Abstract
This study was conducted to identify the association of coding variations in the HSPA8 gene with heat stress in two different breeds of sheep. All the coding regions of the HSPA8 gene of Awassi and Arabi sheep were covered by amplifying nine exons. A single-strand conformation polymorphism (SSCP) was utilized to assess the genetic variations in both breeds. The possible association of the observed genotypes with rectal temperature (RT), respiratory rate (RR), and heat tolerance coefficient (HTC) was analyzed in different seasons. While all the coding regions of both sheep were monomorphous, a remarkable heterogeneity was observed in exon 4, of which two SSCP patterns, a normal TT and a mutant TG, were detected. The TG genotype was characterized by a missense variant of T177P with frequencies of 77% in Awassi and 54% in Arabi. Cumulative in silico tools indicated extremely deleterious consequences for T177P on protein structure, function, and stability. Results indicated that sheep with the TT genotype had significantly (P < 0.05) lower RT, RR, and HTC values than sheep with the TG genotype. Therefore, a significant association of T177P with a lower tolerance of Awassi to higher temperature conditions was revealed. In conclusion, the identified T177P may have damaging effects in the HSPA8, which affects the ability of Awassi sheep to cope up with elevated temperatures compared with Arabi sheep. This manuscript describes a novel description of a highly deleterious missense variant in the HSPA8 gene that may reduce the ability of sheep to withstand high-temperature conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
When a sheep is exposed to a high temperature, remarkable changes occur in several of its biochemical and physiological parameters (Marai et al. 2007; Almeida et al. 2019). These dynamic modulations are associated with changes in vaginal and rectal temperature, heartbeats, respiration rate, and several hematological parameters (Zachut et al. 2020). Most of these changes are a manifestation of the dynamic actions of heat shock proteins (HSPs) (Archana et al. 2017). HSPs represent an evolutionarily conserved family of proteins that have been categorized into several versatile members encoded from a multitude of highly conserved HSP loci (Bai et al. 2019). HSPs are induced in response to several biological stress as a primary intracellular defense system in many living organisms (Dubey et al. 2015; Sailo et al. 2015). However, the syntheses of HSPs are mainly temperature-dependent and are released in response to environmental and oxidative stress (Ikwegbue et al. 2017). The main functions of HSPs have been associated with their participation in the resistance of animals to highly variable environmental and physiological insults. It is well known that HSPs undergo their scheduled tasks by aiding the correct folding of nascent polypeptides and preventing proteins misfolding without being a part of the final structure of the protein (Multhoff 2007). This function is represented by the ability of HSPs to act as molecular chaperons which block the aggregation of unfolded proteins to aid the cell to sustain conditions of elevated temperature (Zhang et al. 2015). Such protective functions have been assumed to be taken place by such proteins to prevent lethal damage to the stressed cells (Ikwegbue et al. 2017). There are several members of the HSPs family with a wide range of molecular masses. HSP family is made of many members divided into a variety of groups based on size, sequence, structure, and function, such as HSP10, HSP40, HSP60, HSP70, HSP90, HSP100, and HSP110 (Nikbin et al. 2014; Jyotiranjan et al. 2017). Due to the highly dynamic nature of the HSP family, sufficient information on the structure and mode of action of several molecules within this family are lacking. One of these non-adequately characterized HSPs is a heat shock protein family A (HSP70) member 8, which is encoded by the HSPA8 gene. This gene is positioned in chromosome 15 and is made of 9 exons in sheep. While the exon 1 is non-coding, the rest 8 exons constitute a whole HSPA8 protein made of 650 amino acids of 71Kd. In cattle, a high degree of genetic variation in HSPs was recorded during higher temperature seasons particularly in tropical countries (Kerekoppa et al. 2015; Verma et al. 2016). HSP70 gene expression has been elevated as a response to heat stress in goats (Shaji et al. 2017). It has been shown that the altered HSP variation is associated with the tolerance of cattle to temperature (Bhat et al. 2016). Moreover, HSPA8 gene polymorphism is suggested as a biological marker for assessing the impact of heat stress in different breeds of cattle prevailing in different climate conditions (Charoensook et al. 2012). However, few reports are available in sheep, which described HSPA8 gene association with heat stress conditions (Singh et al. 2017), while other reports are either being confined to the promoter or 5′ flanking regions of HSPA8 (Marcos-Carcavilla et al. 2010; Salces-Oritz et al. 2015). On the other hand, the rapid revolution in the computational biology of the missense variants predictive consequences has not been exploited in the interpretation of this genotype–phenotype association (Al-Shuhaib et al. 2018). Taking the previously mentioned information into consideration, this study aims to identify the potential association between the polymorphism of the entire coding regions of the HSPA8 gene with the adaptation of two Iraqi breeds of sheep in the Middle Euphrates regions.
