Abstract
A fully automated isotopic profile deconvoluted chromatogram (IPDC) algorithm (Fakouri Baygi et al. in Anal Chem 91:15509–15517, 2019) and user interactive HaloSeeker 1.0 (Léon et al. in Anal Chem 91:3500–3507, 2019) were compared to test the efficacy of these two computationally enhanced non-targeted screening (CENTS) tools in isolating unknown Br/Cl compounds using high-resolution mass spectrometry (HRMS) data. HaloSeeker depends on a user to monitor the performance of the peak picking algorithm and assign molecular formulas for each isotopic signature in an ergonomic interface. Alternatively, the IPDC algorithm automatically assigns and ranks candidate molecular formulas within a set of search criteria. Both CENTS tools were evaluated using fish and sediment data acquired at 22,000 (mHRMS) and > 100,000 (uHRMS) mass resolutions, respectively. The IPDC algorithm detected 85% of compounds detected by HaloSeeker as the first candidate compound in the sediment sample, with fewer false positives. In the sediment data, the IPDC algorithm detected several compounds such as clofoctol and chlorinated paraffins that were not reported using HaloSeeker 1.0. Upon further inspection, these compounds were isolated by the HaloSeeker program, but not reported by the user. HaloSeeker detected all significant and insignificant chemical ionization products (relative to IPDC), but additional false positives were isolated in the mHRMS polychlorinated biphenyl reference standard and trout sample. HaloSeeker detected 62% of the legacy contaminant features isolated by the IPDC algorithm in the fish data (mHRMS). The comparison of these two CENTS tools demonstrates that matrix complexity and mass resolution of the HRMS platform are the key factors when choosing automated and user interactive CENTS tools.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
High-resolution mass spectrometry (HRMS) instruments have been widely used in the past decade to identify unknown compounds in environmental samples (Fakouri Baygi et al. 2019; Léon et al. 2019; Schymanski et al. 2014; Knolhoff et al. 2014). The platforms are designed to support different ionization techniques to support a wide range of applications. The resolution of these instruments typically ranges between 10,000 and 1,000,000 FWHM (full width at half maximum) introducing significant variability when comparing non-targeted screening workflows results for environmental matrices (Fakouri Baygi et al. 2019; Knolhoff et al. 2014; Hernández et al. 2012). Therefore, newly developed computational enhanced non-targeted screening (CENTS) tools should be flexible with respect to HRMS capabilities and matrix complexity (data structure) when determining their ability to isolate unknown compounds.
A number of CENTS tools employ isotopic patterns to predict molecular formulas of unknown compounds based on measured masses (Fakouri Baygi et al. 2019; Léon et al. 2019). Notably, natural stable isotopes of bromine (79Br, 81Br) and chlorine (35Cl, 37Cl) produce easily distinguishable isotopic signatures in mass spectra. Br/Cl isotopic signatures have driven the development of many screening software packages to isolate Br/Cl compounds in several research fields including food safety, metabolomics, and environmental analyses (Fakouri Baygi et al. 2019; Knolhoff et al. 2014; Cariou et al. 2016; Schymanski et al. 2015). Léon et al. (2019) presented HaloSeeker 1.0 as a user-friendly software and evaluated its performance using a reference halogenated standard mixture and Seine River sediment samples analyzed at 140,000 FWHM mass resolution (m/z = 200) on an ultra-HRMS (uHRMS) instrument. Fakouri Baygi et al. (2019) demonstrated carbon, bromine, chlorine, and sulfur dominated isotopic profiles of the majority of environmentally relevant organic contaminants in lake trout samples from Lake Michigan. Using this concept, the authors developed an isotopic profile deconvoluted chromatogram (IPDC) algorithm to resolve unknown molecular formulas based on isotopic profile seeds (Fakouri Baygi et al. 2019) followed by chromatographic reconstruction. The IPDC algorithm was able to isolate low concentration halogenated components in complex biological samples using a medium-HRMS (mHRMS) instrument with 22,000 FWHM dynamic resolution (Fakouri Baygi et al. 2019).
HaloSeeker 1.0 is a web application running in an R environment and integrates a SQLite database for data storage (Léon et al. 2019). HaloSeeker was designed to isolate Br/Cl mass spectrometry signatures in uHRMS data sets through four successive steps. A graphical user interface (GUI) is also provided, displayed in a web browser on the local host, and a workflow starting from proprietary raw data to a Microsoft Excel file with annotated signals. HaloSeeker converts vendor specific data files to an open format (.mzXML) and then integrates signals as features (peaks) by employing the xcms R package (Tautenhahn et al. 2008) (F0 step). The peaks are paired into clusters to retrieve isotopic profiles (F1 step). Then, halogenated ion ratios are used to eliminate less likely features (F2 step). Eventually, the user assigns molecular formulas (and optionally a compound) based on features that passed “polyhalogenated” ion ratio rules (F2+ step) using the Rdisop R package (adapted to consider isotopes rather than chemical elements in its alphabet). HaloSeeker integrates the seven golden rules (Kind and Fiehn 2007; Morikawa and Newbold 2003) to predict formulas by decomposing m/z of base peaks (most intense isotopologue in a cluster). It also takes advantage of the enviPat package to compute theoretical isotopic profile and a matching score. Ultimately, HaloSeeker 1.0 can also compare clusters to previously annotated data (or theoretical compound manually added) to help the user in the assignment. One of major objectives of HaloSeeker software was to present a user-friendly GUI with several graphical tools for manual data exploration (Léon et al. 2019).
