Abstract
In this study, we are reporting bacterial diversity in crude oil-contaminated soil of Assam, India. Integration of physiological community profiling, culture-dependent and culture-independent (metagenome) approaches, was employed to obtain a complete picture of the total bacterial diversity. Samples collected from 10 sites contaminated with crude oil ranging from 0.22 to 89.36% were analysed, and altogether 160 culturable bacteria were isolated (117 Gram-positive and 43 Gram-negative bacteria). Molecular identification showed the predominance of genera Lysinibacillus, Alcaligenes, Bacillus, Clostridium, Enterobacter and Pseudomonas. Conversely, denaturing gradient gel electrophoresis (DGGE) profiles of 16S rDNA phylotypes showed the predominance of Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas. Both culture-dependent and culture-independent approaches resulted in 11 genera of which Bacillus and Pseudomonas were the key inhabitants creating most favourable bacterial milieu in the presence of crude oil contaminants. Metabolic fingerprints data depicted in PCA plot demonstrated that sites CTF-D-1 and Core-10 were most diverged. It was further confirmed that variations of bacterial species dominance in different sites were due to origin of hydrocarbon contamination. We here claim that the present findings is a first-hand report on combined physiological community profiling, culture-based and culture-independent approaches in assessing total bacterial diversity in crude oil-contaminated soil of Assam.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The inherent problem associated with hydrocarbon contamination caused by anthropogenic activities of crude oil drilling, exploration, refining, transport and storage has led to making these compounds recalcitrant environmental pollutants (Allen et al. 2007; Paisse et al. 2008; Mnif et al. 2009; Lu et al. 2012). Studies have reported that hydrocarbon-contaminated environments are inhabited by microorganisms which selectively grow and utilize petroleum hydrocarbons as carbon source for growth and development (Liu and Liu 2013; Beazley et al. 2012; Bargiela et al. 2015). Earlier studies also established that the presence of hydrocarbons determines the dominance of specific microbes (Vila et al. 2010; Hazen et al. 2010). Various reports on variations of bacterial diversity in the presence of hydrocarbons in an environment have shown that the presence of hydrocarbon contaminants often leads to selective enrichment of hydrocarbon-utilizing bacteria (Abed et al. 2002; Saul et al. 2005; Hamamura et al. 2006). Thus, determination of community structure of hydrocarbon-contaminated sites is required for getting first-hand information on microbial diversity (Albokari et al. 2015).
Conventionally, characterization of microbial community in contaminated soil was limited to culturing microorganisms from environmental samples termed as culture-dependent approach. Hydrocarbon-contaminated soils appear to be more favourable to conventional culture-dependent sampling than other soils; the reason being hydrocarbon contamination often leads to a shrink in microbial diversity (Bell et al. 2013; Saul et al. 2005; Kim et al. 2015; Abbasian et al. 2016). Consequently, a minimal sampling effort is required to isolate a representative fraction of the active community inhabiting those sites. In culture-dependent methods, only a small part approximately >3% of the microorganisms that inhabit soil contaminated with hydrocarbons could be cultured in the laboratory, and the rest >97% remain unreported (Macnaughton et al. 1999; Keller and Zengler 2004; Achtman and Wagner 2008). Hydrocarbon contaminants have been reported to restrain growth of certain responsive microbial groups (Santos dos et al. 2011) and selectively trigger subgroups like that of the Actinobacteria and Proteobacteria (Bell et al. 2013, 2014; Jung et al. 2016). Selectivity of growth media, lack of knowledge of the actual conditions under which most bacteria grow in soil, problems with soil sampling, recognition of small cell size bacteria and bacterial viability are critical limitations when trying to gain an understanding of bacterial diversity and community structure in soil. The understanding of microbial diversity has been extended by culture-independent studies, which involve comparative analysis of gene sequences directly from DNA extracted from the environmental samples. It seems likely that the uncultured majority provides a vast, unexploited reservoir of useful enzymes and biological probes. Therefore, integration of culture-dependent and culture-independent (metagenome) approaches is essential to get a complete picture of total bacterial diversity (Vaz-Moreira et al. 2011; Beazley et al. 2012; Mason et al. 2012; Liu and Liu 2013; Kimes et al. 2013; Stefani et al. 2015). Approaches for predicting total bacterial diversity include PCR amplification of 16S rRNA genes from cultured isolates (Aislabie et al. 2004; Saul et al. 2005; Roy et al. 2014); 16S rRNA V3 region amplified directly from soil DNA, followed by denaturing gradient gel electrophoresis (DGGE) profiling and sequencing specific amplicon (Muyzer et al. 1998; MacNaughton et al. 1999; Vinas et al. 2005; Greer et al. 2010), phylogenetic analysis and community-level physiological profiling through carbon utilization pattern (Mishra and Nautiyal 2009; Nautiyal 2009). Integration of both culture-dependent and culture-independent methods is crucial in getting a complete representation of total bacterial diversity.
Crude oil-contaminated areas in Assam are stretched to a large extent, have persisted for long duration and are mostly untapped (Gogoi et al. 2003; Yenn et al. 2014; Roy et al. 2014). Thus, these sites have a high likelihood of harbouring large diversity of unidentified bacteria that may have the ability to grow under hydrocarbon stressed and their utilization as bio-formulations may restore the environment. Recently, Li et al. (2015) and Howard and Howard (2016) had shared a vision to collaborate all environmental researchers towards a common goal, the “New Silk Road” for both beneficially and sustainably minimizing environmental degradation. In the present study, we attempt to determine the soil physicochemical profiles, soil enzymes and bacterial diversity of 10 hydrocarbon-contaminated sites of Duliajan and Jorajan districts in Assam state. To the best of our knowledge, this is the first report where we demonstrate the bacterial diversity associated with hydrocarbon-contaminated soil sites of Duliajan and Jorajan districts in Assam for its further exploitation as a source for bioremediation purposes.
