Abstract
The hippocampus segmentation plays a crucial role in analyzing brain activities, which is a widely used biomarker for Alzheimer’s disease, epilepsy, and schizophrenia diagnosis. The automatic segmentation of hippocampus is a complex task, because of low signal contrast, small structural size, and insufficient image resolution. The automatic hippocampus segmentation utilizing magnetic resonance imaging (MRI) is effective in clinical diagnosis and neuro-science research. In this manuscript, a new automatic segmentation model in MRI is implemented for hippocampus segmentation. Initially, the MRI brain images are collected from Neuroimaging Tools and Resources Collaboratory (NITRC) and the Open Access Series of Imaging Studies (OASIS) databases. Further, color normalization technique is employed for improving the visual ability, and reduce the impulse noise and machinery noise (mechanical and electrical noises) in the image. Finally, the Fuzzy C Means-based Level Set Local Ternary Pattern with Enhanced Edge Indicator (FCM-LSLTPEEI) is proposed for hippocampus segmentation. The proposed model combines FCM and EEI functions in the LSLTP model, where the important phase is to adapt the EEI function effectively with the LSLTP. The experimental outcomes revealed that the proposed FCM-LSLTPEEI model obtained 98.90% and 99.01% of accuracy on the OASIS and NITRC databases, which are superior compared to the traditional models.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
In recent times, hippocampus segmentation has gained more attention among the researcher’s community, because it plays a crucial role in disease monitoring and diagnosis [1]. Schizophrenia, Alzheimer’s disease and epilepsy are neuro-degenerative disorders, which are characterized by defective brain cells like amyloid plaques, and neuro fibrillary tangles [2, 3]. The shrinkage of the hippocampus in the brain is the major physical characteristic, those who suffer from Schizophrenia, Alzheimer’s disease and epilepsy [4], whereas the hippocampus is the thin brain area, which is responsible for storing memories. Therefore, a precise and accurate brain volume measurement is important to understand the nature of brain problems, rigorously [5]. Compared to other imaging modalities such as ultrasound, histopathology, X-ray, and computed tomography, the MRI is the extensively utilized neuro-imaging modality for clinical valuation of the human brain [6].
The MRI modality is effective in understanding the structural abnormalities related to brain disorders, where the manual/handcrafted hippocampus segmentation consumes more time and requires expert physicians in this field. Hence, the expensiveness of the handcrafted segmentation prompted the exploration of automatic hippocampus segmentation algorithms [7, 8]. In recent decades, several automated segmentation algorithms have been developed for hippocampus segmentation such as FCM, K-means clustering, superpixel clustering, and level set approach. However, the conventional segmentation algorithms suffer from three major problems such as requiring previous knowledge about the MRI brain images, sensitive to machinery noise, and increased computational cost [9, 10]. Therefore, a novel automatic hippocampus segmentation model is proposed in this manuscript, and its major contributions are determined as follows:
-
This manuscript aims to propose a superior segmentation model for improving the accuracy of hippocampus segmentation.
-
The input MRI brain images are acquired from two online databases like NITRC and OASIS.
-
The color normalization technique is applied for enhancing the visibility level of MRI brains and for decreasing the unwanted distortions such as impulse noise and machinery noises (mechanical and electrical noises) in the brain images.
-
The hippocampus segmentation is accomplished utilizing FCM-LSLTPEEI. Initially, the optimal clusters are determined by the FCM clustering technique. Then, hippocampus segmentation is accomplished using a level set algorithm with LTP descriptor and EEI function. The inclusion of the LTP descriptor and EEI function in the level set algorithm effectively excludes the extraneous boundary points and preserves the real boundaries for achieving better segmentation results.
-
At last, the proposed FCM-LSLTPEEI model’s performance is validated by means of accuracy, Jaccard coefficient, sensitivity, and dice coefficient.
This manuscript is structured in the following manner: Articles on the topic “hippocampus segmentation” are reviewed in Sect. “Literature Review.” The detailed investigation of the FCM-LSLTPEEI model is denoted in Sect. “Methodology” and the experimental examination of the FCM-LSLTPEEI model is stated in Sect. “Experimental Results.” The conclusion of the FCM-LSLTPEEI model is given in Sect. “Conclusion.”