The IPDC algorithm initially was developed to deconvolute biologically complex HRMS data (Fakouri Baygi et al. 2020). The IPDC algorithm employs a novel approach to incorporate chromatographic analysis with the mass spectrometry parameters to compensate for mass measurement deviations from lower mass resolution platforms and non-Gaussian peak shapes that may be missed by other generic peak picking algorithms. The IPDC algorithm applies a completely different approach relative to HaloSeeker and does not implement the xcms package (Tautenhahn et al. 2008) or depends on the H/Cl mass defect graphical view. Instead, the IPDC algorithm screens for isotopic profiles of a large number of molecular formulas (up to 108) in each chromatogram scan using a computationally efficient method (Fakouri Baygi et al. 2019). Next, the IPDC algorithm uses chromatographic (peak shape) analyses to isolate true positives. The main objective of the IPDC algorithm is to minimize time-consuming manual post-processing workloads. Hence, the IPDC algorithm was designed to offer robust nonlinear data reduction filters (e.g., machine learning classifiers) and simple linear data reduction filters (e.g., intensity thresholds) to further reduce false positive detection depending on the complexity of the matrix. The IPDC algorithm results are presented in m/z-retention time plots to visualize important information such as chromatographic separation of isomeric and coeluting features (Fakouri Baygi et al. 2019).
Although the IPDC algorithm and HaloSeeker were established based on isotopic profile detection, they have fundamental differences in their peak identification workflows and their noise (i.e., electronic noise and non-halogenated false positives) removal approaches. These differences can result in significant variability in their performance isolating Br/Cl components in HRMS data depending on the level of matrix complexity and analytical platform mass resolution. HaloSeeker was already compared with similar user-interactive software packages such as Marine Halogenated Compound Analysis (MeHaloCoA), Nontarget, and Dynamic Cluster Analysis (DCA) (Roullier et al. 2016; Loos et al. 2012; Andersen et al. 2016) and indicated a superior performance. However, user-interactive software packages have not been compared with fully automated approaches. Automation alters the nature of the postprocessing step in the NTS workflows and allows investigating more features. However, automated and user-interactive CENTS tools have not been compared in the literature to evaluate operational aspects of CENTS tools in the environmental analyses. Therefore, in this work, the constraints and advantages of the fully automated IPDC algorithm (Fakouri Baygi et al. 2019) and user-interactive HaloSeeker 1.0 (Léon et al. 2019) were evaluated on mHRMS and uHRMS data generated for fish and sediment matrices, respectively, to provide comparison analyses using these two CENTS workflows. The current experiments offer insights into the differences between the IPDC algorithm and HaloSeeker 1.0 considering confounding interlaboratory factors such as data complexity, HRMS instrument resolution, and automated versus manual post-processing.
Materials and methods
For this exercise, the data files analyzed in the previous publications (Fakouri Baygi et al. 2019; Léon et al. 2019) were used to further evaluate performances of HaloSeeker 1.0 and the IPDC algorithm. The halogenated standard mixture and the Seine River sediment data files were produced by a liquid chromatography-heated electrospray ionization (LC-HESI) coupled to an Exactive Orbitrap instrument (mass resolution of 140,000 FWHM at m/z 200). These files were used to investigate the IPDC algorithm capabilities on uHRMS data. The halogenated standard mixture contained 0.04 ng/μL of 43 non-labeled compounds including 19 polychlorophenols (PCPs), 19 polybromophenols (PBPs), one hydroxylated polychlorinated diphenyl ether (OH-CDE), three hydroxylated polybrominated diphenyl ethers (OH-BDEs), one mixed halogenated hydroxylated diphenyl ether (OH-XDE), two isotope-labeled hexabromocyclododecane isomers (2H18-α- and γ-HBCDDs), and one isotope-labeled tetrabromobisphenol A (13C12-TBBPA) was used to calibrate the IPDC algorithm. HaloSeeker results for these data files were published by Léon et al. (2019), and the new analysis using IPDC is presented in this study.