Materials and methods
Site description, soil physical and chemical analysis
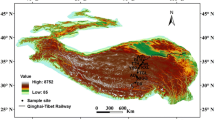
Crude oil-contaminated samples were collected from different locations operated under Oil India limited, Duliajan of Tinsukia District. Sampling was done randomly at ten oil drilling-contaminated sites of Duliajan (27°21′42″ N, 95°19′ 6″ E) and Jorajan (95°11′–95°30′ E; 27° 06′–27° 22′ N) of Dibrugarh and Tinsukia, Assam (Fig. 1). Soil, sludge and mud samples from different locations of crude oil-contaminated sites were collected at random to a depth of 0–10 cm. To represent uniformity, bulk samples were prepared by mixing minimum five to ten samples collected from each location and mixed well. From these homogenous bulk samples, minimum three representative samples were collected and immediately stored at 4 °C and taken to the laboratory for analysis. Samples were analysed for their total hydrocarbon, pH, moisture content, soil organic carbon (SOC), soil enzyme activities and total bacterial count. For total hydrocarbon content, 5 g of contaminated sample was taken in a glass beaker and extracted with diethyl ether. The solvent was then filtered out into another fresh glass beaker. The process was repeated minimum three times or till the soil was found to be free of the oil content. The solvent obtained from all the extraction steps was pooled together. This filtrate was then allowed to dry under room temperature and weighed. The amount of dried filtrate is equal to the total hydrocarbon content in the contaminated sample and expressed as percentage (%). The process was repeated minimum three times. The filtrate obtained was dried and weighed. pH of the contaminated sample was estimated in sample/water suspension (1:2.5) using pH meter (Eutech, Malaysia); moisture content was determined gravimetrically by drying the samples at 70 °C until constant weight. Total SOC was determined according to Walkley and Black (1934) Total organic carbon of the samples was determined according to Walkley and Black (1934). Briefly, to 1 g sample 10 ml of 1 N potassium dichromate and 20 ml of concentrated H2SO4 were added, agitated for about a minute and allowed to stand for 30 min. Following this, 200 ml distilled water, 10 ml orthophosphoric acid and 1 ml diphenylamine indicator were added. The solution was than titrated against standard ferrous ammonium sulphate (FAS) till bluish green colour. Organic carbon is calculated in percentage. Dehydrogenase activity was estimated by using 2,3,5-triphenyl tetrazolium chloride (TTC) red technique (Casid et al. 1964). To 1 g fresh sample, 0.1 gm of CaCO3 and 1 ml of TTC solution were added in 3 replicates each and incubated at 30 °C for 24 h. The resulting slurry was passed through Whatman No. 1 filter paper and extracted with successive aliquots of methanol, adjusted to 50 ml and read at 485 nm using Specord 210 UV–Vis spectrophotometer (Analytikjena). Dehydrogenase activity of the samples per gm was expressed in terms of µg TPF per g dry sample per 24 h. Urease activity of the samples was measured following the method of McGarity and Mayers, (1967) for which to 1 g of sample, 1 ml of toluene was poured and allowed to stand for 15 min. Subsequently, 10 ml buffer (pH 7) and 5 ml of 10% urea solution were added and incubated for 3 h at 37 °C, adjusted to 100 ml with distilled water, mixed thoroughly and filtered through Whatman No. 5 filter paper. From this filtrate, 0.5 ml was taken and made to 30 ml with distilled water and treated with 2 ml phenolate solution and 1.5 ml sodium hypochlorite solution. Absorbance was read at 630 nm. The amount of NH4 +-N released was calculated by a reference calibrated curve and expressed as NH+–N mg per gm dry sample per 3 h. Phosphatase activity was determined as per by Tabatabami and Bremner (1969). To 1 g sample, 4 ml of universal buffer, 0.25 ml of toluene and 1 ml of 0.115 ml p-nitro phenyl phosphate solution were added and incubated at 37 °C for 1 h. One milligram of 0.5 M CaCl2 and 0.5 M NaOH solution were introduced to the mixture. The suspension was filtered through Whatman No. 1 filter paper, and optical density was measured at 430 nm. The phosphatase activity was calculated in terms of concentration of PNP in each sample from a standard curve of PNP in water and expressed as mole of PNP released per g dry sample per hour.
Crude oil fractionation, i.e. asphaltene, resin, wax, aliphatic/aromatic fraction, was separated according to Yenn et al. (2014). To 5 g of crude oil, 150 ml of n-heptane was added and heated under reflux conditions at 100 °C for 1 h in a round-bottom flask. The flask was with stopper and allowed to cool under dark condition for 2 h. The content was then filtered through Whatman No. 42 filter paper without agitation. The residue obtained was asphaltene and was dried and weighed for further experiments. The filtrate then was absorbed in 120 g of silica gel and heated in a water bath for 1 h with continuous stirring. The resulting material was then allowed to stand undisturbed overnight and then filtered. The residue obtained after filtration was collected and washed with distilled water, dissolved in a toluene–methanol (90:10) mixture and dried in a boiling water bath, and the dried fraction was weighed to estimate the percentage of resin. The filtrate was heated in an oven briefly, treated with 5 ml of H2SO4 and then allowed to cool down to room temperature. The wax solution was decanted and washed with warm water and ammonium hydroxide solution several times till the acid in it got removed. Then, the crude wax thus obtained was dissolved in ethylene chloride (CHCl2), cooled to around 32 °C and filtered through a cold filter funnel, and the wax was collected. The filter funnel was washed with hot n-heptane solvent and collected in a weighed flask. The collected material was evaporated and dried to obtain the residual aliphatic and aromatic components.