Literature Review
Carmo et al. [1] have introduced a hippocampus segmentation model named multiple U-Net-based convolutional neural network (CNN) for timely detection of Alzheimer and epilepsy diseases. The quantitative outcomes demonstrated the successfulness of the multiple U-Net-based CNN model in hippocampus segmentation than the conventional U-Net model. Still, the developed model needs a preprocessing step to further improve hippocampus segmentation. Liu and Yan [2] developed a new automated model, which combines lattice Boltzmann (LB) and deep belief network (DBN) for hippocampus segmentation. The developed model attained consistent segmentation results compared to manual segmentation, but its structure was complex.
Shao et al. [3] implemented a classification guided boundary regression (CGBR) model for hippocampal segmentation. Initially, the 3D displacements were predicted from the brain scans, and then the boundary maps were determined using a voting method. Further, the information about longitudinal context and spatial context was combined with the voted hippocampal boundary maps for precise hippocampal segmentation, where the manual intervention was high in the CGBR model that needed to be automated. Shi et al. [4] introduced a generative adversarial networks (GANs) model for hippocampal segmentation. The GANs comprise the adversarial model and U-Net model, which extract local information from the collected brain images and determine the interrelationship between the feature values. The adversarial training smoothens the image edges for precise hippocampal segmentation. In the medical image segmentation, the GANs network structure was unstable.
Vijayalakshmi and Savita [5] introduced an enhanced FCM technique for segmenting hippocampus in MRI images. In the resulting phase, the enhanced FCM technique was tested in light of dice coefficients and Jaccard coefficients. The prior specification of the number of clusters was a main concern in the FCM technique, while implementing image segmentation. Pang et al. [6] presented local linear mapping (LLM) for effective hippocampus segmentation. The LLM model utilizes distance information and labels information of the hippocampus boundary for enhancing the accuracy of segmentation. Additionally, K-means clustering and confidence-based weighted average (CWA) was incorporated with the LLM model for accurate segmentation with limited memory and computational costs.
Jiang et al. [7] integrated level set algorithm and adaptive region growing for hippocampus segmentation in MRI images. In the natural MRI brain images, the edge stopping function was not exactly zero, which may degrade the hippocampus segmentation. Palumbo et al. [8] used two automatic MRI brain segmentation models: statistical parametric mapping (SPM) and free surfer (FS) for hippocampus segmentation. Yang et al. [9] utilized a multi-scale deep CNN model for hippocampal subfields segmentation. The developed multi-scale deep CNN model achieved comparable segmentation accuracy in light of dice coefficient, which was superior compared to the existing models. Nasser et al. [10] introduced U-Net model for hippocampus segmentation. However, the deep CNN and U-Net models were computationally expensive, because they needed an enormous amount of images to attain significant segmentation performance.
By surveying existing literature, the hippocampus has ambiguous boundaries that makes it challenging in segmenting interested regions. Furthermore, raw medical images have varied pixel intensity levels, due to the difference in scanner models and acquisition parameters. In order to address the following concerns, a novel automatic segmentation model, FCM-LSLTPEI, is proposed in this manuscript for precise hippocampus segmentation. The proposed FCM-LSLTPEEI effectively handles fuzzy and complex boundaries in the raw medical image. Additionally, the combination, color normalization with FCM-LSLTPEEI model, fully addresses the problem of inconsistent contrast by clustering image pixels into fuzzy classes based on the intensity value.
Methodology
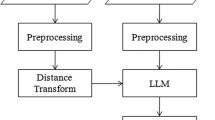
In medical image analysis, the precise hippocampal subfield segmentation in MRI is a challenging process, because of the morphological complexity and small structural size of the hippocampal subfields. The existing segmentation models faced difficulties in obtaining the ideal segmentation results. In this manuscript, the hippocampal subfields segmentation is accomplished using FCM-LSLTPEEI model. In the hippocampus segmentation, the proposed system includes three major phases: image collection: OASIS and NITRC databases, image pre-processing: normalization technique and hippocampal segmentation: FCM-LSLTPEEI model. The working process of the developed framework is mentioned in Fig. 1.