To evaluate the performance of HaloSeeker on mHRMS data, a 20 pg/µL polychlorinated biphenyl (PCB) standard mixture (68B-PAR, Wellington Laboratories, ON, Canada) and biological complex mHRMS data files of Lake Michigan trout analyzed by an atmospheric pressure gas chromatography (APGC) coupled to a Waters Xevo G2-XS quadrupole time-of-flight (QToF) instrument (22,000 FWHM dynamic mass resolution) in negative mode were utilized. A full description of the APGC-QToF instrumentation and the Lake Michigan trout sample preparation was provided by Fakouri Baygi et al. (2019) and Fernando et al. (2018). The IPDC algorithm results for these data files have been previously discussed by Fakouri Baygi et al. (2019), and the new data analyses using HaloSeeker 1.0 are presented in the current inter-comparison study.
Methods and parameters
The same data processing methods described in the previous publications (Fakouri Baygi et al. 2019; Léon et al. 2019) were used in this work. The IPDC algorithm and HaloSeeker use fundamentally different approaches and search parameters. Therefore, the same search criteria should not be used to compare these two workflows. For example, the IPDC algorithm initiates a mass profile matching prior to reconstructing the chromatographic peaks. In this step, the algorithm applies an intensity threshold to the Most Abundant Isotopologue (MAIso) of the isotopic model in individual scans to remove low-level random noise in the m/z screening step. After this step, the IPDC algorithm may employ a filter based on the area or height of reconstructed chromatographic peaks according to the user preference.
HaloSeeker 1.0 applies its threshold criteria to the heights of chromatographic peaks in the peak-picking step (using xcms) prior to isotope matching. After the peak-picking threshold has been applied, HaloSeeker allows users to apply an additional intensity threshold on the cumulative cluster intensity in the H/Cl mass defect plots. These two workflows also use different approaches to measure similarity between experimental mass spectra and theoretical isotopic profile models. The IPDC algorithm employs profile cosine similarity (PCS) and normalized Euclidean mass error (NEME).
where Ii, Mi and N are intensity of the isotopologue, isotopologue mass, and number of isotopologues in the isotopic model. The IPDC algorithm calculates \(\overline{PCS }\) and \(\overline{NEME }\) for an integrated mass spectrum across a chromatographic peak using the Eqs. (1) and (2). HaloSeeker 1.0 uses the following equation to measure profile similarities.
where S is the profile similarity calculated in HaloSeeker. A slight tolerance for profile deviations is beneficial to compensate for inherent signal variability caused by compound and sample properties such as isotope fractionation (Tang and Tan 2018) and matrix interference (Fakouri Baygi et al. 2019). For instance, tolerance of PCS and S to variability in the relative intensity of the MAIso in a Cl4 isotopic profile model is studied in Figure S.1. PCS is significantly less affected by the variation in the relative intensity of the MAIso compared to S. To achieve a score > 95% the PCS (Eq. 2), the relative intensity of the MAIso can vary more ± 50%, whereas the MAIso is limited to ± 1.1% to reach an S > 60% (Eq. 3). Tolerance of PCS to isotopic profile variations is crucial for automated approaches such as the IPDC algorithm (Fakouri Baygi et al. 2019, 2020; Léon et al. 2019; Schymanski et al. 2014, 2015; Knolhoff et al. 2014, 2016; Hernández et al. 2012; Cariou et al. 2016; Tautenhahn et al. 2008; Kind and Fiehn 2007; Morikawa and Newbold 2003; Roullier et al. 2016; Loos et al. 2012; Andersen et al. 2016; Fernando et al. 2018; Tang and Tan 2018).
Approximately 3 million candidate molecular formulas (Table S.1) were employed to process the uHRMS data. The search criteria of the IPDC algorithm are presented in Tables S.2(a–f). To calibrate the data reduction filters of IPDC algorithm, a set of linear cutoffs (Table S.2(g)) were optimized for the uHRMS standard dataset by changing the thresholds until achieving a maximum number of true positive features and minimum number of unknown features. Search criteria of HaloSeeker for mHRMS data are also discussed in the section S.2.
Results and discussion
The IPDC algorithm on uHRMS data
The IPDC algorithm was able to detect 13 PBPs and 14 PCPs out of 19 of each type. The reason for missing a number of PBPs and PCPs is coelution as described by Léon et al. (2019). The IPDC algorithm detected 52 [M − H]−, six [2M − H]− and one [M + NO3]− ion products in the uHRMS data of the halogenated standard mixture. The IPDC algorithm detected an additional 20 halogenated features that were not listed in the halogenated standard mixture including two potential mixed polyhalogenated phenols (PXPs), four OH-CDEs, 13 OH-XDEs, and a polybromoresorcinol (PBR). The IPDC algorithm also isolated 14 additional features using linear data reduction filters. These 14 features satisfied all of the feature qualification metrics shown in Table S.2(g) and should be investigated further. The IPDC algorithm was not able to detect isotope-labeled compounds or those determined to be false halogenated molecular formulas. In the halogenated standard mixture analysis, approximately 88% of molecular formulas determined by the IPDC ranking system were the top candidate without applying any additional conditions.