Enumeration and isolation of bacterial strain
Enumeration of bacterial population was done as described by Roy et al. (2014) and expressed as log10. Briefly, serial dilution technique followed by plating on nutrient agar (NA), Reasoner´s 2A agar (R2A), Pikovskaya, N2-Free media and mineral media M1 with crude oil as carbon source was done for isolating colonies. The incubation temperature was maintained at 30 °C for 24 h. Morphologically distinct colonies were picked and purified in fresh NA plates. The purity was further confirmed microscopically. Morphological and biochemical properties of each isolate were documented as described in Cappuccino and Sherman laboratory manual (1998) and Bergey’s Manual of determinative bacteriology (Weeks and Breed 1957).
Identification of bacterial strains by 16S rDNA sequencing analysis
Bacterial genomic DNA was extracted from the isolates obtained using GenElute bacterial genomic DNA kit (Sigma Aldrich). PCR amplification of the genomic DNA was carried out using the universal 16S rDNA primers fD1 AGAGTTTGATCCTGGCTCAG and rP2 ACGGCTACCTTGTTACGACTT (Weisburg et al. 1991) with reaction conditions set at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 45 s and 72 °C for 1.30 min and then final extension at 72 °C for 10 min. 16S rDNA amplicon from each strain was sequenced, and the data were searched using NCBI-BLAST (http://www.ncbi.nlm.nih.gov/blast) search tool for identification of the strain type.
Soil DNA extraction and 16S rRNA gene PCR amplification
DNA was extracted from approximately 0.5 g of soil using UltraClean™ soil DNA isolation kit (Mo Bio, CA, USA) following the manufacturer’s instructions. Further, the V3 region of 16S rRNA gene from these metagenomic DNA samples was obtained by the partial amplification of the respective gene using specific primers 341f and 518r (Muyzer et al. 1998). Briefly, PCR mixture was carried out in 50 μl volume consisting of 1.0 U Taq polymerase, 1X PCR buffer, 1.5 mM MgCl2, 200 mM of each dNTPs, 10 μM of each primer (forward and reverse) and 20–30 ng of soil DNA. PCR-grade water was used as a negative control in PCR. The resultant amplicons were electrophoresed to check their integrity against 100-bp DNA ladder (Thermo Scientific, USA) using image acquisition software (Universal Hood III, Bio-Rad, USA). Thermal cycling was done in an GENEI-TC3000 thermocycler (GENEI, India) with the conditions given hereunder: 3 min initial denaturation at 94 °C; 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at 58 °C and 60 s elongation at 72 °C; and a 10-min final elongation. PCR amplicons were further subjected for DGGE-based fingerprinting analysis and then sequencing.
DGGE Profiling and 16S rRNA gene sequencing
Gels comprising of 40% (w/v) polyacrylamide (37.5:1 acrylamide/bisacrylamide) were cast using a Universal Mutation Detection System (Dcode™, Bio-Rad, USA) for denaturing gradient gel electrophoresis (DGGE) analysis. The gels had a linear gradient from 40 to 60% denaturant, where 100% denaturing acrylamide contained 7 M urea and 40% (v/v) formamide (Muyzer et al. 1998). Electrophoresis was conducted for 16 h at 60 V at a constant temperature of 60 °C in 7 L of 1 × TAE. The gels were stained in ddH2O containing 0.5 mg l−1 ethidium bromide for 10 min, and the gel images were digitally captured using image acquisition software (UVITEC, GENEI, India). Any bands migrating to the same position in the gel were assumed to be identical amplicons. The DGGE bands were excised from the gel using a sterile scalpel and incubated for 24 h at 4 °C. Further, PCR was done with the same primers devoid of GC-clamp, and the resultant amplicons were outsourced for sequencing (3130xl Applied Biosystems, ABI, USA).
Phylogenetic analysis
To determine evolutionary relationship among the bacterial isolates as well as the uncultured 16S rRNA V3 sequences, the gene sequence were compared against those obtained from GenBank database using NCBI-Blast as mentioned above and Seqmatch tool (Cole et al. 2007). Multiple sequence alignment was carried out using ClustalW (http://www.ebi.ac.uk/clustalw/) sequence alignment tool. Phylogenetic tree was then constructed using the neighbour-joining method of MEGA6 program (Tamura et al. 2013). Branching orders of the trees were ascertained and compared using distance-based neighbour-joining algorithms with Kimura two-parameter corrections to improve the reliability of internal branches (Kimura 1980).
Carbon source utilization pattern-based community-level physiological profiling
Metabolic fingerprint of microbial community were obtained from Biolog Ecoplates (Biolog, Inc, Hayward, CA, USA). Utilization of 31 different carbon sources in triplicate was analysed by inoculating the plates with aqueous preparation of samples. Data were recorded at every 24-h interval till day 7 at 590 nm. The results obtained on 7th day were used for comparison. Microbial activity in each microplate was expressed as average well colour development (AWCD) (Garland 1996). Shannon diversity and evenness indices were calculated following the method as described by Staddon et al. (1997) and Nautiyal (2009).
Statistical analyses
All the data were mean of three independent experiments each. All the data were subjected to analysis of variance (ANOVA) after normalization, and significant differences among the study sites were compared at p < 0.01 according to Tukey’s test. Data related to CFU were converted into logarithmic values, before statistical analysis. Further, to establish the relationship on physiological community profiling of each site based on carbon source utilization pattern, principal component analysis (PCA) was performed. For this, dimension reduction in the Biolog data of 10 soil samples was normalized and transformed as described by Weber et al. (2007). Statistical analysis was performed using IBM SPSS Statistics 20 software (version 16.0).
Results and discussion
Physicochemical properties
A schematic map of the study site is depicted in Fig. 1. Site description and characteristics of samples are summarized in Table 1. The total hydrocarbon content of the contaminated soil was obtained in a range of 0.40–89.36% as revealed by gravimetric assessment. Of the total samples, 90% were of basic pH (7.8–9.1). The sample collected from Core-10 site was found to have significantly different properties as compared to the other study sites. Organic carbon content was found to be high in all the soil samples, ranging from 1.38 (in CTF-D-1) to 6.9 (in CTF-D2-2 and Core-10). Exceptionally low moisture content was found in the core sample compared to the others. Overall, a significantly different moisture content for Core-10 (1.25%), peat water samples PWS-J-4 (52.5%), PWS-DGU-7 (38.8%) and drilling fluid site DF-MJ-3 (30.6%) was observed. The variations in physical and chemical properties of hydrocarbon-contaminated soils were mainly due to the source of hydrocarbon contamination as well as the extent of crude oil contamination.