Materials and methods
Sheep population
The study was conducted according to the regulations of the international recommendations for the care and use of animals (Federation of Animal Science Societies 2010). The sheep experimentations were conducted after obtaining approval from the ethical committee of Al-Qasim Green University (Approval No. 12.10.18). Two breeds of sexually mature and healthy sheep aged between 2 and 3 years were involved in this study, including 75 animals of Awassi (22 rams: 53 ewes) and 75 animals of Arabi (15 rams: 60 ewes). The animals of both breeds had different genetic and geographical backgrounds. Awassi breed prevails in the Middle Euphrates region with summer temperatures of less than 50 °C, while Arabi breed prevails in Southern portions of Iraq with summer temperatures regularly exceeding 50 °C (Suppl. Fig. 1). Both breeds were raised in the Barakat Abu al Fadhl Al-Abbas Station, situated at a longitude of 32.6027° N, the latitude of 44.0197° E, and 32 m above mean sea level (Al-Khafeel co., Karbala province, Iraq) from January to August 2018. Animals were fed ad libitum with seasonal grass, concentrate food about 2.5% of their live body weight daily, comprising a mixture of barley (59%), bran (40%), salt (1%) concentrates, and fresh water.
Thermoregulatory parameters
During the experimental period, the climate of the station is known for being cold and dry during winter and sweltering and arid in the summer with an average daily temperature that reaches 15 °C in winter and extremely hot, around 44 °C in summer. The evaluated thermoregulatory parameters consisted of respiration rate (RR) and rectal temperature (RT). Both parameters were determined twice per day (morning at 8:00 am and afternoon 14:00 pm) for 3 successive days in two seasons, i.e., winter (January and February) and summer (July and August). The specified RT was measured with a sterile digital clinical thermometer inserted about 3-cm deep into the rectum of the animal for 1 min before being recorded (McManus et al. 2015). RR was measured by counting diaphragm movements per minute and recorded as breaths per min. The heat tolerance coefficient (HTC) was calculated based on the heat tolerance index developed by Benezra (1954). The formula is based on both respiration rate and rectal temperature. HTC: RR/27 + RT/39.1.
Genomic DNA extraction
About 2 ml of blood samples were withdrawn from the jugular vein of each analyzed animal and placed in an anticoagulation tube. Subsequently, a rapid manual technique of DNA extraction was used to extract highly purified DNA from both populations included in the study (Al-Shuhaib 2017). Briefly, RBCs were destroyed by mixing blood with distilled water. The pelleted WBCs were re-suspended in a lysis buffer (10 mM Tris-Cl pH 7.7, 0.4 M NaCl, 2 mM EDTA, 0.5% SDS). Proteins were denatured by 6 M NaCl. The supernatant containing DNA was precipitated with absolute ethanol. The precipitated DNA strands were washed with 70% ethanol. DNA was recovered by elution buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). The extracted DNA samples were assessed for quality and purity using Nanodrop μLITE spectrophotometer (Biodrop, UK) and used as a template for polymerase chain reaction (PCR).
Primers design and PCR
Nine pairs of specific PCR oligonucleotides were designed to cover all the coding regions of the HSPA8 gene using the NCBI Primer Blast online server. Since exon 1 is non-coding, the specified designing was extended from exon 2 to exon 9. Except for exon 5 that required two primer pairs to be covered completely, each pair of primer was designed to entirely amplify its particular targeted exon along with its upstream and downstream flanking regions. PCR experiments were conducted using AccuPower® PCR PreMix (Bioneer, Korea), and initiated by denaturation for 5 min, followed by 30 cycles of denaturation (95 °C), annealing (Table 1), and extension (72 °C), for 30 s each, with final extension for 5 min. The specificity of PCR amplicons was confirmed by 1.5% agarose gel electrophoresis before being submitted to SSCP protocols.