Léon et al. (2019) annotated 33 clusters (F2+) using HaloSeeker for features between 1 and 20 min. More than 400 clusters were not investigated after prioritizing intense clusters (> 2 × 106 cumulated intensity) and applying isotopic ratio rules (filter F2+). In this study, the effective retention time range of 4–9 min and 4–13 min was studied for the halogenated standard mixture and the Seine River sediment samples, respectively. A comparison of the results from the IPDC algorithm for the 33 halogenated molecular formulas detected by HaloSeeker (F2+) in the halogenated standard mixture is presented in Table S.3. An RT-m/z plot (spectrogram) of the halogenated standard mixture is shown in Fig. 1, and the results of the IPDC algorithm are presented in the Supporting Information. A summary of the IPDC algorithm and HaloSeeker 1.0 performance on the halogenated standard mixed is shown in Table 1.
Comparison between the isotopic profile deconvoluted chromatogram (IPDC) algorithm and HaloSeeker on the halogenated standard mixture by negative HESI-Orbitrap. The entire points are the IPDC algorithm predictions except purple and blue circles belonging to HaloSeeker (F2+ clusters after applying intensity threshold 2 × 106 AU)
The IPDC algorithm detected 549 and 595 known and unknown features, respectively, compared to 264 clusters (F2+ clusters) detected by HaloSeeker in the Seine River sediment sample shown in Fig. 2. The HaloSeeker user assigned molecular formulas to 99 (96 non-labeled) out of 264 clusters (F2+) in the uHRMS data of the Seine River sediment sample. The IPDC algorithm detected 82 out of 96 non-labeled molecular formulas that HaloSeeker reported in the Seine River sediment sample (Table S.4). The HaloSeeker candidate molecular formula was also the top candidate molecular formula suggested by the IPDC algorithm in 80% of the cases and one of the top two or three for the remaining 20% (Table S.5).
Spectrogram of the Seine River estuary sediment sample in 2002 by negative HESI-Orbitrap using the isotopic profile deconvoluted chromatogram (IPDC) algorithm. The entire points are the IPDC algorithm predictions except purple and blue circles belonging to HaloSeeker (F2+ clusters after applying intensity threshold 2 × 106 AU)
Agreement between the two methods decreased with decreasing algorithm qualification metrics (internal scoring systems). For example, a number of molecular formulas (9 out of 14) that were not detected by the IPDC algorithm fell below the 60% level in the HaloSeeker scoring system. At this threshold, value feature verification by an expert user is critical. This result indicates that the enhanced automation of the IPDC algorithm is more dependent on matching isotopic profiles and chromatogram shapes within search criteria compared to HaloSeeker.
The IPDC algorithm isolated molecular formula of 14 hydroxylated polychlorinated biphenyls (OH-PCBs) congeners, three hydroxylated perchlorinated diethylfuran (OH-CDF) congeners, three OH-BDEs, nine OH-CDEs, two OH-XDEs, four PBPs, and two PCPs that were below the HaloSeeker cumulative intensity threshold based on the H/Cl mass defect plot (< 2 × 106 AU). Moreover, the IPDC algorithm isolated 57 [M − H]−, 153 [M + NO3]−, 227 [M + C2H3O2]−, 25 [M + C2H3O2 + NO3]− ion products for a number of chlorinated paraffin (CP) homologues. HaloSeeker also detected 12 [M + C2H3O2]− ions associated with CPs, but missed the majority of the isomers due to broad peak widths (> 60 s) and abnormal peak shapes or retention time deviations of the resolved isotopologues from the xcms package (< 1 s). The number of isomers that the IPDC algorithm detected for CPs was only an estimate due to the inherent complexity of the CP signatures (e.g., see Figure S.2). [M + NO3]− and [M + C2H3O2]− ionization pathways were consistent with a number of labeled HBCDD ion products in the halogenated standard mixture (Table S.3). A summary of the IPDC algorithm (Fakouri Baygi et al. 2019) and HaloSeeker 1.0 (Léon et al. 2019) performance on legacy contaminant features isolated in the Seine River sediment sample is shown in Table 2.
The ergonomic environment of HaloSeeker facilitates visual inspection of the extracted ion chromatograms (EICs), mass spectra, and other criteria such scoring value or deviation in mass to reduce false positive identifications. The IPDC algorithm was also able to generate EICs of the isolated features to further investigate the algorithm errors due to data quality (e.g., isotopologue fidelity) in some instances. However, the IPDC algorithm does not offer an ergonomic environment similar to HaloSeeker for manual feature evaluation. In its current form, the IPDC algorithm suggests ranked candidate molecular formulas for individual isotopic features. A complete list of candidate molecular formulas detected by the IPDC algorithm for the halogenated standard mixture and the Seine River sediment samples is available in the Supporting Information. Figures 1 and 2 are available in the Supporting Information in a MATLAB format (.fig) for a more detailed data exploration.