Soil enzyme activity
Considerable differences in soil enzyme activity were found among the samples (Fig. 1). Highest amount of dehydrogenase (68.6 ± 0.01 µg/g of fresh soil/h), alkaline phosphatase (8.2 ± 0.01 µg/g of fresh soil/hr) and neutral phosphatase (2.9 ± 0.03 µg/g of fresh soil/h) activity was found for the samples collected from CC1-J-5 site. Nevertheless, urease enzyme activity was found to be higher in site NCC-J-8 (0.002 ± 0.004 µg/g of fresh soil/h). From the observation, it was noticed that compared to dehydrogenase and phosphatase activities, very low urease activities were observed in the hydrocarbon-contaminated soil. In addition, Pearson correlation analysis revealed a significant positive correlation between the bacterial colony counts on crude oil supplemented media (r = 0.6, 0.7) and the total hydrocarbon and organic carbon content (as shown in supplementary table S1). The positive correlation may be due to the selective preference of bacterial growth.
Cultivable bacterial diversity
Using culture-dependent approach, a total of 160 morphologically distinct culturable strains were isolated from the 10 different crude oil-contaminated sites using different culture media (Table 1). Based on Gram staining, biochemical and morphological observation 117 strains were found to be Gram-positive bacteria and the rest 43 strains were identified as Gram-negative bacteria. Highest number of bacterial colonies (n = 52) were identified from the CTF-D-1 site, whereas only 5 isolates could be collected from Core-10 site. The order of bacterial population was highest in site CTF-D-1 followed by DF-MJ-3 > PWS-J-4 > CTF-D2-2, CC2-J-6 > CC1-J-5 > PWS-DGU-7, NCC-J-8 > GDS-J-9 > Core-10, respectively. The variation in the number of isolates was mainly due to the differences in the TPH levels in these sites.
Based on the sequence similarity of 136 strains that were obtained from 160 isolates, we found that the majority of the strains belonged to the phylum Proteobacteria (Betaproteobacteria-2, Gammaproteobacteria-17) and Fermicutes (Bacilli-115, Clostridia-2), under the genus Enterobacter, Pseudomonas, Alkaligenes, Bacillus, Lysinibacillus and Clostridium. List of bacterial strains identified in the present study along with their closest relatives from NCBI database is shown in table S2. From the observation, it was found that, in site CTF-D-1 the presence of genera Enterobacter, Pseudomonas, Bacillus, Lysinibacillus and Clostridium, in site CTF-D2-2 Enterobacter, Bacillus and Alkaligenes, in site DF-MJ-3 Clostridium, Bacillus, Lysinibacillus and Paenibacillus, in sites PWS-J-4, CC2-J-6 and PWS-DGU-7 Pseudomonas, Bacillus and Lysinibacillus, in site CC1-J-5 Pseudomonas, Bacillus, Exiguobacterium and Lysinibacillus, in site NCC-J-8 Bacillus and Lysinibacillus, in site GDS-J-9 Bacillus, Lysinibacillus and Alkaligenes and in site Core-10 Pseudomonas and Bacillus were identified (Table 2). The dominance of genus Enterobacter, Pseudomonas, Bacillus, Lysinibacillus, Alkaligenes and Clostridium could be due to their adaptability to the presence of crude oil and their ability to use petroleum hydrocarbons as carbon source for growth processes.
Culture-independent bacterial community profiling
In order to obtain the uncultured bacterial diversity associated with crude oil-contaminated sites, soil metagenome isolation followed by DGGE of PCR-amplified 16S rDNA fragments was carried out (Fig. 2). All the 10 sites showed prominent DGGE banding profiles, having a total of 33 bands, of which 9 were unique, while the others were present in two or more sites. Highest number of bands (n = 11) were observed in PWS-J-4 site, showing maximum species richness. Dendrogram obtained from clustering of all the sites based on DGGE banding patterns of the 10 sites is shown in Fig. 3. From this analysis, 6 distinct clades were identified with sites CC1-J-5 and CC2-J-6 as two separate branches and CTF-D-1 and CTF-D2-2; PWS-DGU-7 and GDS-J-9; NCC-J-8 and Core-10; and DF-MJ-3 and PWS-J-4 comprising the four clades. From the DGGE profile, 17 distinct bands, referred as operational taxonomic unit (OTU) were successfully sequenced (Fig. 4, Table S3). From this analysis, it was confirmed that the bacterial community mainly originated from the genera Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas. Further, DGGE fingerprints of each site were analysed to identify the species richness and diversity of the crude oil-contaminated sites (Fig. S2). Among the sites, PWS-J-4 was found to have highest bacterial richness (~11) and bacterial diversity (~2.3), whereas CC1-J-5 site was found to have least bacterial diversity as well as bacterial richness. Form the observation, we found that the lowest degree of crude oil contamination showed the maximum species richness and hence the highest diversity.
