Abstract
In recent years, algal research revolves comprehensively in tapping the prospective of various approaches viz. energy sources and nutritional supplements. Researchers worldwide are exploring various aspects of microalgae as they attract the scientific community because of their various unique properties. It has been shown that changing the parameters for algae growth can stimulate these beautiful cells to form substances which are of high value. Biosynthesis of a number of compounds is governed through several enzymatic steps which are further influenced and controlled by the type and concentration of nutrient provided or present in the natural habitat that can act as rate-limiting factor. Algal omics has turned out to be the finest option in recent years for tapping algae as biofuel resource. Genomics and transcriptomics of algae have delivered decisive information to understand lipid biosynthesis. On the other hand, proteomics and metabolomics complement algal omics by offering accurate and useful understandings into the linked physiological settings. Although genomic study reveals many important parameters for various applications using algae, which can be further enhanced when complemented well with the techniques of proteomics, transcriptomics, metabolomics and lipidomics. Combination of datasets from various lipid enhancement approaches can deliver a system-wide impression. These approaches permit closer consideration in the future with an opinion to different practical impacts that are projected in modern era.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
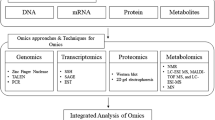
Microalgae are of huge interest while finding alternative options for lipid production and refining it to biodiesel. These can be used for the production of many other useful products like enzymes, medicine, biogas, syngas and biofuels. Microalgae have also been used to produce food supplements like PUFA (DHA, EPA), β-carotene and polysaccharides since late 1990s. Neutral lipids are the storage lipids and are stored as TAGs in thylakoid membrane. These lipid bodies can be used as a source for producing biofuels, whereas structural lipids are used as structural units of membranes such as phospholipids and glycolipids. Environmental stresses such as variation in levels of pH, light intensity, temperature, and salinity are used to improve lipid accumulation in algae (Kiran et al. 2016). Growth rate may get reduced under stress conditions resulting in lower biomass production; therefore, monitoring of optimal growth condition is an important step in enhancement of total lipid content or biodiesel production. Such issues can be monitored through overexpression of key gene involved in lipid biosynthesis using genetic engineering or molecular approach (Levitan et al. 2015; Arora et al. 2018). Lipid synthesis stimulation can be achieved through better understanding of metabolic process and genetic information among the species of concern. In this review, different methods are discussed in context to algal lipids identification and enhancement through proteomics, genomics, transcriptomics and lipidomics. Technologies used in the “omics” provide the tools required to distinguish among DNA, RNA, proteins and other molecules between species and among the individuals of various species. “Omics” provide a huge set of data at different levels of biological organization which uses various techniques to integrate data and obtain a general view of functioning of the whole system. Algae have attracted the scientific world because of many advantages like fast growth rate, minimum land requirement, CO2-capturing potential (Ghosh and Kiran 2017) and wastewater treatment. Transgenic cyanobacteria led to their use as storehouse for various commercial products such as vaccines, lipids and other pharmaceutical products (Akbari et al. 2014).
In-depth knowledge of biological functions in microalgae is mandatory to explore their full application potential. Different approaches are discussed below to understand alternative strategies related to algal lipids opted by different research groups around the globe. These strategies can assist in development of strains with superior productivity, which can pave a path for future sources of renewable energy.
Algal OMICS versus algal lipids
Genomics
Development of various techniques like next-generation sequencing methods, pyrosequencing, sequencing by ligation, real-time sequencing has become the biggest boom in the field of genomics (Metzker 2010). Guillardia theta along with various other species such as Phaeodactylum tricornutum, Thalassiosira pseudonana, Emiliania huxleyi, Aureococcus anophagefferens, Ostreococcus tauri, Micromonas pusilla strain NOUM17, O. lucimarinus and M. pusilla strain CCMP1545 has been successfully sequenced (Parker et al. 2008). Besides the above mentioned, some more species whose genomics studies have been done are shown in Table 1. To understand any metabolic process in biological system, identification or knowledge of functional gene is considered to be the first step which can be achieved through genomics. Complete genome sequence of Chlamydomonas reinhardtii has been published along with genome of organelles (Lefebvre and Silflow 1999; Maul et al. 2002; Merchant et al. 2007). Beside this, red alga, Cyanidioschyzon merolae, has also been studied extensively including its plastid and genome (Ohta et al. 2003). Chances to increase lipid accumulation inside algal cells using genetic engineering tools are very high with the advent of techniques like global transcription machinery engineering which generally incorporates the engineering of transcriptional factors involved in biosynthesis of lipids, and creation of mutant libraries of different strains valuable for desired product synthesis (Alper and Stephanopoulos 2009; Banerjee et al. 2018). Genomics being basis of all other related techniques has been used by many researchers as a tool to increase the rate of lipid biosynthesis and its extraction. For example, neutral lipid increased to 2.5-fold by the overexpression of DGAT-2 gene in Phaeodactylum tricomutum (Niu et al. 2013), whereas in case of Nannochloropsis oceanica there is an increase in neutral lipids by 69% (Li et al. 2016). Similarly, overexpression of NoD12 under the control of promoter induces stress in Nannochloropsis oceanica simultaneously accelerating the production of triacylglycerol and PUFA (polyunsaturated fatty acids) (Kaye et al. 2015).