Data Description
In this manuscript, the effectiveness of FCM-LSLTPEEI model is tested on the OASIS and NITRC databases. The NITRC database has T1-weighted MRI images of fifty subjects in which ten subjects are non-epileptics and the residual forty subjects belong to temporal lobes; here, the term “T1” represents “longitudinal magnetization weighted.” In this database, the hippocampus labels are given to twenty-five subjects for model training. The researchers can submit their segmentation outcome for the residual twenty-five images to validate the model performance. In addition, the OASIS database comprises T1-weighted cross-sectional MRI images of 416 subjects, who belong to the age group of 18 to 96. In this OASIS database, the subjects are right-handed, which includes both women and men. Among 416 subjects, 100 subjects are over 60 age and are diagnosed with moderate Alzheimer’s disease. The collected MRI brain images are specified in Fig. 2.
Image Preprocessing
After image collection, preprocessing is carried out for enhancing the image visibility level by decreasing unwanted distortions in images acquired from OASIS and NITRC databases. The majority of the existing preprocessing techniques are utilized for image reconstruction, noise removal, transforming the image from binary to grayscale, etc. In this particular application, MRI brain images are captured or acquired by using MRI equipment, where the acquired hippocampus images are contaminated with impulse noise and machinery noise. So, the color normalization technique is implemented, which is effective in removing the machinery and impulse noise and improving the image visibility level. In this context, the color normalization technique standardize pixel intensity variations that ensure that the intensity range of the hippocampus region is uniform throughout all images, which is mathematically denoted in Eq. (1). This process makes it easier for the segmentation model (FCM-LSLTPEEI) to delineate and identify the hippocampus precisely.
where\(I\) represents original acquired MRI brain images, \(N\) represents normalized MRI brain images, \({\text{newMax}}-{\text{newMin}}\) specifies altered new image pixel intensity values, and \({\text{Min}}\) and \({\text{Max}}\) represent minimum and maximum pixel intensity values, which ranges from 0 to 255. The sample normalized MRI brain images of OASIS and NITRC databases are represented in Fig. 3.
Hippocampal Segmentation
In this scenario, the normalized MRI brain images are passed into the FCM-LSLTPEEI model for hippocampal segmentation. Firstly, the FCM clustering technique is used for selecting the correct clusters and then the LSLTPEEI is employed on the selected clusters for hippocampal segmentation. The selection of correct clusters improves the segmentation performance in ill-defined portions and enhances the object localization in the complex template. Generally, the FCM clustering technique considers an object as a cluster member with the value of degree of membership (DoM) function. The objective function \({O}_{j}\) is minimized in each iteration of the FCM technique, which is defined in Eq. (2).
where \(D\) indicates data-points of the normalized MRI brain images\(N\), \(C\) represents the number of clusters, \({\delta }_{ij}\) denotes DoM function of \({i}^{{\text{th}}}\) data-points \({x}_{i}\) in the cluster\(j\), \({c}_{j}\) represents center vector of the cluster\(j\), the term \(\Vert {x}_{i}-{c}_{j}\Vert \) estimates the similarity between the data-points \({x}_{i}\) and the center vector of the cluster\(j\), and \(\Vert .\Vert \) indicates an absolute value of an expression. The DoM function of the data points \({x}_{i}\) is determined using Eq. (3).
where \(m\) denotes fuzziness coefficient, and the center vector of the cluster \(j\) is estimated by using Eq. (4). Furthermore, the term \(\Vert {x}_{i}-{c}_{k}\Vert \) computes the similarity between the data points \({x}_{i}\) and \({c}_{k}\).