SSCP
The SSCP-genotyping reactions were performed according to the procedure of Al-Shuhaib et al. (2018), with several optimizations. Briefly, 1 μl of all PCR amplicons were denatured with an equal volume of SSCP loading buffer (95% formamide, 0.05% bromophenol blue, 0.05% xylene cyanol, and 20 mM EDTA, pH 8) for 7 min at 94 °C. PCR amplicons were then placed on ice for at least 10 min and loaded on neutral polyacrylamide gels (0.1-mm gel thickness, 10-cm length, and 20-cm width). Electrophoresis conditions were optimized as described in Table 2. Gels were stained according to the protocol of Byun et al. (2009).
Sequencing
To confirm the observed SSCP banding patterns, two representatives from each detected SSCP banding pattern was sent for sequencing reactions from both termini according to the instruction manual described by Macrogen laboratories (Geumcheon, Seoul, South Korea). The received chromatograms were edited and aligned by the EditSeq tool, ver. 7.1.0 (DNA STAR, Lasergene). The nucleic acid variations were demonstrated by comparing DNA sequencing electropherograms with the deposited ovine HSPA8 DNA sequences (GenBank acc. no. NC_019472.2). The observed variations were visualized and annotated by SnapGene Viewer, ver. 4.0.4. (GSL. Biotech. LLC). The novelty of the observed variants was checked by the deposited variants of the ovine HSPA8 gene database in the Ensemble genome browser 96 (https://asia.ensembl.org/index.html). All the covered coding regions were deposited in the NCBI database and got GenBank accession numbers (MN075929 and MN075930).
In silico prediction
Many computational tools were utilized to assess the consequences of the observed missense variants on the resulting mutant protein structures and functions, namely SIFT (Pauline and Steven 2003), PolyPhen-2 (Adzhubei et al. 2010), Provean (Choi et al. 2012), Panther (Tang and Thomas 2016), SNAP2 (Smigielski et al. 2000), and PhD SNP (Capriotti et al. 2006). Then, the 3D structure of HSPA8 was generated by RaptorX server before and after mutation (Källberg et al. 2012) and validated by verify3D and PROCHECK servers (http://servicesn.mbi.ucla.edu/Verify3D/). The stability of HSPA8 upon mutation was evaluated by I-Mutant2.0 (Capriotti et al. 2005), mCSM (Pires et al. 2012), SDM (Worth et al. 2012), DUET (Pires et al. 2014), and CUPSAT (Parthiban et al. 2006). Subsequently, the mode of action of the observed variant was predicted by MutPred (Pejaver et al. 2017) and Missense 3D (Ittisoponpisan et al. 2019).
Assessment of genetic polymorphism
The allele and genotype frequencies, observed heterozygosity (Ho), expected heterozygosity (He), and effective number of alleles were analyzed using PopGen32 software, v. 1.31 (Yeh et al. 1999). The polymorphism information content (PIC) was calculated using HET software version 1.8 (Ott 2001).
Marker-trait association analyses
Association analyses were performed using SPSS v23.0 (IBM, NY, USA). The significant effect of genotypes, breed, and season on the various variables studied was analyzed with the following general linear model:
where Yijkl is the observation of phenotypic trait, μ is overall mean, Gi is the effect of ith genotype (i = TT, TG), Sj is the effect of jth season (summer, winter), Bk is the effect of kth breed (k = Awassi, Arabi), and eijkl = random error assumed to be NID (0, σ2e). Means were compared using adjusted Tukey’s test with a significance level of (P < 0.05). Preliminary statistical analyses indicated that the interaction effects between the factors investigated were not significant and were excluded from the statistical model. Besides, age and sex were not considered in the final analysis because they did not have a significant influence on the variability of traits in the analyzed populations.
Results
The genetic polymorphism
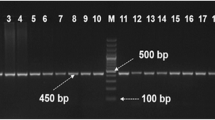
Almost all analyzed exons showed monomorphous SSCP banding patterns with one exception found in exon 4, in which two SSCP variants were detected (Fig. 1).