Homblocks developed by Bi et al. (2018) for creating multiple sequence alignment (MSA) using homologous block searching method was found to be in accordance with that created by conventional methods. This method works by identifying the linearly falling blocks from the genome and conveys important phylogenetically communicative regions for creation of multiple sequence alignment. Homblocks and traditional methods work with similar efficiency except shorter time required by HomBlocks. The utility of this method has been demonstrated by comparing the phylogenetic tree drawn for 41 red algae mitochondrial genomes using both conventional and HomBlocks methods. This kind of study helps us to understand the feasibility of multiple sequence alignment among different species and their comparison using phylogenetic tree and genome sequences. These methods can further be used to synchronize the distribution of different algae species in diverse groups on the basis of their genome construction and phylogenetic similarities. In one of the recent studies, organization of plastid genome in Batrachospermales was studied in order to characterize and compare for establishment of a better infra-ordinal classification system through phylogenetic relationships (Paiano et al. 2018). Red algae top the list among various algae groups in terms of having largest plastid genomes which range from 149,987 nt lengthwise in Cyanidioschyzon merolae (Cyanidiophyceae) to 259,000 nt in Porphyridium sordidum (Porphyridiophyceae) (Lee et al. 2016).
It suggests that algae genomics can be a vital tool in deciding future directions. It can further be used to establish a relation between two unknown species or to classify them in a better way. Known sequences present in genome of any species can be used to produce more beneficial products by up- or downregulation. Genomics gives us a huge insight into the algal classifications, relationships and differences among diverse classes of algae.
Transcriptomics
This is another important molecular tool to study gene expression and the transcriptome produced during lipid biosynthesis. Transcriptomics assists with better ways to understand what is going on at the genomic level; i.e., the transcriptomic sequences are very much efficient in order to obtain the functional genome information of studied microalga strain. There are studies coupling transcriptome sequences with NGS (next-generation sequencing) technique (Parchman et al. 2010). However, problem arises when it comes to improvise transcriptional analysis with model microalgae species lacking gene sequencing data. This is one of the necessary parameters while dealing with genetic modifications of algae for efficient biofuel production (Rismani-Yazdi et al. 2012). A detailed study of transcriptome will explain active regions of specified genome encoding cell, disclosing their constituent molecular secrets and understanding their development and disease-causing genes. The main aim of transcriptomics is to reveal the gene structure, gene expression and their regulation, and function of the product formed along with their dynamic structure and constituents. Profiling techniques for gene expression such as serial analysis of gene expression (SAGE) and suppression subtractive hybridization (SSH) do not require any prior information about the genome or genes to be analyzed. On the other hand, expression of sequencing tag (EST) requires prior information for the gene in question.
Beside these, RNA-Seq (RNA sequencing) is identified as the utmost recent and developed technique which uses deep sequencing technologies for more complex and comprehensive experiments (Penn et al. 2014; Kopf et al. 2015). RNA-Seq is used for both mapping and quantification of transcriptomes. This technique has overcome different methods including hybridization, sequence-based approach like EST and is expected to revolutionize the area of research. Interaction of species can also be analyzed using RNA-sequencing technique. It provides data for both the coding and non-coding RNA (which means no prior reference is required and can be applied to any species). It gives more accurate and sensitive measurement of gene regulation or gene expression (Nagalakshmi et al. 2008). The information of genomic functionality is not always available for all algae species as their sequenced genome might not be available. Thus, this is an important area which needs further research in relation to transcriptome sequencing or expressed sequence tag for microalgae lipids to generate information regarding the genomic functionality or identification of responsible genes which will further help in the determination of functional elements involved in lipid biosynthesis (Andersen and Lubberstedt 2003). Transcriptomics tool in the area of cyanobacterial research has broadened the list of opportunities. It analyzes and compares different cyanobacteria species along with their response to varying environmental conditions and interaction among each other. The biggest obstacle for RNA-Seq is to identify or pursue complex transcriptomic systems and keep a check on changes in expression of rare RNA isoforms for the targeted genes. Studies have identified that targeting sense and antisense RNA can also modify the lipid production pattern. For example in Thalassiosira pseudonana, RNA interference and antisense targeting lipase (Thaps3-264297) increased the lipid content from 2.4- to 3.3-fold (Trentacoste et al. 2013). Transcriptomics studies have revealed many species such as C. reinhardtii (Miller et al. 2010) showing several functional lipase enzymes under nitrogen stress which function together in retrieving fatty acid from membrane in order to synthesize TAG. Li and Ismar (2018) studied five strains of Phaeodactylum tricornutum against grazing treatments through morphological analysis, nutritional analysis and RNA sequencing. RNA sequence libraries for P. tricornutum were constructed through RNA sequence library protocol. In harmful cyanobacterial species, microcystin is found to interfere with defense against oxidative stress with decline in the activity of thioredoxin and peroxiredoxin which are involved in hydrogen peroxide degradation. Total RNA showed difference in gene expression of toxic and nontoxic Microcystis strains (Schuurmans et al. 2018).
Thus, different parameters can be used to manipulate normal cell circumstances in order to see how gene expression or transcripts produced are associated with these conditions. Study of these transcripts helps to identify the probable cause of any phenomenon happening in a cell at molecular level. Proteins are the products formed by the transcribed regions of gene and have specific functions. Thus, study of proteins and transcripts should be done simultaneously in order to have clear understanding of the phenomenon involved in any of the molecular processes resulting in phenotypical fluctuations like changes in morphology of the cells or production of a specific product.
Proteomics
This is an approach or a way to identify the “proteome” which stands for the entire set of proteins available in a given cell or organism. Cell response depends upon the alterations in protein levels and expression of those proteins coding for fatty acids and triacylglycerol biosynthesis during normal and stress conditions. Protein structure, modifications, localization and interaction with other proteins and cofactors can be studied using proteomics. Additionally, expression of functional proteins and some of the epigenetic modifications also come under the main aim of proteomics. Hildebrand et al. (2013) illustrated wide diversification in metabolic pathways followed by different algal classes found through comparative genomics. These kinds of studies based on proteins involved in lipid biosynthesis give an idea of the target gene to be altered/modified for the enhancement of lipid production (Guarnieri and Pienkos 2015). Current research related to the topic deals with the analysis of quantitative changes occurring in protein due to nutrient availability/starvation, changes in environmental condition and various other factors. Recent research shows that the cyanobacterial proteomics had moved from the identification of small set of proteins to a whole genome analysis. Several novel proteins were identified using 2D electrophoresis and mass spectrometry from marine algae such as Botryococcus braunii (Nguyen and Harvey 2003), Dunaliella bardawil (Katz et al. 1995). Proteomics not only handles the identification of proteins involved but it also enlightens the hidden functions including the role in the evolution along with taxonomic studies and biochemical pathways (Kim et al. 2008).