H/Cl mass defect plots are particularly useful graphical visualization tools to highlight homologue series that share similar mass defect values. Léon et al. (Léon et al. 2019) detected 23 molecular formulas with C12HxBryClzO2 structure on the H/Cl mass defect plot as well as 76 molecular formulas with various molecular formula structures. Mass defect values of candidate masses have limited utility when they do not correspond to known molecular formula structures (Fakouri Baygi et al. 2019). For example, Léon et al (2019) missed the [C21H25Cl2O]− ion in the sediment sample with a H/Cl mass defect of 0.5451 (compared to 0.2728 for C12HxBryClzO2), even though HaloSeeker prioritized this feature and displayed it in the H/Cl mass defect plot. Alternatively, the IPDC algorithm automatically detected this molecular formula and predicted clofoctol as the first ranked candidate using the compound library of the EPA chemical dashboard (Williams et al. 2017) embedded in the IPDC algorithm database. The retention time of clofoctol compared well with a neat standard (Figure S.3) and a concentration of 0.03 ng/g was estimated using a global response factor with respect to the internal standard (labeled 2H18-α-HBCDD). This example illustrates potential drawbacks of highly user interactive qualification requirements in CENTS tools and H/Cl mass defect plots with respect to automated user-independent tools.
HaloSeeker 1.0 on mHRMS data
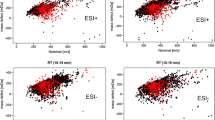
HaloSeeker was able to detect all 35 significant features in the PCB standard mixture in the region of interest (15 min ≤ RT ≤ 51.5 min and 100 ≤ m/z ≤ 600 Da)(Fig. 3). HaloSeeker also detected 74 (15 if RT < 50 min) uncategorized features in the reference PCB standard mainly due to the limitations of the xcms peak picking module that utilized a relatively large mass error (80 ppm) to ensure the detection of consecutive scans of the reference compounds. Using these settings, a number of the uncategorized features were observed that coeluted with the PCB compounds. These features were found to be ion fragments of the PCBs in the solution although their abundances were considerably lower than the dominant [M − Cl + O]−, [M]− and [M − H − Cl + O2]− ions (Fakouri Baygi et al. 2019). The m/z intensity threshold on the MAIso mass (> 500) of the IPDC algorithm was applied to individual scans, and it was too high to detect these ionization products. The IPDC algorithm detected all 35 relevant features without any additional false positives.
Comparison between the isotopic profile deconvoluted chromatogram algorithm and HaloSeeker on the mHRMS data of the PCB standard mixture in negative APGC-QToF. (Adapted with permission from (Fakouri Baygi et al. 2019).
HaloSeeker detected 2468 clusters in the mHRMS data of the Lake Michigan trout sample (Fig. 4). The IPDC algorithm detected 418 known features associated with halogenated legacy contaminants and 437 unknown features. HaloSeeker was able to detect 258 out of 418 and 167 out of 437 known legacy compounds and unknown features that were in common with the IPDC algorithm results, respectively. Annotating molecular formulas on 2468 features using the user interactive interface of HaloSeeker requires a significant amount of effort in the dereplication step due to the wide mass errors associated mHRMS data and uncertainty in candidate molecular formulas for mHRMS data of complex samples (Fakouri Baygi et al. 2019). The linear data reduction techniques of the IPDC algorithm retained 8130 features in the mHRMS data. To remove the false positives in the complex mHRMS data, the IPDC algorithm used a nonlinear machine learning classifier (MLC) in addition to the linear filters. The machine learning classifier (MLC) removed the majority of false positives (> 99%) but may also have removed a fraction (< 15%) of true positives due to systematic errors in the training of the MLC function (Fakouri Baygi et al. 2019) illustrating the tradeoff between automated MLC function for false positive removal and assigning a user to manually check mHRMS data and remove false positives. The development of the MLC false positive removal module using four individual data sets of the PCB standard mixture was described in the previous publication (Fakouri Baygi et al. 2019). Summary results of HaloSeeker performance on the PCB standard mixture and Lake Michigan trout are presented in the Supporting Information. Figures 3 and 4 in the Supporting Information are presented in their MATLAB format (.fig) for detailed explorations.
Comparison between the isotopic profile deconvoluted chromatogram (IPDC) algorithm and HaloSeeker on the mHRMS data of the Lake Michigan trout in negative APGC-QToF. Blue circles, unannotated features by HaloSeeker; golden diamonds, uncategorized features by the IPDC algorithm; rest of the points, legacy contaminants detected by the IPDC algorithm presented by Fakouri Baygi et al. (2019). (Adapted with permission from Fakouri Baygi et al. 2019.
A comparison of performances of each CENTS workflow is presented in Table 3.
An operational comparison between the IPDC algorithm and HaloSeeker 1.0 is presented in Table 4.
Conclusion
The IPDC algorithm and HaloSeeker demonstrated distinct similarities and differences when analyzing uHRMS and mHRMS data. The primary reason for these differences is related to the different objectives of each workflow. The IPDC algorithm attempted to provide a user-independent workflow predicated on computational approaches, while HaloSeeker was developed to provide an ergonomic environment to manually check the quality of each single isotopic profile hit. These differences can be attributed to the CENTS workflow, isotopic profile matching, data reduction and prioritization techniques, and post-processing limitations.