Sequencing reactions confirmed these variations by detecting the presence of a novel T88G only in one of the SSCP variants. According to this T88G nucleic acid substitution, the detected SSCP variants were assigned TT and TG genotypes, respectively. ExPASy translate software indicated a missense nature of this T88G, as it resulted in an amino acid substitution of Thr to Pro at the 177th position of HSPA8 protein. The TG pattern, or T177P, was detected in different frequencies in both investigated populations. Awassi sheep exerted 77% of the TG pattern, while Arabi sheep showed only 54% of the same pattern. This observation entailed a noticeable prevalence of these T177P variants in Awassi sheep and a remarkably lower dominance of the same variant in Arabi sheep. Meanwhile, no heterogenic SSCP banding patterns were observed in the other amplified fragments. However, sequencing reactions identified a total of additional eight variants in several coding and non-coding regions in the HSPA8 gene. The observed variants ranged from intronic variations (62ACT63 insertion and G72T substitution in exon 2), synonymous variants (T114C, A204G, A74G, and T182G in exon 3, exon 4, and exon 9 respectively), and nonsynonymous variants (T77G in exon 8). Furthermore, the sequencing reactions of the exon 5 clarified some unknown amino acid sequences observed in the referring HSPA8 protein (GenBank acc. no. NC_019472.2). This clarification came from the obvious positioning of Val in the 209 amino acid residue that was notified with unknown, or X, in the referring protein. However, all the previously mentioned variations were detected in both studied populations (Table 3).
The current study was concerned only with the nucleic acid substitution T88G or its consequent amino acid substitution T177P as it is the only single nucleotide polymorphism (SNP) detected in different frequencies between Awassi and Arabi investigation breeds (Fig. 2).
Noteworthy, the currently observed nonsynonymous single nucleotide polymorphism (nsSNP) was not previously detected and the available data for the 3D structure of ovine HSPA8 was not adequate. Nevertheless, the scanning of all the possible variations within the HSPA8 gene in the present study provided the first high-quality HSPA8 protein in the ovine species. This is due to the presence of some undetermined amino acid sequences within the deposited protein database that was annotated with “X” in the 209 amino acid position (GenBank acc. no. NC_019472.2). This observation was clarified in the 216-bp amplicons of exon 5 which obviously confirmed the presence of Val in the referred position. Therefore, this study has confirmed that HSPA8 is made of Val instead of X, or unknown amino acid, in the 209th position in the ovine HSPA8 protein.
In silico prediction of T177P
Out of nine detected variants, only T177P was highlighted in the present study, as it was only detected in the TG banding pattern whereas the other 8 variants were found in all the studied animals. Therefore, a series of computational tools were utilized to assess the final consequences of this missense variant on the altered HSPA8 protein. The UniProt accession number for ovine HSP8A is W5NPN4. Only a short-length three-dimensional (3D) structure of HSPA8 protein and its homologous templates were available in protein databank (PDB) entries (http://www.rcsb.org/). Therefore, two 3D protein structures for TT and TG patterns of HSPA8 were built by RaptorX server and 100% of the referring 650 amino acids components of the HSP8A was successfully modeled with only 7 to 8% positions which were predicted to be disordered for TT and TG HSPA8 respectively (Table 4).
Thus, it was found that there was an alteration between HSPA8 of TT pattern and HSPA8 of TG pattern in terms of the presence of altered disordered positions, secondary structure, and solvent accessibility alterations. This observation indicated a noticeable role for T177P in altering the structural characteristics of the TG HSPA8 protein. Interestingly, all the utilized in silico tools were in line with each other in terms of the entire deleterious consequences of T177P on the altered protein. This observation was revealed by several computational tools that were used to assess the potential deleterious effect of a particular nsSNP on protein structure and function, such as SIFT, PolyPhen-2, Provean, Panther, SNAP2, and PhD SNP (Table 5, A). Furthermore, negative deleterious consequences of T177P on HSPA8 stability were predicted by several in silico tools that were utilized to assess the effect of a particular nsSNP on the free energy of the altered protein, including I-Mutant2.0, mCSM, SDM, DUET, and CUPSAT (Table 5, B). Another layer of confirmation came from further analyses of the structural and functional effects of T177P on the altered HSPA8. Several negative functional alterations were predicted by the MutPred server, such as altered metal binding, relative loss of solvent accessibility, and altered transmembrane characteristics, while remarkable damage in the structural variables was changed as predicted by Missense 3D server (Table 5, C).