Among different omics technologies, proteomics is the perfect choice for the analysis of various biological processes at protein level. MS (mass spectrometry) is most ubiquitous to measure endogenous proteins, whereas array-based systems (such as yeast two-hybrid assay which is used to study protein–protein interactions) or structural/imaging tools are non-MS technologies. Mass spectrometry-based techniques such as Tandem-MS and gel-based technique became a prime tool in the area of proteomics for analysis of whole protein profile and also protein dynamics quantitatively. It generates huge quantity of data which enables the researchers for analyzing their result with the existing knowledge. LMPD (Lipid MAPS Proteome Database) helps in the identification of proteins involved in lipid biosynthesis along with its metabolism as LMPD consists of lipid-associated protein sequence and annotations. Similar to the above database, there are many other bioinformatics tools which help to analyze or compare proteins of interest. In the field of molecular biology, proteomics has gained its importance to work on high-yield approaches in protein expression analysis of cells or organisms. Algae are exposed to various changes in environment such as alteration in light intensity, availability/unavailability of various macro- or micronutrients, temperature, salinity, drought, presence of heavy metals that can cause vigorous modifications in their metabolism and various pathways enabling the organism to adjust to the given conditions. Such changes can be analyzed and evaluated at protein level for better understanding of the mechanism involved in sustenance and protection of these microbes. Different tools have been used for identification and quantification of protein in proteome-based experiments.
For the past few years, 2D gel electrophoresis has been identified as the most preferred and appropriate method for the separation of proteins based on their charge and mass. The advantage of using 2D-PAGE is that thousands of polypeptides can be analyzed in a single run but sometimes analysis of acidic, basic, hydrophobic or hydrophilic proteins which are low in abundance can be difficult. For the quantitative analysis, gel can be stained with different types of dyes such as coomassie blue (Wittig et al. 2007), SYPRO Ruby (Fulda et al. 2006) or radioactive labeling followed by autoradiography (Guikema and Sherman 1982). Differential expression of proteins between cell types can also be analyzed through 2D gel electrophoresis. Much soluble protein that is present in cyanobacterial compartments like cytoplasm or periplasm has been quantified through this technique, for example, in Synechocystis sp. strain PCC 6803 (Huang et al. 2006). It is an accurate, powerful and highly used method of differential proteomics analysis.
Proteins undergo various types of modifications such as posttranslational modifications (PTM), methylation, phosphorylation and functional group attachment which further enhance the complexity of proteomics. From the last 15 years, research in the area of proteomics has been broadened due to advancement in mass spectroscopy technologies. Ionization methods like matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) with the invention of MS made the study of protein structures more simple allowing scientists to obtain mass protein “fingerprints” for matching with proteins and peptides available in the databases such as Swiss Prot, NCBI, EMBL to predict and identify unknown proteins (Cui et al. 2013). Quantization of target proteins (in terms of both relative and absolute quantities) has become possible with the use of isotopic tagging. Mass analyzer is considered to be the main component of the mass spectrometry. Different types of mass analyzers have been discovered such as time-of-flight (TOF) chamber, quadrupole (Q), ion traps (IT) including Orbitrap having different designs, shapes and characteristics with their own merits and demerits. Proteomics experiments can be further optimized by connecting the high-pressure liquid chromatography (HPLC) resulting in the additional separation capability with mass spectrometry (LC–MS or LC–MS/MS). Proteomics studies have identified several proteins which are involved in the enhancement of lipids or TAGs. Lipid droplet analysis found around 16 proteins necessary for lipid synthesis, among which there is a new protein identified as MLDP (major lipid droplet protein) playing an important role in lipid droplet synthesis (Guzmán et al. 2011). There is a correlation found between protein profile of Cylindrotheca closterium and production of fucoxanthin because fucoxanthin is found to be bounded to proteins forming a FCP complex (Wang et al. 2018).
Proteomics gives a chance to understand how proteins associated with different biomolecules can be upregulated or downregulated under varying conditions affecting the production of a specific product necessary for cell survival. Proteins are the ground-level molecules which execute the functions attributed to them by the transcripts from which they are formed. With the modification of produced transcripts, the protein may get modified or may not even get formed. Gene expression, transcripts and proteins are interdependent on each other for proper cell functioning. It is really important to understand how proteins are associated with the functioning of each cell part for further up- or downregulation according to the need of hour. Commercialization of many algae products can be given a boon with controlled regulation of these interdependent terms in order to increase the production of specified products. Genomics, transcriptomics, proteomics and metabolomics have their own importance in order to get a better understanding of the cell functionality and modifications.
Metabolomics
Metabolites ought to be produced through different metabolic pathways, and their levels of production relate biological systems with those of genetic- or environment-induced changes. Metabolomics assists in the study of alterations in metabolic pathways of microalgae leading to improved production of specified products. This approach gives an opportunity to take control of the cell’s machinery responsible for production of various compounds and subsequently derive the specific products of interest using the control obtained through transgenes or mutagenesis. Microalgae represent a diverse group of organisms which are very sensitive to slight changes in their surroundings, and these fluctuations in their environment are reflected through the variations in their cellular metabolism.