In FCM clustering technique, the fuzziness coefficient \(m\) determines the cluster tolerance, as mentioned in Eqs. (3) and (4). The higher value of fuzziness coefficient \(m\) denotes the larger overlap between the clusters \(C\). The higher value of \(m\) utilizes more data points \({x}_{i}\), and the DoM function is neither 0 nor 1. The DoM function validates the number of iterations completed by the FCM clustering technique. In this scenario, the term “accuracy” \(a\) is computed utilizing the DoM function from the current iteration \(k\) to the succeeding iterations \(k+1\). The term \(a\) is estimated using Eq. (5).
where\(\Delta \) indicates higher vector value, and \({\delta }_{ij}^{k}\) and \({\delta }_{ij}^{k+1}\) represents DoM function of present iteration \(k\) and the succeeding iterations \(k+1\). The selected clusters are three, which is stated in Fig. 4.
After finding the optimal clusters, the level set algorithm is applied to the selected clusters for segmenting the hippocampal with the benefit of active contour. The level set formulation should be smoother, during the level set evolutions. In this manner, the level set has fulfilled the condition of the signed distance function (SDF),\(\left|\nabla \mathrm{\varnothing }\right|=1\), where the SDF \(z=\mathrm{\varnothing }\left(x,y\right)\) represents the image surface. The time dependent level set formulation \(\mathrm{\varnothing }\left(t,x,y\right)\) is employed for rendering the level set solution instead of parametric characterization of active contours. The term \(\mathrm{\varnothing }\left(t,x,y\right)\) is utilized for facilitating the evolution of the active contours by determining the zero-level-set \(\Gamma \left(t\right)\), where the conditions of the term \(\mathrm{\varnothing }\left(t,x,y\right)\) is indicated in Eq. (6).
However, the time variable \(t\) in the level set formulation leads to higher dimensions, so an additional computation is included with the practical advantages. By estimating the level set formulation value \(\mathrm{\varnothing }\), the interface \(\Gamma \left(t\right)\) is obtained that manages the topological variations in the implicit interface \(\Gamma \left(t\right)\). The revolution of level set formulation \(\mathrm{\varnothing }\) vs time \(t\) is derived using the chain rule, as mentioned in Eqs. (7) and (8).
, whereas
Equation (7) is rewritten as represented in Eq. (9). Hence, the polar co-ordinates are the suitable description for the contour revolution, and it is denoted in Eq. (10). Particularly, the evolution \(\mathrm{\varnothing }\) is calculated using the numerical level set evolution equation, which is stated in Eq. (11).
where \({F}_{n}\) specifies normal forces that include external force (artificial momentum or image gradient) and internal force (area, contour length, mean curvature, etc.), \(\left|\nabla \mathrm{\varnothing }\right|\) denotes normal direction and \({\mathrm{\varnothing }}_{0}(x,y)\) represents initial contour. The driving force \(F\) is controlled by the EEI function \(g\) for evolving the initial level set contour toward the local optimal solution. In addition, the EEI function \(g\) emphasizes the gray level difference between the directions x and y. The EEI function includes other two directions like − 45 and + 45 degrees for depressing the image orientation effect that is mathematically specified in Eqs. (12), (13), and (14).
where
where \({N}_{{G}_{\sigma ,x}}^{{{\prime}}}\) and \({N}_{{G}_{\sigma ,y}}^{{{\prime}}}\) measure the diagonal angles of 45° and 135°, where these angels help in excluding the extraneous boundary points that preserve the real boundaries. The term \({g}^{{{\prime}}}\) is used for differentiating the positive variational boundaries, still, the MRI brain images suffer from the pathology and physiology complexities, so the LTP descriptor is employed to deal with incompleteness and imprecision. In this segmentation model, the LTP descriptor is employed for regularizing the dynamic interface in the level set algorithm. However, the segmented image of the FCM-LSLTPEEI model is indicated in Fig. 5, and the steps followed in the developed framework is mentioned below.
Steps followed in the developed framework
Step 1: Preprocessing: Employed a standard preprocessing technique: color normalization for enhancing the contrast and visibility level of brain images.
Step 2: FCM: It allocates every pixel to a cluster with a DoM function. The number of clusters is determined based on the nature of the image data. The FCM performs clustering on the preprocessed grayscale image. Based on the pixel intensity value, FCM allocates pixels to one of the clusters.