The cumulative computational tools indicated an obvious deleterious effect of T177P on the altered HSPA8 protein, which was detected in different proportions in the analyzed breeds, 77% in Awassi and 54% in Arabi. This observation signified more deleterious consequences of T177P in Awassi than Arabi breeds (Fig. 3).
Assessment of T177P polymorphism
Genotype and allele frequencies and Hardy–Weinberg test of HSP70 gene in the analyzed populations of Awassi and Arabi sheep are presented in Table 6.
The observed value of chi-square means for both Arabi and Awassi indicated that the population under study was not in Hardy–Weinberg equilibrium (HWE), which was significant at P ˂ 0.05. According to the classification of PIC (low polymorphism, if PIC value < 0.25; median polymorphism if 0.25 < PIC value < 0.5, and high polymorphism if PIC value > 0.5) (Trakovická et al. 2013), the present analyses showed moderate levels of polymorphism information content within the ovine HSPA8 gene in both analyzed breeds.
Association of T177P with measured heat tolerance traits
Higher adaptability of Arabi to heat stress tolerance was observed as Arabi exhibited lower (P < 0.05) RT and RR than Awassi sheep (Table 7). Again, HTC values of this study in Arabi gave lower scores than Awassi signifying the superiority of Arabi in terms of adaptability to heat stress.
The average values of RT, RR, and HTC were found to be significantly different (P < 0.05) between summer and winter seasons. The mean RT, RR, and HTC values were higher in summer compared to winter, indicating noticeable stress conditions in summer for both analyzed populations.
The RT and RR values of sheep with TG genotype were higher (P < 0.05) than sheep with TT genotype, indicating a clear superiority of TT genotype over the TG genotype in terms of adaptability to heat stress. This observation was confirmed by HTC values of both detected genotypes, as sheep with TG genotype showed higher values of HTC than sheep with TT genotype.
Discussion
In this study, all the coding regions of the HSPA8 gene were screened by PCR-SSCP to identify the possible causative SNP(s) behind the noticeable living variations observed between Awassi and Arabi breeds. The present study describes a crucial T177P variant as a highly deleterious nsSNP observed in Awassi breed with high frequency. The higher prevalence of this deleterious SNP could be one of the many factors responsible for the lower adaptability of the Awassi breed. This observation was confirmed by the experiments conducted in this study. This is due to the remarkable differences in the frequency of T177P between both involved breeds; 77% in Awassi and 54% in Arabi. However, it has been reported that Awassi sheep breed displays low reproductive performance, as compared with European breeds (Talafha and Ababneh 2011; Al-Saaidi et al. 2018). A major cause of the low productivity of Awassi sheep may result from the effects of higher temperatures at the mating season (Salman et al. 2009). In contrast to Awassi, the lower frequency of the same T177P has therefore been associated with the high adaptability of Arabi to higher temperatures. Unfortunately, Arabi breed is rarely documented and no thermoregulatory studies have been performed on this breed for comparison with our findings. However, two widely used physiological variables were utilized in this study, RT and RR. When an animal gives lower values of RT, RR, and HTC after being exposed to heat stress, greater adaptability was indicated (McManus et al. 2015). However, lower values of RT, RR, and HTC have been indicated for better tolerance of an animal to heat stress conditions (Bhat et al. 2016). Arabi breed exhibited lower RT, RR, and HTC than Awassi breed, indicating a higher tolerance of Arabi than Awassi to heat stress. As long as the values of RT, RR and HTC for TT genotype were found to be lower than those detected in TG genotype, the deleterious T177P SNP was suggested as one of the causative agents for the lower tolerance of TG genotype to heat stress.