Clustering based on DGGE banding pattern of crude oil-contaminated sites under study. a DGGE gel showing banding profiles of 16S rRNA-V3 gene region isolated from the each soil DNA; b dendrogram obtained from clustering of DGGE bands of the 10 soil samples. A total of six distinct clades with sites CC1-J-5 and CC2-J-6 as two separate branches; CTF-D-1 and CTF-D2-2; PWS-DGU-7 and GDS-J-9; NCC-J-8 and Core-10; and DF-MJ-3 and PWS-J-4 comprising the four clades were identified
Phylogenetic tree showing the relationships among the 16S rRNA-V3 gene sequences of DGGE bands obtained from the 10 crude oil-contaminated soils. Single OTUs identified from the sequenced data of amplified 16S rRNA-V3 gene region revealed the occurrence of genera Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas in the contaminated sites
Carbon source utilization pattern-based community-level physiological profiling
Evaluation of community-level physiological profiling based on carbon utilization pattern, a significantly different Shannon diversity (p < 0.05), Simpson diversity index and McIntosh evenness was observed in CTF-D-1 and Core-10 than in the other study site (Table 3 ). Principal component analysis (PCA) also showed no distinct association (Fig. 5). It was found that Core-10 (V10) was most diverse, indicating the presence of significantly different microbial community. Similar to culture-dependent and culture-independent approach, in physiological community profile we observed that source and crude oil contamination affected the overall microbial community-level physiological activities.
Principal component analysis (PCA) based on metabolic fingerprint obtained from 10 crude oil-contaminated sites. From the PCA plot, it was seen that site Core-10 was most diverse at 45.35 and 13.9% on first 2 principle components, indicating that this had significantly different bacterial phyla. Site CTF-D-1 also had a diverse pattern as depicted by their physiological profile. Again sites CC2-J-6 and GDS-J-9, CC1-J-5 and NCC-J-8, PWS-J-4 and PWS-DGU-7 were seen to have metabolic fingerprint pattern similar to each other but different from the rest. Sites CTF-D2-2 and DF-MJ-3 were seen to be different from each other in bacterial community structure and also to the others
Among other ecological niche crude oil-contaminated environment is one of the ecologically degraded environments which harbour numerous bacteria (Aislabie et al. 2004; Saul et al. 2005). The microbes inhabited in crude oil-contaminated environments determine the fate of crude oil in nature. Therefore, assessment of diversity and species richness of microbial population in crude oil-degraded environment is being given utmost important (Hazen et al. 2010; Beazley et al. 2012; Liu and Liu 2013). Assam houses many crude oil-contaminated environments which have emerged as major matter of concern. In our previous study, we have compared bacterial diversity in different locations of Sivasagar and Jorhat districts and maximum of oil-contaminated soil environment was found to contain 15.0–56.0% (Roy et al. 2014; Yenn et al. 2014; Das et al. 2015), whereas in the present study, the origin and nature of contamination were quite different (Table 1). Overall hydrocarbon content in the present study was fairly high and ranged from 0.04 to 89.36% which was due to bottom water tank sludge, drilling fluid, drilling sites, etc. The highest soil organic carbon (6.99%) was found in mud sample, whereas lowest (1.38%) was seen for oil sludge collected from pit, Duliajan. The contaminated samples were alkaline in nature except for the Core-10 sample collected from a depth of 3600 m. Biological activities of the present soils were very low. This wide variation in physical, chemical and biological properties of crude oil-contaminated soil of present study was due to source of hydrocarbon contamination and hydrocarbon load. Soil biological activity, including soil enzymatic activity, has been reported to be influenced by a range of environmental stresses and contaminants (Alrumman et al. 2015).
The natures of contamination of crude oil in different soil environment were due to nature of the operational contamination. Overall hydrocarbon content in the present study was also quite high and ranges from 0.04 to 89.36% which was due to bottom water tank sludge, drilling fluid, drilling sites, etc. Studies on special environmental conditions of crude oil contamination soils reported that there were overall less microbial diversity and species richness in oil-contaminated soil (Aislabie et al. 2004; Saul et al. 2005) which is in agreement with the present findings. Hydrocarbon contaminants have also been reported to restrain growth of certain responsive microbial groups (Santos dos et al., 2011) and selectively trigger subgroups like that of the Actinobacteria and Proteobacteria in contaminated soils (Bell et al. 2013, 2014), hence affecting the microbial diversity. In the present study, we were able to isolate 160 morphologically distinct bacteria and 16S rDNA analysis confirmed that the isolates belonged to the genus Bacillus, Alkaligenes, Enterobacter, Clostridium, Exoguibacterium and Pseudomonas. The highest number of population isolated was from CTF-D-1 followed by TFD2-2 > DF-MJ-3 > PWS-J-4 > CC1-J-5 > CC2-J-6 > PWS-DGU-7 and NCC-J-8 > GDS-J-9 > Core-10, respectively. We are also reporting for the first time the presence of Exiguobacterium sp. from CCJ2-J-6 which might be due to drilling sludge and other stress. To forecast any relationship between microbial population and hydrocarbon, we found that except TPH with microbial population in M1 media, no other parameters showed significant positive correlation (Table S1). The positive correlation between TPH and M1 media is due to crude oil-specific growth. Interestingly, previous study conducted by Margesin et al. (2003) also showed no correlation between concentration of TPH in soil environment and microbial population.
From the DGGE banding profile, 17 distinct bands were successfully sequenced. BLAST-N analyses revealed prevalence of genus Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas in the samples. Proteobacteria was reported to typically dominate microbial assemblage in hydrocarbon-contaminated soils (Greer et al. 2010). Several previous studies reported Pseudomonas and Acenitobacter in various hydrocarbon-contaminated environments using culture-dependent (Belhaj et al. 2002; Bhattacharya et al. 2003) and culture-independent (Kaplan and Kitts 2004; Margesin et al. 2003; Roling et al. 2004) approaches. The presence of Spingomonas and Raslstonia in contaminated sites was also reported by various researchers (Juhasz and Naidu 2000; Zocca et al. 2004). We also found many of the unidentified clones having >99% sequence similarity to the bacterial clones (Accession nos. KC884580.1, KC884570.1, KC521869.1 and EU491331.1) (Table S3 and Fig. 4) that were reported by researchers from other oil-contaminated sites including Dagang oil fields of China (Li et al. 2006). Interestingly, we observed a disparity between culture-based techniques to that of uncultured diversity assessment which was in agreement with the earlier reports (Lagier et al. 2012; Shade et al. 2012) and contrary to many reports (Lagier et al. 2012; Carraro et al. 2011; Vaz-Moreira et al. 2011). Hamamura et al. (2006) in their study also reported cultivation of numerous HCB from oil-contaminated soil which did not show relationship with the results obtained using culture-independent molecular techniques. Here the difference in culture-based techniques to that of uncultured diversity assessment is mainly due to the lack of ability of some of the bacteria to grow under laboratory condition. This lack of ability may be as a result of inability of providing optimum conditions for the indigenous to grow in laboratory media plates.