One of the natural mechanisms for the alteration of normal metabolic pathway and diverging toward enhanced lipid production is stress induction. Stress conditions can be given by deprivation of growth nutrients or by varying other environmental conditions (Ratledge 2004). Qualitative and quantitative analysis of metabolites present in the targeted species is considered to be main application of metabolomics (Potvin and Zhang 2010). Such analysis can be done through high-performance liquid chromatography/thin-layer chromatography (HPLC/TLC), nuclear magnetic resonance (NMR), and gas chromatography–mass spectrometry (GC–MS). Besides having highly sophisticated technologies, it is quite difficult to identify metabolome because of the complex nature of molecule with a wide range of concentrations. This has made a great impact on current and future metabolome research to analyze the extracted lipid which is one of the algae metabolites using HPLC (Jones et al. 2012). Metabolomic profile of secondary metabolites can be generated to observe the effect of nitrates and phosphates as done for three freshwater cyanobacterial strains, namely Oscillatoria sp. UIC 10045, Nostoc sp. UIC 10110 and Scytonema sp. UIC 10036 (Crnkovic et al. 2018). Natural products prepared from the secondary metabolites having diverse biological properties can be obtained from cyanobacteria. These secondary metabolites can be further purified and commercialized for various applications. XCMS online (an online platform to study the untargeted metabolomics data) can be used for metabolomics analysis of the data obtained from LC–MS.
With deep knowledge of the conditions affecting production of these metabolites and the molecular mechanisms involved, chances and success of commercialization can be greatly enhanced. Therefore, it is necessary to understand the physiological as well as genetic parameters in order to get the desired product in bulk without having a negative impact.
Lipidomics
Lipidomics is an emerging field in biology which enables researchers to analyze quality and quantity of lipid bodies present in cells. To achieve higher lipid productivity, there is need to know the pathways and factors affecting lipid biosynthesis, which can be unfolded nowadays using lipidomic studies. The exact knowledge and detailed composition of lipid are necessary for the production of various products such as biofuels, PUFA.
Extracting lipids from cells is a challenge in itself because method used for extraction can influence the yield and nature. After extraction, lipids are converted into fatty acid methyl esters to prevent oxidation. Folch et al. (1957) and Bligh and dyer (1959) are the most accepted and widely used methods for extraction of lipids. In recent years, various other methods have been derived from these two methods. A one-step rapid extraction method has also been optimized showing fivefold higher yield as compared to conventional methods (Axelsson and Gentili 2014). Solvents play an important role in the process of lipid extraction. Generally, a mixture of polar and nonpolar solvents is used to extract high amount of lipids from cells such as hexane, chloroform, chloroform–methanol in different ratios. The mixture of chloroform–methanol (2:1) after ultrasonication gave 19% yield of total lipids (Dos Santos et al. 2015). Supercritical carbon dioxide extraction, pressurized liquid extraction and sonication are different approaches which can be used for the efficient lipid extraction from microalgae.
Gravimetric is the most commonly used method which is able to depict the quantity of lipids extracted but not the quality of lipids such as carbon chain length, degree of unsaturation, etc. Additionally, large amount of sample is required for accurate and reliable results. The gravimetric method has been overcome by colorimetric approaches such as sulpho-phospho-vanillin (SPV) method which is able to detect lower concentrations of lipids even from wet algae. SPV reagent does not react with carbohydrate, protein or glycerol which makes it more reliable (Byreddy et al. 2016). Fluorescence techniques such as fluorescent dye (Nile red) staining of lipid droplets in algal cells have also been evolved with time which is a noninvasive technique. Nile red is fluorescence dye which binds specifically to polar and nonpolar lipid bodies. It gives yellow fluorescence when bound to neutral lipids like TAGs and red fluorescence when bound to phospholipids. This method is used to visualize lipid bodies inside algal cells. Size of lipid bodies can also be measured using confocal microscopy. Optical density of 0.8 to 1 is considered best for NR analysis, whereas high O.D. results in fluorescence quenching (Gusbeth et al. 2016). Another limitation is photooxidization of dyes due to which properties of lipids like degree of saturation, melting temperature, etc. cannot be predicted. To overcome these limitations, researchers have come up with another noninvasive technique that is in vivo analysis of cells using Raman spectroscopy. It enables us to quantify lipid bodies as well as unsaturation, chain length, Tm, etc. Single-cell laser-trapping Raman spectroscopy (LTRS), coherent anti-stroke Raman scattering (CARS), surface-enhanced Raman spectroscopy (SERS), resonance Raman spectroscopy (RRS) and confocal Raman spectroscopy (CRM) are few variants of RS used for lipid analysis (Sharma et al. 2015). Beside above-mentioned methods, various spectroscopic methods such as NIR, FTIR, MRS, NMR can also be used for lipid detection as well as quantification in vivo (Yan 2015).
Various fatty acids have different roles in cellular functions and metabolism. GC–MS is most commonly used technique to identify different fatty acids. Different fatty acids quantified from various algal species quantified using GC–MS are presented in Table 2. Diverse ionic liquid columns have been tested for separation of FAMEs and FAEEs for better peak resolution and retention time which was not achieved using normal polyethylene glycol (PEG) column. Cis and trans forms of fatty acids were also resolved using ionic columns (Weatherly 2016). FI-MS studies helped in annotating PUFA from biodiesel FAs. A vacuum UV detector has also been tried in place of FID during GC analysis to deconvolute FAs which were co-eluting for the rapid differentiation between cis and trans isoforms of FAs (Furuhashi et al. 2016; Fan et al. 2016).