Clear computational confirmation of the extremely deleterious consequences of T177P has given a further indication for the mechanism of this nsSNP in this association. All of the utilized in silico tools have collectively given the same deleterious predictions for the T177P, signifying that the alteration of HSPA8 with T177P missense variant caused several damaging aspects which may negatively affect its capability to undertake its scheduled task of protecting sheep from higher temperature conditions (Berihulay et al. 2019). So, a higher frequency of T177P in a sheep indicates a lower tolerance to higher temperature conditions. Therefore, the utilization of in silico computations predicted a series of deleterious effects of T177P on the structure, function, and stability of the altered HSPA8 protein. In another word, the currently implemented computational tools explained one of the reasons for the superiority of Arabi on Awassi breed with regard to heat tolerance as the substitution of Thr to Pro in the 177th amino acid residue reduced the adaptability of an animal to higher temperature stress. Therefore, the observed data of this study suggested a clear superiority of the sheep with TT genotype to cope up with heat stress conditions. Nevertheless, alteration in gene expression could not be eliminated from this explanation.
Conclusion
Two genotypes were detected in the HspA8 gene, TT genotype, and TG genotype. For the individuals with TG genotype, the amino acid Thr was substituted with Pro at the 177th position. In silico tools predicted a clear deleterious role played by T177P in HSPA8 protein in terms of structure, function, and stability. The measured temperature tolerance stress conditions indicated better thermotolerance performance for individuals with TT genotype than individuals with TG genotype. Furthermore, in silico computations have confirmed the above-stated role of T177P in ovine thermotolerance adaptability to a hot climate. This utilization of cumulative computations provided an interpretation for the possible role of this SNP in the reduced tolerance of Awassi sheep to high-temperature stress.
References
Adzhubei, I.A., Schmidt, S., Peshkin, L., Ramensky, V.E., Gerasimova, A., Bork, P., Kondrashov, A.S., Sunyaev, S.R., 2010. A method and server for predicting damaging missense mutations. Nature Methods, 7, 248–249
Almeida, A.M., Zachut, M., Hernández-Castellano, L.E., Šperanda, M., Gabai, G., Mobasheri, A. 2019. Biomarkers of fitness and welfare in dairy animals: healthy living. Journal of Dairy Research, 86(4), 379–387.
Al-Saaidi, J.A.A., Khudair, K.K., Khafaji, S.S., 2018. Reproductive fecundity of Iraqi Awassi ewes immunized against synthetic inhibin-α subunit or steroid-free bovine follicular fluid. Asian Australian Journal of animal sciences, 31(8), 1169
Al-Shuhaib, M.B.S., Al-Kafajy, F.R., Badi, M.A., AbdulAzeez, S., Marimuthu, K., Al-Juhaishi, H.A.I., Borgio, J.F., 2018. Highly deleterious variations in COX1, CYTB, SCG5, FK2, PRL and PGF genes are the potential adaptation of the immigrated African ostrich population. Computers in Biology and Medicine, 100, 17–26
Al-Shuhaib, M.B.S., 2017. A Universal, rapid, and inexpensive method for genomic DNA isolation from the whole blood of mammals and birds. Journal of Genetics, 96(1), 171–176
Archana, P.R., Aleena, J., Pragana, P., Vidya, M.K., Abdul Niyas, P.A., Bagath, M., Krishnan, G., Manimaran, A., Beena, V., Kurien, E.K., Sejian, V., Bahtta, R., 2017. Role of heat shock proteins in livestock adaptation to heat stress. Journal of Dairy, Veterinary & Animal Research, 5, 00127
Bai, J., Liu, X-N., Lu, M-X., Du, Y-Z., 2019. Characterization of genes encoding small heat shock proteins from Bemisia tabaci and expression under thermal stress. PeerJ, 7, e6992
Benezra, M.V., 1954. A new index for measures the adaptability of cattle to tropical condition. Journal of Animal Science, 13, 1015