The PCA plot of carbon source utilization pattern by microbial community of soil samples: sites CTF-D-1 and Core-10 was seen to be most diverse and was also found to have the highest pH of 9.11 and lowest pH of 3, respectively, as compared to the rest and hence different microbial community inhabiting these sites. Previous studies have also reported bacterial community composition to be associated with the quality of soil organic matter, geographical region and environmental factors such as temperature, nutrient availability, soil pH (Wallenstein et al. 2007; Coolen et al. 2011; Lauber et al. 2009; Shen et al. 2013; Bartram et al. 2014). High moisture content of CTF-D-1 (66.6%) and significantly low moisture content of Core-10 (1.25%) as compared to the other sample were another factor that can be responsible for variation in community structures of these sites. Soil water content determines microbial community structure, and Zhou et al. (2002) suggested that free water connecting soil particles may influence diversity patterns, by controlling nutrient availability and cell movement. Low soil moisture was reported to decrease microbial activity by reducing diffusion of soluble substrates, microbial mobility and intracellular water potential (Zhou et al. 2002).
The current study determined the diversity of indigenous bacteria in the crude oil-contaminated sites of Duliajan and Jorajan, Assam. Bacterial isolates obtained from culture-based approach showed predominance of Lysinibacillus, Alcaligenes, Bacillus, Clostridium, Enterobacter and Pseudomonas in these study sites. Culture-independent DGGE technique confirmed the presence of genera Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas. For the first time, we are reporting the presence of genus Exoguibacterium in crude oil-contaminated soil of Assam. PCA analysis revealed CTF-D-1 and Core-10 to be most diverged, indicating significant bacterial phyla, while sites CC2-J-6 and GDS-J-9 were found to have closely similar bacterial phyla. Both culture-dependent and culture-independent approach resulted in a total of 11 genera. The present findings is the first-hand report on total bacterial diversity in crude oil-contaminated soil of Assam, which can be assisted in further exploitation of these for bioremediation purposes.
Conclusion
Pollution by hydrocarbons may stimulate the growth of hydrocarbon-utilizing/hydrocarbon-degrading bacteria and alter the structure of microbial communities in the contaminated area. Thus, classification of the organisms that participate in hydrocarbon biodegradation is important for assessing and developing in situ bioremediation strategies. Assessment of diversity and species richness of microbial population in crude oil-degraded environment is being given utmost importance. The present study demonstrated the assessment of bacterial population of crude oil-contaminated samples collected from different locations operated under Oil India limited, Duliajan (27°21′42″ N, 95°19′ 6″ E) and Jorajan (95°11′ - 95°30′E; 27° 06′ - 27° 22′ N) of Dibrugarh and Tinsukia, Assam. Here, we report 160 morphologically distinct bacteria which could be isolated using conventional culturing techniques of which 117 strains were found to be Gram-positive bacteria and the rest 43 strains were identified as Gram-negative bacteria; majority of the strains were seen to belong to the phylum Proteobacteria (Betaproteobacteria, Gammaproteobacteria and Fermicutes (Bacilli, Clostridia), under the genus Enterobacter, Pseudomonas, Alkaligenes, Bacillus, Lysinibacillus and Clostridium. Moreover, an alternative approach, the denaturing gradient gel electrophoresis (DGGE) profiles of 16S rDNA phylotypes showed the predominance of Sphingomonas, Ralstonia, Sphingobium, Massilia, Acinetobacter and Pseudomonas. Both culture-dependent and culture-independent approaches resulted in 11 genera of which Bacillus and Pseudomonas were the key inhabitants creating most favourable bacterial milieu in the presence of crude oil contaminants. Previous studies have reported that there was an overall less microbial diversity and species richness in crude oil-contaminated environments which is in agreement with the present findings. The highest bacterial population was obtained from CTF-D-1 followed by CTF-D2-2 > DF-MJ-3 > PWS-J-4 > CC1-J-5 > CC2-J-6 > PWS-DGU-7 and NCC-J-8 > GDS-J-9 > Core-10, respectively. This study also reports for the first time Exiguobacterium sp. from site CCJ2-J-6 to inhabit crude oil-contaminated areas of Assam. Currently, hydrocarbonoclastic bacteria are being given much importance by researchers and have been found to be efficient alternatives in controlling the fate of natural and anthropogenic crude oil seepage. Hence, increased interest is being paid in studying the diversity of indigenous bacteria capable of degrading hydrocarbon contaminants. This study thus helped in the identification of the principal bacterial species inhabiting the contaminated sites that might be potential in degradation of the contaminants, and these results will prove to be significant for chalking-out ways of most favourable in situ bioremediation strategies.