CENTS workflows
The IPDC algorithm was designed to be sensitive to all isotopic signatures, and this design enables it to screen for non-Br/Cl as well as Br/Cl compounds, while HaloSeeker was designed exclusively to screen for Br/Cl compounds. For instance, Singh et al. (2019) successfully detected perfluoroalkyl substances (PFAS) breakdown products as compounds with considerably less distinguishable isotopic profiles in mHRMS data solely using the IPDC algorithm. The polyhalogenated ratio rules of HaloSeeker discriminate against sulfur isotopic signatures, while the IPDC algorithm is able to detect sulfur signatures without any extra steps. However, the IPDC algorithm in its current state is limited to screening for < 108 molecular formulas (~ 3 × 106 in this work) in each single run, while HaloSeeker is more flexible with regards to the number of candidate molecular formulas. Both the IPDC algorithm and HaloSeeker are able to facilitate cataloging m/z, RT, and peak areas of unknown features for further investigation.
The xcms package (Tautenhahn et al. 2008) has been used in many metabolomics (Aggio et al. 2011) and environmental analyses software packages (Léon et al. 2019; Aggio et al. 2011) due of its simplicity, efficiency, and variety of useful tools in different applications. However, the xcms package (Tautenhahn et al. 2008) is not able to differentiate isomeric features that are not well separated chromatographically and requires detection of the same m/z (with a tolerance) in consecutive scans. The IPDC algorithm has a chromatography analysis toolbox consisting of missing consecutive scan interpolation, peak smoothing, isomeric feature detection, peak shape evaluation and automated peak fronting/tailing resolution that was adjusted for poor chromatography conditions. These major differences allow the IPDC algorithm to potentially resolve overlapping peaks grouped by the xcms package and detect distorted chromatographic peaks. For example, the IPDC algorithm was able to resolve α and β isomers of HBCDD in the Seine River sediment samples while xcms detected only one feature due to overlapping peaks. Despite these limitations in the xcms package, HaloSeeker was able to pair the isotopic profile of the HBCDD in the detected chromatography peaks. Nevertheless, both xcms and chromatography analysis toolbox of the IPDC algorithm may not function ideally with poor chromatographic peak shapes.
Isotopic profile matching errors
The IPDC algorithm and HaloSeeker were designed to match theoretical isotopic profiles on HRMS data and depend on an accurate calculation of isotopic profiles. Reliable matching of theoretical isotopic profiles on experimental spectra is complicated, and MS data mass resolution needs to be considered when neighboring isotopologues in an isotope model are present within the instrument mass resolution (Fakouri Baygi et al. 2019, 2016). Isotopic profiles are not unique signatures, and interferences such as overlaying isotopic profiles of [M]+ and [M ± H]+ ions in chemical ionization methods can significantly alter the characteristics of isotopic profiles even in neat reference standards (Fakouri Baygi et al. 2019). Moreover, isotopic profiles cannot reduce the complexity of the unknown identification process (Fakouri Baygi et al. 2019). HaloSeeker provides a flexible approach to isotopic profile variability in some instances. For example, HaloSeeker detected two dichlorophenol congeners in the uHRMS data of the Seine River sediment sample even though some characteristic 13C isotopologues for the IPDC algorithm were missing in the mass spectra. Conversely, the IPDC algorithm depends on a confident match of isotopic profiles that is both a strength and drawback for the IPDC algorithm. Parameters such as PCS and NEME enable the IPDC algorithm to boost tolerance to isotopic profile variability, but if an isotopologue in the isotopic model is not matched in the m/z screening step, the IPDC algorithm ignores the entire isotopic profile in the mass spectra and later attempts to interpolate a value for the missing chromatogram scan on the chromatogram peak construction step. Alternatively, a confident isotopic profile reduces false positive identifications and increases the identification confidence, especially when false positive detection is a serious concern (Fakouri Baygi et al. 2019). Approximately 85% of the feature detected by HaloSeeker in the uHRMS data from the Seine River sediment passed the isotopic profile requirements of the IPDC algorithm.
Data reduction and prioritization techniques
The IPDC algorithm is able to apply automated data reduction filters to assess whether a feature may represent a true positive or not. Hence, when a feature does not match true positive qualification metrics, the IPDC algorithm removes it as a false positive. On the other hand, HaloSeeker uses a different approach and attempts to preserve all these questionable features for the user’s assessment. Hence, HaloSeeker was designed to offer multiple prioritization options to allow users to adjust their preferred prioritization according to their needs and level of expertise and then manually annotate features in an ergonomic environment.