Berihulay, H., Abeid, A., He, X., Jiang, L., Ma, Y., 2019. Adaptation mechanisms of small ruminants to environmental heat stress. Animals, 9, 75
Bhat, S., Kumar, P., Kashyap, N., Deshmukh, B., Dige, M.S., Bhushan, B., Chauhan, A., Kumar, A., Singh, G., 2016. Effect of heat shock protein 70 polymorphism on thermotolerance in Tharparkar cattle. Veterinary World, 9(2), 113–17
Byun, S.O., Fang, Q., Zhou, H., Hickford, J.G.H., 2009. An effective method for silver-staining DNA in large numbers of polyacrylamide gels. Analytical Biochemistry, 385, 174–175
Capriotti, E., Calabrese, R., Casadio, R., 2006. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics, 22, 2729–2734
Capriotti E., Fariselli P., Casadio R., 2005. I-Mutant 2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Research, 33, W306–W310
Charoensook, R., Gatphayak, K., Sharifi, A.R, Chaisongkram, C., Brenig, B., Knorr, C., 2012. Polymorphisms in the bovine HSP90AB1 gene are associated with heat tolerance in Thai indigenous cattle. Tropical Animal Health and Production, 44, 921–928
Choi, Y., Sims, G.E., Murphy, S., Miller, J.R., Chan, A.P., 2012. Predicting the functional effect of amino acid substitutions and indels. PLOS ONE 7, e46688
Dubey, A., Prajapati, K.S., Swamy, M., Pachauri, V., 2015. Heat shock proteins: a therapeutic target worth to consider. Veterinary World, 8(1), 46–51
Federation of Animal Science Societies, 2010. Guide for the care and use of agricultural animals in research and teaching. Champaign, IL 61822, 3rd edition
Ikwegbue, P.C., Masamba, P., Oyinloye, B.E., Kappo, A.P., 2017. Roles of heat shock proteins in apoptosis, oxidative stress, human inflammatory diseases, and cancer. Pharmaceuticals (Basel), 23;11(1)
Ittisoponpisan, S., Islam, S.A., Khanna, T., Alhuzimi, E., David, A., Sternberg, M.J.E., 2019. Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? Journal of Molecular Biology,431(11), 2197–2212
Jyotiranjan, T., Mohapatra, S., Mishra, C., Dalai, N., Kundu, A.K., 2017. Heat tolerance in goat- a genetic update. The Pharma Innovation Journal, 6(9), 237–245
Källberg, M., Wang, H., Wang, S., Peng, J., Wang, Z., Lu, H., Xu, J., 2012. Template-based protein structure modeling using the RaptorX web server, Nature Protocols, 7, 1511
Kerekoppa, R.P., Rao, A., Basavaraju, M., Geetha, G.R., Krishnamurthy, L., Rao, T.V.L.N. Das, D.N., Mukund, K., 2015. Molecular characterization of the HSPA1A gene by single strand conformation polymorphism and sequence analysis of Holstein-Friesian crossbreed and Deoni cattle raised in India. Turkish Journal of Veterinary and Animal Science, 39, 128–133
Marai, I.F.M., El-Darawany, A.A., Fadiel, A., Abdel-Hafez, M.A.M. 2007. Physiological traits as affected by heat stress in sheep—a review. Small ruminant research, 71(1–3), 1–12
Marcos-Carcavilla, A., Moreno, C., Serrano, M., Laurent, P., Cribiu, E.P., Andreoletti, O., Ruesche, J., Jean-Louis, W., Calvo, J.H., Moazami-Goudarzi, K., 2010. Polymorphisms in the HSP90AA1 5′ flanking region are associated with scrapie incubation period in sheep. Cell Stress and Chaperones, 15, 343–349
McManus, C., Bianchini, E., Paim, T.P., Lima, F.G., Braccini Neto, J., Castanheira, M., Esteves, G.I.F., Cardoso, C., Dalcin, V.C., 2015. Infrared thermography to evaluate heat tolerance in different genetic groups of lambs. Sensors, 15, 17258–17273
Multhoff, G., 2007. Heat shock protein 70 (Hsp70): membrane location, export and immunological relevance. Methods, 43(3), 229–37.
Nikbin, S., Panandam, J.M., Yaakub, H., Murugaiyah, M., Sazili, A.Q., 2014. Novel SNPs in heat shock protein 70 gene and their association with sperm quality traits of Boer goats and Boer crosses. Animal Reproduction Science, 146, 176–181
Ott, J., 2001. Program Het version 1.8. Utility programs for analysis of genetic linkage. Rockefeller University. New York
Parthiban, V., Gromiha, M.M., Schomburg, D., 2006. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Research, 34, W239–42.