References
Abbasian F, Lockington R, Mallavarapu M, Naidu R (2016) The biodiversity changes in the microbial population of soils contaminated with crude oil. Curr Microbiol. doi:10.1007/s00284-016-1001-4
Abed RM, Safi NM, Koster J, De Beer D, El-Nahhal Y, Rullkotter J, Garcia-Pichel F (2002) Microbial diversity of a heavily polluted microbial mat and its community changes following degradation of petroleum compounds. Appl Environ Microbiol 68:1674–1683
Achtman M, Wagner M (2008) Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol 6:431–440
Aislabie J, Balks M, Foght J, Waterhouse E (2004) Hydrocarbon spills on Antarctic soils: effects and management. Environ Sci Technol 38:1265–1274
Albokari M, Mashhour I, Alshehri M, Boothman C, Al-Enezi M (2015) Characterization of microbial communities in heavy crude oil from Saudi Arabia. Ann Microbiol 65:95–104
Allen J, Atekwana E, Atekwana E, Duris J, Werkema D, Rossbach (2007) The microbial community structure in petroleum-contaminated sediments corresponds to geophysical signatures. Appl Environ Microbiol 73:2860–2870
Alrumman SA, Standing DB, Paton GI (2015) Effects of hydrocarbon contamination on soil microbial community and enzyme activity. J King Saud Univ Sci 27:31–41
Bargiela R, Mapelli F, Rojo D, Chouaia B, Tornes J, Borin S et al (2015) Bacterial population and biodegradation potential in chronically crude oil-contaminated marine sediments are strongly linked to temperature. Sci Rep 5:11651
Bartram AK, Jiang X, Lynch MD, Masella AP, Nicol GW, Dushoff J, Neufeld JD (2014) Exploring links between pH and bacterial community composition in soils from the craibstone experimental farm. FEMS Microbiol Ecol 87:403–415
Beazley MJ, Martinez RJ, Rajan S, Powell J, Piceno YM, Lauren MT (2012) Microbial community analysis of a coastal salt marsh affected by the Deepwater Horizon oil spill. PLoS ONE 7:e41305
Belhaj A, Desnoues N, Elmerich C (2002) Alkane biodegradation in Pseudomonas aeruginosa strains isolated from polluted zone, identification of alkB and alkB-related genes. Res Microbiol 153:339–344
Bell TH, Yergeau E, Maynard C, Juck D, Whyte LG, Greer CW (2013) Predictable bacterial composition and hydrocarbon degradation in Arctic soils following diesel and nutrient disturbance. ISME J 7:1200–1210
Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E et al (2014) Linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J 8:331–343
Bhattacharya D, Sarma PM, Krishnan S, Mishr S, Lal B (2003) Evaluation of genetic diversity among Pseudomonas citronellolis strains isolated from oily sludge contaminated sites. Appl Environ Microbiol 69:1435–1441
Carraro L, Maifreni M, Bartolomeol I, Martino ME, Novelli E, Frigo F (2011) Comparison of culture-dependent and-independent methods for bacterial community monitoring during Montasio cheese manufacturing. Res Microbiol 162:231–239
Casid LE, Klein DA, Santoro T (1964) Soil dehydrogenase activity. Soil Sci 98:371–376
Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM (2007) The ribosomal database project RDP-II, introducing myRDP space and quality controlled public data. Nucleic Acids Res 35:D169–D172
Coolen MJL, van de Giessen J, Zhu EY, Wuchter C (2011) Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ Microbiol 13:2299–2314
Das D, Baruah R, Sarma Roy A, Singh AK, Deka Boruah HP, Kalit J, Bora TC (2015) Complete genome sequence analysis of Pseudomonas aeruginosa N002 reveals its genetic adaptation for crude oil degradation. Geno 105:182–190
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem 28:213–221
Gogoi B, Dutta N, Goswami P, Mohan TA (2003) Case study of bioremediation of petroleum-hydrocarbon contaminated soil at a crude oil spill site. Adv Environ Res 7:767–782
Greer CW, Whyte LG, Niederberger TD (2010) Microbial communities in hydrocarbon-contaminated temperate, tropical, alpine, and polar soils. In: Timmis KN (ed) Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, pp 2313–2328
Hamamura N, Olson SH, Ward DM, Inskeep WP (2006) Microbial population dynamics associated with crude oil biodegradation in diverse soils. Appl Environ Microbiol 72:6316–6324
Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N (2010) Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208
Howard KWF, Howard KK (2016) The new “Silk Road Economic Belt” as a threat to the sustainable management of Central Asia’s transboundary water resources. Environ Earth Sci 75:976
Juhasz AL, Naidu R (2000) Bioremediation of high molecular weight polycyclic aromatic hydrocarbons, a review of the microbial degradation of benzo[a]pyrene. Int Biodeterior Biodegrad 45:57–88
Jung J, Philippot L, Park W (2016) Metagenomic and functional analyses of the consequences of reduction of bacterial diversity on soil functions and bioremediation in diesel-contaminated microcosms. Sci Rep 6:23012
Kaplan CW, Kitts CL (2004) Bacterial succession in petroleum land treatment unit. Appl Environ Microbiol 70:1777–1786
Keller M, Zengler K (2004) Tapping into microbial diversity. Nat Rev Microbiol 2:141–150
Kim S-J, Kweon O, Sutherland JB, Kim H-L, Jones RC, Burback BL, Graves SW, Psurny E, Cerniglia CE (2015) Dynamic response of Mycobacterium vanbaalenii PYR-1 to BP deepwater horizon crude oil. Appl Environ Microbiol 81:4263–4276
Kimes NE, Callaghan AV, Aktas DF, Smith WL, Sunner J, Golding B, Drozdowska M, Hazen TC, Suflita JM, Morris PJ (2013) Metagenomic analysis and metabolite profiling of deep-sea sediments from the Gulf of Mexico following the deepwater horizon oil spill. Front Microbiol 4:50
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C (2012) Microbial culturomics, paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193
Lauber CL, Hamady M, Knight R, Fierer N (2009) Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75:5111–5120
Li H, Yang S, Mu B, Rong Z, Zhang J (2006) Molecular analysis of the bacterial community in a continental high-temperature and water-flooded petroleum reservoir. FEMS Microbiol Lett 257:92–98
