cone attraction to netrin-1 is converted to repulsion by laminin-1. Nature. 1999;401(6748):69–73.
Jung H, Yoon BC, Holt CE. Axonal mRNA localization and local protein synthesis in nervous system assembly, maintenance and repair. Nat Rev Neurosci. 2012;13(5):308–24.
Shigeoka T, Lu B, Holt CE. Cell biology in neuroscience: RNA-based mechanisms underlying axon guidance. J Cell Biol. 2013;202(7):991–9.
Nédelec S, Peljto M, Shi P, Amoroso MW, Kam LC, Wichterle H. Concentration-dependent requirement for local protein synthesis in motor neuron subtype-specific response to axon guidance cues. J Neurosci. 2012;32(4):1496–506.
Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26(16):3729–36.
Wang J, Boja ES, Tan W, Tekle E, Fales HM, English S, et al. Reversible glutathionylation regulates actin polymerization in A431 cells. J Biol Chem. 2001;276(51):47763–6.
Karakozova M, Kozak M, Wong CCL, Bailey AO, Yates JR, Mogilner A, et al. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313(5784):192–6.
Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, et al. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol Biol Cell. 2010;21(8):1350–61.
• Kalinski AL, Sachdeva R, Gomes C, Lee SJ, Shah Z, Houle JD, et al. mRNAs and protein synthetic machinery localize into regenerating spinal cord axons when they are provided a substrate that supports growth. J Neurosci. 2015;35(28):10357–70. One of the key differences between the PNS neurons that regenerate and the CNS neurons that do not is that only PNS growth cones exhibit local translation. However, when CNS neurons are grown in a condition that promotes regeneration, local translation is upregulated, suggesting that the lack of local translation may contribute to the inability of CNS neurons to regenerate.
Court FA, Hendriks WTJ, MacGillavry HD, Alvarez J, van Minnen J. Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J Neurosci. 2008;28(43):11024–9.
Court FA, Midha R, Cisterna BA, Grochmal J, Shakhbazau A, Hendriks WT, et al. Morphological evidence for a transport of ribosomes from Schwann cells to regenerating axons. Glia. 2011;59(10):1529–39.
Gumy LF, Yeo GSH, Tung Y-CL, Zivraj KH, Willis D, Coppola G, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17(1):85–98.
Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol Cell Neurosci. 2006;32(1–2):37–48.
Li C, Bassell GJ, Sasaki Y. Fragile X mental retardation protein is involved in protein synthesis-dependent collapse of growth cones induced by Semaphorin-3A. Front Neural Circuits. 2009;3:11.
Hancock ML, Preitner N, Quan J, Flanagan JG. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Soc Neurosci. 2014;34(1):66–78.
•• Colak D, Ji S-J, Porse BT, Jaffrey SR. Regulation of axon guidance by compartmentalized nonsense-mediated mRNA decay. Cell. 2013;153(6):1252–65. Axons that traverse complex pathways often navigate by an initial attraction to intermediate targets, followed by subsequent repulsion from the same targets. In commissural neurons, this involves axon-specific mRNA isoforms that are uniquely subject to a decay signal released by its intermediate target, the floor-plate of the midline.
Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130(1):179–91.
Sheng Z-H. Mitochondrial trafficking and anchoring in neurons: new insight and implications. J Cell Biol. 2014;204(7):1087–98.
Taylor AM, Berchtold NC, Perreau VM, Tu CH, Jeon N, Cotman CW. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J Neurosci. 2009;29(15):4697–707.
Yoon BC, Jung H, Dwivedy A, O’Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148(4):752–64.
• Kar AN, Sun C-Y, Reichard K, Gervasi NM, Pickel J, Nakazawa K, et al. Dysregulation of the axonal trafficking of nuclear-encoded mitochondrial mRNA alters neuronal mitochondrial activity and mouse behavior. Dev Neurobiol. 2014;74(3):333–50. One of the first studies to identify a behavioral phenotype resulting from the axon-specific loss of a gene, Cytochrome C oxidase IV (COXIV).
Sherff CM, Carew TJ. Coincident induction of long-term facilitation in aplysia: cooperativity between cell bodies and remote synapses. Science. 1999;285(5435):1911–4.
Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, et al. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell. 1997;91(7):927–38.
Liu K, Hu JY, Wang D, Schacher S. Protein synthesis at synapse versus cell body: enhanced but transient expression of long-term facilitation at isolated synapses. J Neurobiol. 2003;56(3):275–86.