In addition to this, thin-layer chromatography (TLC) has also been used which provides high resolution for different classes of lipids. In a recent study, urea and AgNO3 coated plates were used to separate branched chain fatty acids leading to better resolution than normal TLC or GC method (Yan 2015). HPLC is also used in lipidome studies, for example. In chromerid algae, more than 250 analytes were separated and identified using Orbitrap mass analyzer (Jouhet et al. 2017). A variant of HPLC, i.e., UPLC, helped in identification of lipids in Chlamydomonas sp. (Tomvcala et al. 2017; Sharma et al. 2015). Integrated chromatography–mass spectrophotometry are hyphenated techniques which are always better than independent versions for the quantitative scanning of all existing fatty acids in sample, whereas it does not identify lipid class to which fatty acids are associated. Development of highly sophisticated instruments has led to the development of new approaches for lipid research. Area of lipidomics has taken its height with the development of various ionization techniques such as electrospray ionization, desorption electrospray ionization and matrix-assisted laser desorption ionization (EI) (Lingwood et al. 2011). Mass spectrometers with different analyzers like Orbitrap, quadrupole (Q) and time of flight (TOF) can be used for the analysis of polar lipid. These efforts collectively added momentum to the field of lipidomics. Based on the functional group/structural features, lipids are identified by MS in positive or negative mode. For example, neutral lipids are always identified as positive mode which means as a protonated molecule [M + H]+, ammonium adducts [M + NH4]+ or alkali cations [Na+ or Li+], [M + Na]+ whereas SQDGs are mainly detected as negative ions [M-H] (Gonen et al. 2005).
Each MS configuration has its own advantages and disadvantages whether it is of high resolution like quadrupole-time of flight (Q-TOF), MALDI-TOF or low-resolution techniques such as triple quadrupole, ion trap, Fourier transform ion cyclotron resonance mass spectrometry (FTICR-MS) in terms of functions, scanning speed and peak resolution. Lipids can further be explored using either electrospray ionization (ESI)-QQQ-MS or ESI-QTOF-MS or shotgun-MS approach.
Collision-induced dissociation produces fragments of different lipid classes having diverse characteristics, which can be identified through neutral loss or precursor ion scanning after infusing directly. In this, the lipid of certain interest is added as internal standard to lipid extracts of biological samples (Han and Gross 2005).
Tandem mass spectroscopy is a vital part of shotgun lipidomics and generates high mass accuracy and high mass resolutions in the form of peaks. Quadrupole-time-of-flight instrument can also play a significant role for lipid profiling of any specific lipid class extracted from sample of interest (Krank et al. 2007; Mcdonald et al. 2007).
Lipids synthesis takes place in plastid, where acetyl co-A is formed from pyruvate which is a product of glycolysis. Acetyl co-A is the building block for lipid biosynthesis, and the key enzyme for this pathway is acetyl co-A carboxylase which converts acetyl co-A into malonyl co-A, which is an irreversible and a rate-limiting reaction. The ACCase enzyme requires biotin for its activity (Roessler et al. 1994). Further synthesis of fatty acid chain occurs via three main steps: condensation, reduction and dehydration. These reactions form C16:0 ACP or C18:0 ACP with the help of enzyme fatty acid synthase (FAS). C18:1 is often converted into C18:1 by the enzyme Δ-9 SAD activity (Slabas and Fawcett 1992). Another major step in lipid synthesis is termination of chain elongation. The enzymes responsible for this step are acyl-transferase and acyl-ACP thioesterase. Both the enzymes have different roles in lipid synthesis; viz., products of thioesterase are free fatty acids which are destined for TAG, a storage form of lipids in algal cells, whereas acyl-transferase enzyme forms glycolipids. Two types of thioesterase are present in plastids, FatA and FatB, which selectively convert FA-ACPs into free fatty acids. FatA is specific for unsaturated, while FatB is associated with the saturated fatty acids (Sivakumar et al. 2012).
Desaturation is another step of lipid biosynthesis which includes various desaturases. Their nomenclature is based on the position of C where they introduce the double bonds like Δ-12, Δ-6, Δ-15, etc. Three enzymes, i.e., SAD (DΔ9), FAD2 (DΔ12) and FAD3 (Δ-15), work in a sequential way to produce PUFAs. PUFA synthesis occurs in the walls of ER (Wallis and Watts 2002). Triacylglycerols are the molecules of interest for biodiesel feed. The formation of TAG in algal cells occurs either via Kennedy pathway or via PC-DAG pathway. The pathways depend on two major enzymes DGAT and PGAT, respectively (Merchant et al. 2012).
Lipid accumulation is most likely to occur in the presence of high NADPH, due to high energy requirement by enzymes like elongases and synthesases. NADPH is produced by the conversion of malate and pyruvate by the activity of malate enzyme which is a light-dependent process. In a study, activity of enzymes associated with lipid synthesis with respect to light was studied and it was found that during dark period, acetyl co-A broke down into citrate using ATP-citrate lyase which decreases the NADPH production and thus reduces the lipid accumulation in algal cells (Bellou and Aggelis 2013). Co-expression of genes involved in fatty acid synthesis and TCA cycles has also been found to be expressed together, and metabolites of TCA cycle can also be utilized in FA synthesis pathways (Mühlroth 2013). Regulation of fatty acid synthesis is an important phenomenon. Lipid accumulation is dependent on TAG biosynthesis and the stability of TAGs inside cells. In Chlamydomonas, CHT7 is found to promote TAG accumulation under nitrogen-depleted conditions and even it maintains high levels of TAGs up to 48 h of nitrogen repletion. It also downregulates the genes associated with cellular quiescence following better cell growth in depleted conditions (Tsai et al. 2014). Additionally, engineered transcriptional regulator assists in effective carbon partitioning for biosynthesis of lipids along with high biomass productivity (Chen et al. 2018).
Omics can also help to solve the problems related to lipid enhancement. The limitations can be overcome by studying the metabolism pathways. These pathways are interdependent and have positive or negative effect on each other. In a transcriptomic study, P. tricornutum showed co-expression of 106 genes related to TCA cycle and PUFA synthesis (Mühlroth 2013). This knowledge helps us in diversion of carbon partitioning more toward lipid synthesis. Proteomics can also help in studying the activity of different enzymes, proteins and transcription factors. Transcription factors play major role in regulation of gene expression of various enzymes responsible for metabolite production. N− and N+ cultures of N. gladiana revealed 20 TFs which were affecting the carbon partitioning into lipid. Mutants of TFs were developed using CRISPER-Cas9, and they showed 40–50% lipid carbon partitioning as compared to WT 20% (Ajjawi et al. 2017). Isoforms of various enzymes and their effects can be studied using omics techniques.