To maximally take advantage of an HRMS instrument mass resolution power, the automated molecular formula prediction built into the IPDC algorithm utilizes mass measurement precision as a criterion to minimize false positives that do not correspond to any realistic molecular formulas. The IPDC algorithm includes sets of diverse and effective data reduction techniques, and unlike HaloSeeker, there was no need for users to apply a high intensity threshold or omit monohalogenated features to mitigate the number of features for the manual post-processing. Generally, CENTS workflows that implement the polyhalogenated ratio rule eliminate monohalogenated compounds (Léon et al. 2019; Zhang et al. 2019). The IPDC algorithm demonstrated its capabilities to preserve low abundant and monohalogenated features using variety of linear and nonlinear reduction techniques. The IPDC algorithm indicated that only simple linear cutoffs were sufficient to remove false positives in uHRMS halogenated standard mixture data without losing the true positives feature. HaloSeeker was not able to offer effective prioritization methods to moderate the number of hits in the mHRMS data of the trout for manual investigation within an acceptable time window. However, Fakouri Baygi et al. (2019) indicated that a nonlinear data reduction filter (e.g., machine learning classifier) was able to remove the majority of false positives in mHRMS data at the expense of a small fraction of true positives. Generally, the IPDC algorithm detected a higher ratio of known features to unknown features for uHRMS and mHRMS data compared to the HaloSeeker results.
Post-processing limitations
The IPDC algorithm was able to automatically predict molecular formulas that were consistent with those manually assigned with the help of HaloSeeker in significantly less time and effort in 85% of cases. Fully automated methods may have systematic errors, but results are consistent regardless of possible user bias. The automated approach demonstrated that the IPDC algorithm is suitable for time sensitive projects or when manual post-processing labor/expertise is limited with a defined error margin (< 15%). The IPDC algorithm presented 85% similarity to the expert-inspected HaloSeeker results plus additional features that were not selected in the data prioritization of HaloSeeker in the uHRMS data of the sediment and standard samples. The GUI of HaloSeeker allows users to manually interpret spectra anomalies and facilitates circumventing the defined feature qualification metrics by an expert user in an ergonomic environment. For example, approximately 20% of the candidate molecular formulas for the uHRMS data of the sediment sample suggested by HaloSeeker exhibited scores or lacked characteristic isotopologues that required user interpretation/confirmation. However, user errors also need to be considered when evaluating user-interactive software packages such as HaloSeeker when several days were needed to process one complex uHRMS data file by a non-expert user. The IPDC algorithm may use a library of known legacy contaminants to assign molecular formulas on known features. This option facilitates targeting unknown compounds in matrices that were dominated by known legacy compounds.
The intrinsic differences between automated and user-interactive methodologies were studied in this work and illustrated advantages and disadvantages of each CENTS workflow. Advances in computational power and instrumental precision will enable the IPDC algorithm and HaloSeeker to become more efficient and employed in many advanced research applications such as hybrid targeted and non-targeted analysis (Crimmins et al. 2018) where datasets have very many isotopic profiles and CENTS tools necessitate to benefit from more automations (Cariou et al. 2016).
References
Aggio R, Villas-Bôas SG, Ruggiero K (2011) Metab: an R package for high-throughput analysis of metabolomics data generated by GC-MS. Bioinformatics 27(16):2316–2318
Andersen AJC, Hansen PJ, Jørgensen K, Nielsen KF (2016) Dynamic cluster analysis: an unbiased method for identifying A+ 2 element containing compounds in liquid chromatographic high-resolution time-of-flight mass spectrometric data. Anal Chem 88:12461–12469
Cariou R, Omer E, Léon A, Dervilly-Pinel G, Le Bizec B (2016) Screening halogenated environmental contaminants in biota based on isotopic pattern and mass defect provided by high resolution mass spectrometry profiling. Anal Chim Acta 936:130–138
Crimmins BS, McCarty HB, Fernando S, Milligan MS, Pagano JJ, Holsen TM, Hopke PK (2018) Commentary: integrating non-targeted and targeted chemical screening in Great Lakes fish monitoring programs. J Great Lakes Res 44:1127–1135
Fakouri Baygi S, Crimmins BS, Hopke PK, Holsen TM (2016) Comprehensive emerging chemical discovery: novel polyfluorinated compounds in Lake Michigan trout. Environ Sci Technol 50:9460–9468
Fakouri Baygi S, Fernando S, Hopke PK, Holsen TM, Crimmins BS (2019) Automated isotopic profile deconvolution for high resolution mass spectrometric data (APGC-QToF) from biological matrices. Anal Chem 91:15509–15517
Fakouri Baygi S, Fernando S, Hopke PK, Holsen TM, Crimmins BS (2020) Decadal differences in emerging halogenated contaminant profiles in Great Lakes Top Predator Fish. Environ Sci Technol 54:14352–14360