Step 3: Level set initialization: Initialize level set functions for every cluster by utilizing an active contour methodology; here, every level set function states a boundary or contour.
Step 4: Enhanced edge indicator: In this step, compute enhanced edge indicator maps that highlight texture boundaries and edges in the image.
Step 5: Level set evolution: Evolve level set functions utilizing the enhanced edge indicator map and the FCM derived information. The level set functions are iteratively updated based on terms like regional regularizers, edge information, and other constraints.
Step 6: Post processing: Performed post-processing for refining the segmented interested region. The post-processing includes filling holes and eliminating small regions.
Step 7: Evaluation: The proposed FCM-LSLTPEEI model’s efficacy is validated using following performance metrics such as Jaccard coefficient, sensitivity, accuracy, and dice coefficient.
The accurate segmentation of hippocampus in MRI images enables researchers and clinicians in monitoring disease progression over time. Additionally, the precise hippocampus segmentation assists doctors in planning surgical interventions like removal of brain tumor and excisions for epilepsy treatment. Furthermore, it helps surgeons in minimizing damage to healthy tissue and leads to improved surgical results.
Experimental Results
The proposed segmentation model: FCM-LSLTPEEI has been executed utilizing the MATLAB 2020a tool on a computer with 128 GB random access memory, Windows 11 (64 bit) operating system and 8 TB hard drive. In the context of image segmentation, the FCM-LSLTPEEI model is validated individually on 316 and 50 MRI scans acquired from OASIS and NITRC databases. Additionally, the proposed FCM-LSLTPEEI models effectiveness is analyzed by means of Jaccard coefficient, sensitivity, accuracy and dice coefficient. The possible variations in the segmented region are quantified by means of dice coefficient value, which is an overlap measure between two MRI brain images. The dice coefficient value ranges from zero to one, where one denotes overlap and zero represents no overlap between MRI brain images (A and B). The mathematical formula of the dice coefficient is denoted in Eq. (15).
The Jaccard coefficient value is mathematically stated in Eq. (16). In the hippocampus segmentation, if the proposed FCM-LSLTPEEI model matches exactly with the ground truth, then the Jaccard coefficient value is 1, or-else 0 (no overlap).
Additionally, the sensitivity determines the percentage of the active positives, which are precisely identified. The performance measure: accuracy reports the percentage of the image pixels, which are precisely classified. The mathematical depiction of sensitivity and accuracy is specified in Eqs. (17) and (18).
where False Positive (FP) indicates that the background pixels are incorrectly recognized as hippocampus pixels, False Negative (FN) represents that the hippocampus pixels are incorrectly recognized as background pixels, True Positive (TP) states that the hippocampus pixels are correctly recognized as hippocampus pixels, and True Negative (TN) specifies that the background pixels are correctly recognized as background pixels.
Performance Evaluation of OASIS Database
The FCM-LSLTPEEI model’s effectiveness is validated using the OASIS database. Additionally, the experimental examination is performed in two ways with preprocessing and without preprocessing technique. Table 1 represents the result of the proposed and the existing segmentation algorithms without using the color normalization technique. Table 1 shows that the FCM-LSLTPEEI model’s effectiveness is validated by comparing its result with existing segmentation algorithms: k-means clustering, FCM, superpixel clustering, LSLTP, Level Set-based Local Gabor Transitional Pattern (LSLGTP), and FCM-LSLTP in light of Jaccard coefficient, dice coefficient, sensitivity, and accuracy. By investigating Table 1, the proposed FCM-LSLTPEEI model attained maximum result in the hippocampus segmentation with the Jaccard coefficient of 93.68%, dice coefficient of 92.45%, accuracy of 90.22%, and sensitivity of 91.08%. These outcomes are significant in comparison with the existing segmentation algorithms like k-means clustering, FCM, superpixel clustering, LSLTP, LSLGTP, and FCM-LSLTP. Hence, the graphical comparison of FCM-LSLTPEEI model without normalization technique on OASIS database is indicated in Fig. 6.