Casadio A, Martin KC, Giustetto M, Zhu H, Chen M, Bartsch D, et al. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell. 1999;99(2):221–37.
Beaumont V, Zhong N, Fletcher R, Froemke RC, Zucker RS. Phosphorylation and local presynaptic protein synthesis in calcium-and calcineurin-dependent induction of crayfish long-term facilitation. Neuron. 2001;32(3):489–501.
Till SM, Li H-L, Miniaci MC, Kandel ER, Choi Y-B. A presynaptic role for FMRP during protein synthesis–dependent long-term plasticity in Aplysia. Learn Mem. 2011;18(1):39–48.
Zhang X, Poo M. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36(4):675–88.
Huang C-C, Hsu K-S. Local protein synthesis and GABAB receptors regulate the reversibility of long-term potentiation at murine hippocampal mossy fibre–CA3 synapses. J Physiol. 2004;561(Pt 1):91–108.
Yin HH, Davis MI, Ronesi JA, Lovinger DM. The role of protein synthesis in striatal long-term depression. J Neurosci. 2006;26(46):11811–20.
Schacher S, Wu F, Panyko JD, Sun Z-Y, Wang D. Expression and branch-specific export of mRNA are regulated by synapse formation and interaction with specific postsynaptic targets. J Neurosci. 1999;19(15):6338–47.
Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured aplysia neurons. Neuron. 2006;49(3):349–56.
• Taylor AM, Wu J, Tai H-C, Schuman EM. Axonal translation of β-catenin regulates synaptic vesicle dynamics. J Neurosci. 2013;33(13):5584–9. Synaptogenic signals induce the presynaptic accumulation and translation of the RNA encoding β-catenin, a critical regulator of synaptic morphology and function. This translation impacts the organization of the synaptic vesicle pool.
Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, et al. Role of β-catenin in synaptic vesicle localization and presynaptic assembly. Neuron. 2003;40(4):719–31.
Bolshakov VY, Siegelbaum SA. Regulation of hippocampal transmitter release during development and long-term potentiation. Science. 1995;269(5231):1730–4.
Sebeo J, Hsiao K, Bozdagi O, Dumitriu D, Ge Y, Zhou Q, et al. Requirement for protein synthesis at developing synapses. J Neurosci. 2009;29(31):9778–93.
Hsiao K, Bozdagi O, Benson DL. Axonal cap-dependent translation regulates presynaptic p35. Dev Neurobiol. 2014;74(3):351–64.
Kesavapany S, Lau KF, McLoughlin DM, Brownlees J, Ackerley S, Leigh PN, et al. p35/cdk5 binds and phosphorylates beta-catenin and regulates beta-catenin/presenilin-1 interaction. Eur J Neurosci. 2001;13(2):241–7.
Flentke GR, Garic A, Hernandez M, Smith SM. CaMKII represses transcriptionally active β-catenin to mediate acute ethanol neurodegeneration and can phosphorylate β-catenin. J Neurochem. 2014;128(4):523–35.
Pratt T, Davey JW, Nowakowski TJ, Raasumaa C, Rawlik K, McBride D, et al. The expression and activity of β-catenin in the thalamus and its projections to the cerebral cortex in the mouse embryo. BMC Neurosci. 2012;13(1):20.
Kundel M, Jones KJ, Shin CY, Wells DG. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent Β-catenin mRNA translation in developing hippocampal neurons. J Neurosci. 2009;29(43):13630–9.
Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mrnas linked to synaptic function and autism. Cell. 2011;146(2):247–61.
O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485(7397):246–50.
De Boulle K, Verkerk AJMH, Reyniers E, Vits L, Hendrickx J, Van Roy B, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3(1):31–5.
Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10(2):149–59.
Ben-Yaakov K, Dagan SY, Segal-Ruder Y, Shalem O, Vuppalanchi D, Willis DE, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012;31(6):1350–63.
Ji S-J, Jaffrey SR. Intra-axonal translation of SMAD1/5/8 mediates retrograde regulation of trigeminal ganglia subtype specification. Neuron. 2012;74(1):95–107.