Conclusion
Algal biofuel is one of the renewable sources of fuel and falls under third- and fourth-generation biofuel feedstocks which has the potential to combat against the depleting fossil fuels for energy demand. In comparison with plants, alga as a feedstock for biodiesel production holds a major advantage in the renewable energy field as it requires less land and produces more biomass. Three strategies could be potentially applied for the enhancement of lipid production in microalgae such as biochemical engineering approach, genetic engineering approach and transcriptomic approach. Biochemical approach depends upon exposure to physiological stress using conventional methods such as nutrient-derived stress to channel metabolic flux and determination of factors which play major role during all these processes leading to lipid accumulation and production of other beneficial products. Genetic engineering approach and transcriptomic engineering approach are the long-term perspectives. Scientific world has recognized omics and has reconnoitered it enormously. Still lot is left there to explore through the omics approach. Such technology has been applied in many different fields. This review highlights multidisciplinary scenario for the comparative analysis of lipid molecules which seems to be an important candidate as renewable source of energy. Beside genomics, transcriptomics, metabolomics and proteomics, lipidomics has been highlighted much effectively in recent years. Lipidomics is indispensable tool for the system biology research. Exploring metabolic pathways of algae for enhanced lipid production, comprehensive screening of desired molecule and TAG profiling at molecular level are some of the future challenges for fixing the algae as a better alternative source of biofuel production. Optimization of biomass production and recovery of secondary metabolite are quite necessary for the potential application as a final marketable product. Further, for optimization and improvisation of algal systems, integration of various strategies needs to be assured. There is need to highlights that the integration of such systems would help and compliment research on microalgal applications. People working in the area of physiology, chemistry, biology and engineering have to work together to overcome and improve the algal systems. Omics can revolutionize the algal lipid field with higher lipid productivity and techniques supporting analysis and production.
References
Ajjawi I, Verruto J, Aqui M, Soriaga LB, Coppersmith J, Kwok K, Peach L, Orchard E, Kalb R, Xu W, Carlson TJ, Francis K, Konigsfeld K, Bartalis K, Schultz A, Lambert W, Schuwartz AS, Brown R, Moellering ER (2017) Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat Biotechnol 35(7):647–652
Akbari F, Eskandani M, Khosroushahi AY (2014) The potential of transgenic green microalgae; a robust photobioreactor to produce recombinant therapeutic proteins. World J Microbiol Biotechnol 30:2783–2796
Alper H, Stephanopoulos G (2009) Engineering for biofuels: exploiting innate microbial capacity or importing biosynthetic potential? Nat Rev Microbiol 7:715–723
Andersen JR, Lubberstedt T (2003) Functional markers in plants. Trends Plant Sci 8:554–560
Arora N, Pienkos PT, Pruthi V, Poluri KM, Guarnieri MT (2018) Leveraging algal omics to reveal potential targets for augmenting TAG accumulation. Biotechnol Adv 36(4):1274–1292
Axelsson M, Gentili F (2014) A single-step method for rapid extraction of total lipids from green microalgae. PLoS ONE 9(2):e89643
Bahl J, Lau MC, Smith GJ, Vijaykrishna D, Cary SC, Lacap DC, Lee CK, Papke RT, Warren-Rhodes KA, Wong FK (2011) Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat Commun 2:163
Banerjee A, Banerjee C, Negi S, Chang JS, Shukla P (2018) Improvements in algal lipid production: a systems biology and gene editing approach. Crit Rev Biotechnol 38(3):369–385
Bellou S, Aggelis G (2013) Biochemical activities in Chlorella sp. and Nannochloropsis salina during lipid and sugar synthesis in a lab-scale open pond simulating reactor. J Biotechnol 164(2):318–329
Bi G, Mao Y, Xing Q, Cao M (2018) HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics 110(1):18–22
Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J (2010) The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22:2943–2955
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Canad J Biochem Physiol 37(8):911–917
Byreddy AR, Gupta A, Barrow CJ, Puri M (2016) A quick colorimetric method for total lipid quantification in microalgae. J Microbiol Methods 125:28–32
Capell T, Christou P (2004) Progress in plant metabolic engineering. Curr Opin Biotechnol 15:148–154
Chen X, Hu G, Liu L (2018) Hacking an algal transcription factor for lipid biosynthesis. Trends Plant Sci 23(3):181–184
Crnkovic CM, May DS, Orjala J (2018) The impact of culture conditions on growth and metabolomic profiles of freshwater cyanobacteria. J Appl Phycol 30(1):375–384
Cui Y, Zheng G, Li X, Lin H, Jiang P, Qin S (2013) Cloning and characterization of a novel diacylglycerol acyltransferase from the diatom Phaeodactylum tricornutum. J Appl Phycol 25:1509–1512
Dos Santos RR, Moreira DM, Kunigami CN, Aranda DAG, Teixeira CMLL (2015) Comparison between several methods of total lipid extraction from Chlorella vulgaris biomass. Ultrason Sonochem 22:95–99
Dunahay TG, Jarvis EE, Dais SS, Roessler PG (1996) Manipulation of microalgal lipid production using genetic engineering. Appl Biochem Biotechnol 57:223–231
Ejsing CS et al (2013) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proceed Nat Acad Sci 106:2136–2141