The result of the FCM-LSLTPEEI model with normalization technique on the OASIS database is given in Table 2. By viewing Tables 1 and 2, the FCM-LSLTPEEI model with normalization technique attained better segmentation performance compared to the FCM-LSLTPEEI model without normalization technique. As represented in Table 2, the FCM-LSLTPEEI model with the normalization technique achieved a maximum Jaccard coefficient of 97.90%, dice coefficient of 98.94%, accuracy of 98.90%, and sensitivity of 97.90% in the hippocampus segmentation. In addition, the achieved results are higher compared to the existing segmentation algorithms such as k-means clustering, FCM, superpixel clustering, LSLTP, LSLGTP and FCM-LSLTP. The graphical illustration of FCM-LSLTPEEI model with normalization technique on OASIS database is represented in Fig. 7.
Performance Evaluation of NITRC Database
Similar to the previous database, the proposed FCM-LSLTPEEI model’s performance is validated with and without employing the normalization technique. The experimental outcome of the FCM-LSLTPEEI model without normalization technique on the NITRC database is given in Table 3 by means of Jaccard coefficient, dice coefficient, accuracy, and sensitivity. As seen in Table 3, the proposed FCM-LSLTPEEI model has achieved 90.57% of Jaccard coefficient, 91.32% of dice coefficient, 90.16% of accuracy, and 89.03% of sensitivity, which are higher related to the comparative algorithms such as k-means clustering, FCM, superpixel clustering, LSLTP, LSLGTP, and FCM-LSLTP in the hippocampus segmentation. Hence, the graphical comparison of FCM-LSLTPEEI model without the normalization technique on NITRC database is depicted in Fig. 8.
In this scenario, the proposed FCM-LSLTPEEI model with the normalization technique has obtained higher segmentation results on the NITRC database by means of Jaccard coefficient, dice coefficient, accuracy, and sensitivity. By inspecting Table 4, the FCM-LSLTPEEI model with the normalization technique has attained 97.22% of Jaccard coefficient, 99.01% of accuracy, 98.59% of dice coefficient, and 97.22% of sensitivity in the hippocampus segmentation. The proposed FCM-LSLTPEEI model with normalization technique achieves several edge maps, which are highly close to the interested region boundaries for obtaining a better segmentation performance. Hence, the graphical comparison of FCM-LSLTPEEI model with normalization technique on NITRC database is specified in Fig. 9.
Comparative Analysis
The analysis between the FCM-LSLTPEEI model and the comparative models is specified in Table 5. Liu and Yan [2] implemented a novel model on the basis of LB and DBN for hippocampus segmentation. In the resulting phase, the presented model obtained 84% of dice coefficient on the OASIS database. Palumbo et al. [8] developed two automatic MRI brain segmentation models: SPM and FS for hippocampus segmentation. The empirical investigation showed that the presented model attained 80% of the dice coefficient on the OASIS database. Related to the existing models, the FCM-LSLTPEEI model attained 97.90% of the dice coefficient value.
Nasser et al. [10] presented two CNN models based on U-Net: Hippocampus Segmentation Multi-classes (HSMC) and Hippocampus Segmentation Single Entity (HSSE) for segmenting hippocampal sub-regions, and hippocampus as a single entity. In this study, the implemented model’s performance was validated on the NITRC database. As seen in Table 6, the proposed FCM-LSLTPEEI model attained high segmentation results compared to the HSSE and HSMC models using performance measures like sensitivity, accuracy, and dice coefficient.
Conclusion
In this manuscript, the FCM-LSLTPEEI model is developed for hippocampus segmentation. The proposed system comprises three phases such as image collection, image pre-processing, and hippocampus segmentation. After acquiring MRI brain images from NITRC and OASIS databases, color normalization is employed for improving image quality and eliminating noise. Finally, the FCM-LSLTPEEI model is implemented for hippocampus segmentation. With the assistance of the FCM and EEI function in the LSLTP, the proposed model provides the edge maps close to the interested region boundaries to obtain a better segmentation performance. The proposed FCM-LSLTPEEI model achieved 98.90% and 99.01% of segmentation accuracy on the OASIS and NITRC databases. Here, the achieved results are significant in comparison with the conventional models such as k-means clustering, FCM clustering, superpixel clustering, FCM-LSLTP, LSLTP and LSLGTP models. In the future work, a new deep learning-based classification technique can be proposed for hippocampus subtypes/subfields classification.