• Ying Z, Misra V, Verge VMK. Sensing nerve injury at the axonal ER: activated Luman/CREB3 serves as a novel axonally synthesized retrograde regeneration signal. Proc Natl Acad Sci USA. 2014;111(45):16142–7. Identifies Luman/CREB3 as a transcription factor that is locally synthesized and activated in response to axonal injury, acting as a retrogradely-trafficked signal to regulate regeneration. This study provides the first pieces of evidence directly linking an axonally-synthesized transcription factor to regenerative axon outgrowth.
Mandolesi G, Madeddu F, Bozzi Y, Maffei L, Ratto GM. Acute physiological response of mammalian central neurons to axotomy: ionic regulation and electrical activity. FASEB J. 2004;18(15):1934–6.
Cho Y, Cavalli V. HDAC5 is a novel injury-regulated tubulin deacetylase controlling axon regeneration. EMBO J. 2012;31(14):3063–78.
Cho Y, Sloutsky R, Naegle KM, Cavalli V. Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155(4):894–908.
• Cho Y, Park D, Cavalli V. Filamin A is required in injured axons for HDAC5 activity and axon regeneration. J Biol Chem. 2015;290(37):22759–70. Although histone deacetylases are classically associated with regulating chromatin in the nucleus, HDAC5 also plays a role in axons. Specifically, HDAC5 interacts with axonally-synthesized Filamin A after injury to modify the microtubule cytoskeleton.
Hanz S, Perlson E, Willis D, Zheng J-Q, Massarwa R, Huerta JJ, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40(6):1095–104.
Perry RB, Doron-Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, et al. Subcellular knockout of importin β1 perturbs axonal retrograde signaling. Neuron. 2012;75(2):294–305.
Yudin D, Hanz S, Yoo S, Iavnilovitch E, Willis D, Gradus T, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59(2):241–52.
Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45(5):715–26.
Perlson E, Michaelevski I, Kowalsman N, Ben-Yaakov K, Shaked M, Seger R, et al. Vimentin binding to phosphorylated Erk sterically hinders enzymatic dephosphorylation of the kinase. J Mol Biol. 2006;364(5):938–44.
Koyuncu OO, Perlman DH, Enquist LW. Efficient retrograde transport of pseudorabies virus within neurons requires local protein synthesis in axons. Cell Host Microbe. 2013;13(1):54–66.
De Vos KJ, Grierson AJ, Ackerley S, Miller CCJ. Role of axonal transport in neurodegenerative diseases. Annu Rev Neurosci. 2008;31:151–73.
Millecamps S, Julien J-P. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci. 2013;14(3):161–76.
Iqbal K, Liu F, Gong C-X, Alonso ADC, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol (Berl). 2009;118(1):53–69.
Liu Y, Yoo M-J, Savonenko A, Stirling W, Price DL, Borchelt DR, et al. Amyloid pathology is associated with progressive monoaminergic neurodegeneration in a transgenic mouse model of Alzheimer’s disease. J Neurosci. 2008;28(51):13805–14.
Um JW, Kaufman AC, Kostylev M, Heiss JK, Stagi M, Takahashi H, et al. Metabotropic glutamate receptor 5 is a coreceptor for Alzheimer Aβ oligomer bound to cellular prion protein. Neuron. 2013;79(5):887–902.
Poon WW, Carlos AJ, Aguilar BL, Berchtold NC, Kawano CK, Zograbyan V, et al. β-Amyloid (Aβ) oligomers impair brain-derived neurotrophic factor retrograde trafficking by down-regulating ubiquitin C-terminal hydrolase, UCH-L1. J Biol Chem. 2013;288(23):16937–48.
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–93.
DeMattos RB. Apolipoprotein E dose-dependent modulation of beta-amyloid deposition in a transgenic mouse model of Alzheimer’s disease. J Mol Neurosci MN. 2004;23(3):255–62.
Shulman JM, Imboywa S, Giagtzoglou N, Powers MP, Hu Y, Devenport D, et al. Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2014;23(4):870–7.
Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8.
Acknowledgements
Work in the Akins lab is funded by NIMH Grant R00MH90237. We thank Ryan Y. Korsak for the artwork in our figure. Michael R. Akins reports grants from NIMH during the conduct of study.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection on Neurogenetics and Psychiatric Genetics.
Rights and permissions
About this article
Cite this article
Korsak, L.I.T., Mitchell, M.E., Shepard, K.A. et al. Regulation of Neuronal Gene Expression by Local Axonal Translation. Curr Genet Med Rep 4, 16–25 (2016). https://doi.org/10.1007/s40142-016-0085-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40142-016-0085-2