Pauline, C.N., Steven, H., 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Research, 31, 3812–3814
Pejaver, V., Urresti, J., Lugo-Martinez, J., Pagel, K.A., Lin, G.N., Nam, H., Mort, M., Cooper, D.N., Sebat, J., Iakoucheva, L.M., Mooney, S.D., Radivojac, P., 2017. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv, 134981
Pires, D.E., Ascher, D.B., Blundell, T.L., 2014. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Research, 42, W314–W319
Pires, D.E.V., Ascher, D.B., Blundell, T.L., 2012. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics, 30, 335–342.
Sailo, L., Gupta, I.D., Verma, A., Singh, A., Chaudhari, M.V., Das, R. Upadhyay, R.C., Goswami, J., 2015. Single Nucleotide Polymorphism in HSP90AB1 gene and its association with thermo-tolerance in Jersey cross-breed cows. Animal Science Reporter, 9(2), 43–49
Salces-Oritz, J., Gonzalez, C., Martinez, M., Mayoral, T., Calvo, J.H., Serrano, M.M., 2015. Looking for adaptive footprints in the HSP90AA1 ovine gene. BMC Evolutionary Biology, 15, 7
Salman, A.D., Ibrahim, H.K., Al-Khalisy, A.F., Ibrahim, R.A., 2009. Effect of body condition and supplementary feeding on the reproductive performance of Awassi ewes. The Iraqi Journal of Veterinary Medicine, 33(2), 138–145
Shaji, S., Sejian, V., Bagath, M., Manjunathareddy, G.B., Kurein, E.K., Varma, G., Bhatta, R., 2017. Summer season related heat and nutritional stresses on the adaptive capability of goats based on blood biochemical response and hepatic HSP 70 gene expression. Biological Rhythm Research, 48(1), 65–83
Singh, K.M., Singh, S., Ganguly, I., Nachiappan, R.K., Ganguly, A., Venkataramanan, R., Chopra, A., Narula, H.K., 2017. Association of heat stress protein 90 and 70 gene polymorphism with adaptability traits in Indian sheep (Ovis aries). Cell Stress Chaperones, 22(5), 675–684
Smigielski, E.M., Sirotkin, K., Ward, M., Sherry, S.T., 2000. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Research, 28, 52–355
Talafha, A.Q., Ababneh, M.M., 2011. Awassi sheep reproduction and milk production. Tropical animal health and production, 43(7), 1319–1326
Tang, H., Thomas, P.D., 2016. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics, 32, 2230–2
Trakovická, A., Moravčíková, N., Kasarda, R., 2013. Genetic polymorphisms of leptin and leptin receptor genes in relation with production and reproduction traits in cattle. Acta Biochimica Polonica, 60, 783–60
Verma, N., Gupta, I.D., Verma, A., Kumar, R., Das, R., Vineeth, M.R., 2016. Novel SNPs in HSPB8 gene and their association with heat tolerance traits in Sahiwal indigenous cattle. Tropical Animal Health and Production, 48(1), 175–180
Worth, C.L., Preissner, R., Blundell, T.L., 2012. SDM-a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Research, 39, W215–W222.
Yeh, F.C., Yang, R., Boyle, T., 1999. POPGENE: Version 1.31. Microsoft window-based freeware for population genetic analysis, University of Alberta. Edmonton
Zachut, M., Speranda, M., de Almeida, A., Gabai, G., Mobasheri, A., Hernandez-Castellano, L.E., 2020. Biomarkers of fitness and welfare in dairy cattle: healthy productivity. Journal of Dairy Research, 87(1), 4–16.
Zhang, K. Ezemaduka, A.N., Wang, Z., Hu, H., Shi, X., Liu, C., Lu, X., Fu, X., Chang, Z., Yin, C.C., 2015. A novel mechanism for small heat shock proteins to function as molecular chaperones. Scientific Reports, 5, 8811
Funding
This research was partially funded by Al-Qasim Green University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 918 kb)
Rights and permissions
About this article
Cite this article
Al-Thuwaini, T.M., Al-Shuhaib, M.B.S. & Hussein, Z.M. A novel T177P missense variant in the HSPA8 gene associated with the low tolerance of Awassi sheep to heat stress. Trop Anim Health Prod 52, 2405–2416 (2020). https://doi.org/10.1007/s11250-020-02267-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11250-020-02267-w