Fernando S, Renaguli A, Milligan MS, Pagano JJ, Hopke PK, Holsen TM, Crimmins BS (2018) Comprehensive analysis of the Great Lakes top predator fish for novel halogenated organic contaminants by GC× GC-HR-ToF mass spectrometry. Environ Sci Technol 52:2909–2917
Hernández F, Sancho JV, Ibáñez M, Abad E, Portolés T, Mattioli L (2012) Current use of high-resolution mass spectrometry in the environmental sciences. Anal Bioanal Chem 403:1251–1264
Kind T, Fiehn O (2007) Seven golden rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinform 8:105
Knolhoff AM, Callahan JH, Croley TR (2014) Mass accuracy and isotopic abundance measurements for HR-MS instrumentation: capabilities for non-targeted analyses. J Am Soc Mass Spectrom 25:1285–1294
Knolhoff AM, Zweigenbaum JA, Croley TR (2016) Nontargeted screening of food matrices: development of a chemometric software strategy to identify unknowns in liquid chromatography–mass spectrometry data. Anal Chem 88:3617–3623
Léon A, Cariou R, Hutinet S, Hurel J, Guitton Y, Tixier C, Munschy C, Antignac JP, Dervilly-Pinel G, Le Bizec B (2019) HaloSeeker 1.0: a user-friendly software to highlight halogenated chemicals in nontargeted high-resolution mass spectrometry data sets. Anal Chem 91:3500–3507
Loos M, Hollender J, Schymanski E, Ruff M, Singer H (2012) Bottom-up peak grouping for unknown identification from high-resolution mass spectrometry data. Presented at ASMS 2012, Vancouver, Canada, May 20−24, 2012
Morikawa T, Newbold BT (2003) Analogous odd-even parities in mathematics and chemistry. Chemistry 12:445–449
Roullier C, Guitton Y, Valery M, Amand S, Prado S, Robiou du Pont T, Grovel O, Pouchus YF (2016) Automated detection of natural halogenated compounds from LC-MS profiles–application to the isolation of bioactive chlorinated compounds from marine-derived fungi. Anal Chem 88:9143–9150
Schymanski EL, Singer HP, Longrée P, Loos M, Ruff M, Stravs MA, Ripollés Vidal C, Hollender J (2014) Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ Sci Technol 48:1811–1818
Schymanski EL, Singer HP, Slobodnik J, Ipolyi IM, Oswald P, Krauss M, Schulze T, Haglund P, Letzel T, Grosse S, Thomaidis NS (2015) Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal Bioanal Chem 407:6237–6255
Singh RK, Fernando S, Baygi SF, Multari N, Thagard SM, Holsen TM (2019) Breakdown products from perfluorinated alkyl substances (PFAS) degradation in a plasma-based water treatment process. Environ Sci Technol 53:2731–2738
Tang C, Tan J (2018) Simultaneous observation of concurrent two-dimensional carbon and chlorine/bromine isotope fractionations of halogenated organic compounds on gas chromatography. Anal Chim Acta 1039:172–182
Tautenhahn R, Boettcher C, Neumann S (2008) Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform 9:504
Williams AJ, Grulke CM, Edwards J, McEachran AD, Mansouri K, Baker NC, Patlewicz G, Shah I, Wambaugh JF, Judson RS, Richard AM (2017) The CompTox chemistry dashboard: a community data resource for environmental chemistry. J Cheminform 9(1):61
Zhang X, Di Lorenzo RA., Helm PA, Reiner EJ, Howard PH, Muir DC, Sled JG, Jobst KJ (2019) Compositional space: a guide for environmental chemists on the identification of persistent and bioaccumulative organics using mass spectrometry. Environ Int 132:104808
Acknowledgements
The U.S. Environmental Protection Agency Great Lakes Fish Monitoring and Surveillance Program provided partial supported this project under contract GL-96594201-1. We also wish to thank the Program Managers, Elizabeth Murphy and Brian Lenell, and many people who assisted in sample collection and processing. Although the research described in this article has been funded wholly or in part by the United States Environmental Protection Agency, it has not been subjected to the Agency’s required peer and policy review and therefore, does not necessarily reflect the views of the Agency and no official endorsement should be inferred. The authors express their acknowledgments to European Union’s Horizon 2020 research and innovation programme HBM4EU under Grant Agreement No. 733032 for their financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interests.
Additional information
Editorial responsibility: Hari Pant.
Supplementary Information
The Supporting Information contains a brief description of search criteria used in the IPDC algorithm and HaloSeeker, comparisons of results of the IPDC algorithm with HaloSeeker on the halogenated standard mixture and the Seine River sediment samples, clofoctol retention time comparison with a neat standard, complete results of the IPDC algorithm and HaloSeeker for the halogenated standard mixed and the Seine River sediment samples, complete results of HaloSeeker and the IPDC algorithm for the PCB standard mixture and Lake Michigan trout samples. (.pdf & .zip).
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fakouri Baygi, S., Hutinet, S., Cariou, R. et al. Comparison between automated and user-interactive non-targeted screening tools: isotopic profile deconvoluted chromatogram (IPDC) algorithm and HaloSeeker 1.0. Int. J. Environ. Sci. Technol. 19, 10535–10546 (2022). https://doi.org/10.1007/s13762-021-03878-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-021-03878-y