Fan H, Smuts J, Bai L, Walsh P, Armstrong DW, Schug KA (2016) Gas chromatography–vacuum ultraviolet spectroscopy for analysis of fatty acid methyl esters. Food Chem 194:265–271
Folch J, Lees M, Sloane SGH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509
Fulda S, Mikkat S, Huang F, Huckauf J, Marin K, Norling B, Hagemann M (2006) Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6(9):2733–2745
Furuhashi T, Nakamura T, Fragner L, Roustan V, Schon V, Weckwerth W (2016) Biodiesel and poly-unsaturated fatty acids production from algae and crop plants—a rapid and comprehensive workflow for lipid analysis. Biotechnol J 11:1262–1267
Ghosh A, Kiran B (2017) Carbon concentration in algae: reducing CO2 from exhaust gas. Trends Biotechnol 35(9):806–808
Gonen T, Cheng Y, Sliz P, Hiroaki Y, Fujiyoshi Y, Harrison SC, Walz T (2005) Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438:633–638
Guarnieri MT, Pienkos PT (2015) Algal omics: unlocking bioproduct diversity in algae cell factories. Photosynth Res 123:255–263
Guikema J, Sherman L (1982) Protein composition and architecture of the photosynthetic membranes from the cyanobacterium, Anacystis nidulans R2. Biochimica et Biophysica Acta (BBA)-Bioenergetics 681(3):440–450
Gusbeth CA, Eing C, Göttel M, Sträßner R, Frey W (2016) Fluorescence diagnostic for lipid status monitoring of microalgae during cultivation. Int J Renew Energy Biofuels 2016:1–12
Guzmán HM, de la Jara Valido A, Duarte LC, Presmanes KF (2011) Analysis of interspecific variation in relative fatty acid composition: use of flow cytometry to estimate unsaturation index and relative polyunsaturated fatty acid content in microalgae. J Appl Phycol 23:7–15
Han X, Gross RW (2005) Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev 24:367–412
Hildebrand M et al (2013) Metabolic and cellular organization in evolutionarily diverse microalgae as related to biofuels production. Curr Opinion Chem Biol 17:506–514
Huang F, Fulda S, Hagemann M, Norling B (2006) Proteomic screening of salt-stress-induced changes in plasma membranes of Synechocystis sp. strain PCC 6803. Proteomics 6(3):910–920
Jones J, Manning S, Montoya M, Keller K, Poenie M (2012) Extraction of algal lipids and their analysis by HPLC and mass spectrometry. J Am Oil Chem Soc 89(8):1371–1381
Jouhet J, Lupette J, Clerc O, Magneschi L, Bedhomme M, Collin S, Roy S, Maréchal E, Rébeillé F (2017) LC-MS/MS versus TLC plus GC methods: consistency of glycerolipid and fatty acid profiles in microalgae and higher plant cells and effect of a nitrogen starvation. PLoS ONE 12:e0182423
Kalayasiri P, Jeyashoke N, Krisnangkura K (1996) Survey of seed oils for use as diesel fuels. J Am Oil Chem Soc 73:471–474
Katz A, Jimenez C, Pick U (1995) Isolation and characterization of a protein associated with carotene globules in the alga Dunaliella bardawil. Plant Physiol 108:1657–1664
Kaye Y et al (2015) Metabolic engineering toward enhanced LC-PUFA biosynthesis in Nannochloropsis oceanica: overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG. Algal Res 11:387–398
Khozin-Goldberg I, Cohen Z (2006) The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochem 67:696–701
Kim GH, Shim JB, Klochkova TA, West JA, Zuccarello GC (2008) The utility of proteomics in algal taxonomy: Bostrychia radicans/B. moritziana (Rhodomelaceae, Rhodophyta) as a model study. J Phycol 44:1519–1528
Kiran B, Pathak K, Kumar R, Deshmukh D, Rani N (2016) Influence of varying nitrogen levels on lipid accumulation in Chlorella sp. Int J Env Sci Technol 13:1823–1832
Klopfenstein W (1982) Estimation of cetane index for esters of fatty acids. J Am Oil Chem Soc 59:531–533
Kopf M, Klahn S, Scholz I, Hess WR, Voß B (2015) Variations in the non-coding transcriptome as a driver of inter-strain divergence and physiological adaptation in bacteria. Sci Rep 5:9560
Krank J, Murphy RC, Barkley RM, Duchoslav E, McAnoy A (2007) Qualitative analysis and quantitative assessment of changes in neutral glycerol lipid molecular species within cells. Methods Enzym 432:1–20
Krisnangkura K (1986) A simple method for estimation of cetane index of vegetable oil methyl esters. J Am Oil Chem Soc 63:552–553
Lee J, Cho CH, Park SI, Choi JW, Song HS, West JA, Yoon HS (2016) Parallel evolution of highly conserved plastid genome architecture in red seaweeds and seed plants. BMC Biol 14(1):75
Lefebvre PA, Silflow CD (1999) Chlamydomonas: the cell and its genomes. Genet Soc Am 151:9–14
Levitan O, Dinamarca J, Zelzion E, Lun DS, Guerra LT, Kim MK, Kim J, Van Mooy BA, Bhattacharya D, Falkowski PG (2015) Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proc Natl Acad Sci 112:412–417
Li S, Ismar SM (2018) Transcriptome, biochemical and growth responses of the marine phytoplankter Phaeodactylum tricornutum Bohlin (Bacillariophyta) to copepod grazer presence. Cell Physiol Biochem 46(3):1091–1111
Li DW et al (2016) A type 2 diacylglycerol acyltransferase accelerates the triacylglycerol biosynthesis in heterokont oleaginous microalga Nannochloropsis oceanica. J Biotechnol 229:65–71
Lingwood D, Binnington B, Róg T, Vattulainen I, Grzybek M, Coskun Ü, Lingwood CA, Simons K (2011) Cholesterol modulates glycolipid conformation and receptor activity. Nat Chem Biol 7:260–262
Maul JE, Lilly JW, Cui L, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome islands of genes in a sea of repeats. Plant Cell 14:2659–2679
McDonald JG, Thompson BM, McCrum EC, Russell DW (2007) Extraction and analysis of sterols in biological matrices by high performance liquid chromatography electrospray ionization mass spectrometry. Methods Enzy 432:145–170
Merchant SS et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–250
Merchant SS et al (2012) TAG, You’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotechnol 23(3):352–363