Data Availability
The datasets generated during and/or analyzed during the current study are available in the NITRC database available link: https://www.nitrc.org/projects/hippseg_2011/, OASIS database available link: https://www.oasis-brains.org/
References
D. Carmo, B. Silva, C. Yasuda, L. Rittner, R. Lotufo, A.D.N. Initiative, Hippocampus segmentation on epilepsy and Alzheimer’s disease studies with multiple convolutional neural networks. Heliyon 7(2), e06226 (2021). https://doi.org/10.1016/j.heliyon.2021.e06226
Y. Liu, Z. Yan, A combined deep-learning and Lattice Boltzmann model for segmentation of the hippocampus in MRI. Sensors 20(13), 3628 (2020). https://doi.org/10.3390/s20133628
Y. Shao, J. Kim, Y. Gao, Q. Wang, W. Lin, D. Shen, Hippocampal segmentation from longitudinal infant brain MR images via classification-guided boundary regression. IEEE Access 7, 33728–33740 (2019). https://doi.org/10.1109/ACCESS.2019.2904143
Y. Shi, K. Cheng, Z. Liu, Hippocampal subfields segmentation in brain MR images using generative adversarial networks. Biomed. Eng. Online 18, 5 (2019). https://doi.org/10.1186/s12938-019-0623-8
S. Vijayalakshmi, Savita, An automated enhanced FCM based hippocampus segmentation. Ann. Rom. Soc. Cell Biol. 25(1), 1861–1871 (2021)
S. Pang, J. Jiang, Z. Lu, X. Li, W. Yang, M. Huang, Y. Zhang, Y. Feng, W. Huang, Q. Feng, Hippocampus segmentation based on local linear mapping. Sci. Rep. 7, 45501 (2017). https://doi.org/10.1038/srep45501
X. Jiang, Z. Zhou, X. Ding, X. Deng, L. Zou, B. Li, Level set based hippocampus segmentation in MR images with improved initialization using region growing. Comput. Math. Methods Med.. Math. Methods Med. 2017, 5256346 (2017). https://doi.org/10.1155/2017/5256346
L. Palumbo, P. Bosco, M.E. Fantacci, E. Ferrari, P. Oliva, G. Spera, A. Retico, Evaluation of the intra-and inter-method agreement of brain MRI segmentation software packages: a comparison between SPM12 and FreeSurfer v6. 0. Phys. Med. 64, 261–272 (2019). https://doi.org/10.1016/j.ejmp.2019.07.016
Z. Yang, X. Zhuang, V. Mishra, K. Sreenivasan, D. Cordes, CAST: a multi-scale convolutional neural network based automated hippocampal subfield segmentation toolbox. Neuroimage 218, 116947 (2020). https://doi.org/10.1016/j.neuroimage.2020.116947
S. Nasser, M. Naoui, G. Belalem, S. Mahmoudi, Semantic Segmentation of Hippocampal Subregions with U-Net Architecture. Int. J. E-Health Med. Commun. (IJEHMC) 12(6), 1–20 (2021). https://doi.org/10.4018/IJEHMC.20211101.oa4
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
I/We declare that the work submitted for publication is original, previously unpublished in English or any other language(s), and not under consideration for publication elsewhere.
Consent for Publication
I certify that all the authors have approved the paper for release and are in agreement with its content.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kaka, J.R., Prasad, K.S. Hippocampus Segmentation Using Fuzzy C Means-Based Level Set Local Ternary Pattern with Enhanced Edge Indicator. J. Inst. Eng. India Ser. B 105, 565–574 (2024). https://doi.org/10.1007/s40031-024-00989-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40031-024-00989-1