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11:31
Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zauner S, Cornish A, Liu B, Bullard B (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen-deprivation predict diversion of metabolism. Plant Physiol. https://doi.org/10.1104/pp.110.165159
Mühlroth A (2013) Pathways of lipid metabolism in marine algae, co-expression network, bottlenecks and candidate genes for enhanced production of EPA and DHA in species of Chromista. Mar Drugs 11(11):4662–4697
Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320:1344–1349
Nguyen RT, Harvey HR (2003) Preservation via macromolecular associations during Botryococcus braunii decay: proteins in the Pula Kerogen. Organ Geochem 34(10):1391–1403
Niu YF, Zhang MH, Li DW, Yang WD, Liu JS, Bai WB, Li HY (2013) Improvement of neutral lipid and polyunsaturated fatty acid biosynthesis by overexpressing a type 2 diacylglycerol acyltransferase in marine diatom Phaeodactylum tricornutum. Mar Drugs 11(11):4558–4569
Ohta N et al (2003) Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res 10:67–77
Paiano MO, Del Cortona A, Costa JF, Liu SL, Verbruggen H, De Clerck O, Necchi O Jr (2018) Organization of plastid genomes in the freshwater red algal order Batrachospermales (Rhodophyta). J Phycol 54(1):25–33
Palenik B et al (2007) The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc Natl Acad Sci 104:7705–7710
Parchman TL, Geist KS, Grahnen JA, Benkman CW, Buerkle CA (2010) Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genom 11:180
Parker MS, Mock T, Armbrust EV (2008) Genomic insights into marine microalgae. Annu Rev Genet 42:619–645
Penn K, Wang J, Fernando SC, Thompson JR (2014) Secondary metabolite gene expression and interplay of bacterial functions in a tropical freshwater cyanobacterial bloom. ISME J 8:1866–1878
Potvin G, Zhang Z (2010) Strategies for high-level recombinant protein expression in transgenic microalgae: a review. Biotechnol Adv 28:910–918
Ramos MJ, Fernández CM, Casas A, Rodríguez L, Pérez Á (2009) Influence of fatty acid composition of raw materials on biodiesel properties. Biores Technol 100:261–268
Ratledge C (2004) Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochemie 86:807–815
Rismani-Yazdi H, Haznedaroglu BZ, Hsin C, Peccia J (2012) Transcriptomic analysis of the oleaginous microalga Neochloris oleoabundans reveals metabolic insights into triacylglyceride accumulation. Biotechnol Biofuels 5:74
Roessler PG, Bleibaum JL, Thompson GA, Ohlrogge JB (1994) Characteristics of the gene that encodes acetyl-CoA carboxylase in the Diatom Cyclotella crypticaa. Ann NY Acad Sci 721:250–256
Schuurmans JM, Brinkmann BW, Makower AK, Dittmann E, Huisman J, Matthijs HC (2018) Microcystin interferes with defense against high oxidative stress in harmful cyanobacteria. Harmful Algae 78:47–55
Sharma SK et al (2015b) An integrative Raman microscopy-based workflow for rapid in situ analysis of microalgal lipid bodies. Biotechnol Biofuels 8:165
Sivakumar G et al (2012) Integrated green algal technology for bioremediation and biofuel. Biores Technol 107:1–9
Slabas AR, Fawcett T (1992) The biochemistry and molecular biology of plant lipid biosynthesis. Plant Mol Biol 19(1):169–191
Tomvcala A, Kyselová V, Schneedorferová I, Opekarová I, Moos M, Urajová P, Kručinská J, Oborník M (2017) Separation and identification of lipids in the photosynthetic cousins of Apicomplexa Chromera velia and Vitrella brassicaformis. J Sep Sci 40(17):3402–3413
Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH (2013) Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci 110(49):19748–19753
Tsai CH et al (2014) The protein compromised hydrolysis of triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc Natl Acad Sci 111(44):15833–15838
Tsugawa H (2015) MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods 12:523
Wallis JG, Watts JL (2002) Polyunsaturated fatty acid synthesis: what will they think of next? Trends Biochem Sci 27(9):467–473
Wang S, Verma SK, Said IH, Thomsen L, Ullrich MS, Kuhnert N (2018) Changes in the fucoxanthin production and protein profiles in Cylindrotheca closterium in response to blue light-emitting diode light. Microb Cell Fact 17(1):110
Weatherly CA (2016) Analysis of long-chain unsaturated fatty acids by ionic liquid gas chromatography. J Agric Food Chem 64:1422–1432
Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6(7):1215–1225
Worden AZ et al (2009) Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324:268–272
Yan Y (2015) Combined urea-thin layer chromatography and silver nitrate-thin layer chromatography for micro separation and determination of hard-to-detect branched chain fatty acids in natural lipids. J Chromatogr 1425:293–301
Acknowledgements
The authors acknowledge funding support provided by DBT, Govt. of India (Project No. BT/PR13276/PBD/26/466/2015). Authors also acknowledge MHRD and UGC for providing fellowship support to Mr. Vishal Anand, Ms. Kanchan Samadhiya and Mr. Mrinal Kashyap. The funding organizations have not played any role in study design, decision to publish or preparation of the manuscript. Authors are thankful to Prof. Pradeep Mathur, Director, IIT Indore, for encouraging research and providing necessary facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest involved with this.
Additional information
Editorial responsibility: Shahid Hussain.
Rights and permissions
About this article
Cite this article
Anand, V., Kashyap, M., Samadhiya, K. et al. Strategies to unlock lipid production improvement in algae. Int. J. Environ. Sci. Technol. 16, 1829–1838 (2019). https://doi.org/10.1007/s13762-018-2098-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-018-2098-8