Li P, Qian H, Howard KWF, Wu J (2015) Building a new and sustainable “Silk Road economic belt” Environ. Earth Sci 74:7267–7270
Liu Z, Liu J (2013) Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the deepwater horizon oil spill. Microbiol Open 2:492–504
Lu Z, Deng Y, Van Nostrand JD, He Z, Voordeckers J, Zhou A (2012) Microbial gene functions enriched in the Deepwater Horizondeep-sea oil plume. ISME J 6:451–460
MacNaughton SJ, Stephen JR, Venosa AO, Davis GA, Chang Y, White DC (1999) Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol 65:3566–3574
Margesin R, Labbe D, Schinner F, Greer CW, Whyte LG (2003) Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine Alpine soils. Appl Environ Microbiol 69:3085–3092
Mason OU, Hazen TC, Borglin S, Chain PS, Dubinsky EA, Fortney JL (2012) Metagenome, metatranscriptome and single-cell sequencing re-veal microbial response to Deepwater Horizon oil spill. ISME J 6:1715–1727
McGarity JW, Mayers MG (1967) A survey of urease activity in soils of northern New South Wales. Plant Soil 27:217–238
Mishra A, Nautiyal CS (2009) Functional diversity of the microbial community in the rhizosphere of chickpea grown in diesel fuel spiked soil amended with Trichoderma ressei using sole-carbon source utilization profiles. World J Microbiol Biotechnol 25:1175–1180
Mnif S, Chamkha M, Sayadi S (2009) Isolation and characterization of a Halomonas sp. strain C2SS100 a hydrocarbon degrading bacterium under hypersaline condition. J Appl Microbiol 107:785–794
Muyzer G, Brinkhoff T, Nubel U, Santegoeds C, Schafer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans ADL, Van Elsas JD, De Bruijn FT (eds) Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–27
Nautiyal CS (2009) Self-purificatory Ganga water facilitates death of pathogenic Escherichia coli 157, H7. Curr Microbiol 58:25–29
Paisse S, Coulon F, Goni-Urriza M, Peperzak L, McGenity T, Duran R (2008) Structure of bacterial communities along a hydrocarbon contamination gradient in coastal sediment. FEMS Microbiol Ecol 66:295–305
Roling WFM, Milner MG, Jones DM, Fratepietro F, Swannell RP, Daniel F, Head IM (2004) Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl Environ Microbiol 70:2603–2613
Roy AS, Baruah R, Borah M, Singh AK, Deka Boruah HP, Saikia N (2014) Bioremediation potential of native hydrocarbon degrading bacterial strains in crude oil contaminated soil under microcosm study. Int Biode Biodeg 94:79–89
Santos dos HF, Cury JC, do Carmo FL, Santos dos AL, Tiedje J, van Elsas JD et al (2011) Mangrove bacterial diversity and the impact of oil contamination revealed by pyrosequencing: bacterial proxies for oil pollution. PLoS ONE 6:e16943
Saul D, Aislabie J, Brown C, Harris L, Foght J (2005) Hydrocarbon contamination changes the bacterial diversity of soil from around Scott Base, Antarctica. FEMS Microbiol Ecol 53:141–155
Shade A, Hogan CS, Klimowicz AK, Linske M, McManus PS, Handelsman J (2012) Culturing captures members of the soil rare biosphere. Environ Microbiol 14:2247–2252
Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211
Staddon WJ, Duchesne LC, Trevors JT (1997) Microbial diversity and community structure of post disturbance forest soils as determined by sole-carbon-source utilization patterns. Microb Ecol 34:125–130
Stefani FOP, Bell TH, Marchand C, de la Providencia IE, Yassimi AE, St-Arnaud M, Hijri M (2015) Culture-dependent and independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PLoS ONE 106:e0128272
Tabatabami A, Bremner JM (1969) Use of p-nitrophenyl phosphate for assay of soil phosphataseactivity. Soil Biol Biochem 1:301–307
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Vaz-Moreira I, Egas C, Nunes OC, Manaia CM (2011) Culture-dependent and culture-independent diversity surveys target different bacteria, a case study in a freshwater sample. Antonie Van Leeuwenhoek 100:245–257
Vila J, Nieto M, Mertens J, Springael D, Grifoll M (2010) Microbial community structure of a heavy fuel oil-degrading marine consortium, linking microbial dynamics with polycyclic aromatic hydrocarbon utilization. FEMS Microbiol Ecol 73:349–362
Vinas M, Sabate J, Espuny MJ, Solanas AS (2005) Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl Environ Microbiol 71:7008–7018
Walkley A, Black IA (1934) An examination of the Degtjareff method for determining organic carbon in soils: effect of variations in digestion conditions and of inorganic soil constituents. Soil Sci 63:251–263
Wallenstein MD, McMahon S, Schimel J (2007) Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol Ecol 59:428–435
Weber P, Grove J, Gehder M, Anderson W, Legge R (2007) Data transformations in the analysis of community-level substrate utilization data from microplates. J Microbiol Methods 69:461–469
Weeks OB, Breed RS (1957) In: Bergey’s manual of determinative bacteriology, 7th edn. Williams and Wilkins, Baltimore, p 320
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703
Yenn R, Borah M, Deka Boruah HP, Baruah R, Saikia N, Sahu OP, Tamuli AK (2014) Phytoremediation of abandoned crude oil contaminated drill sites of Assam with the aid of a hydrocarbon-degrading bacterial formulation. Int J Phytorem 9:909–925
Zhou J, Xia B, Treves DS, Wu LY, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM (2002) Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68:326–334
Zocca C, Di S, Visentini F, Vallini G (2004) Gregorio biodiversity amongst cultivable polycyclic aromatic hydrocarbon-transforming bacteria isolated from an abandoned industrial site. FEMS Microbiol Lett 238:375–382
Acknowledgements
Authors acknowledge Oil India Limited (OIL), Duliajan, for their support in procuring crude oil and contaminated samples. Authors also thank Directors, CSIR-NEIST and CSIR-NBRI, for their support. Reshita Baruah acknowledges Department of Science and Technology (DST), Govt. of India, for providing INSPIRE fellowship (IF120020). Financial support from Council of Scientific and Industrial Research for projects BSC0109 and BSC117 is duly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Editorial responsibiility: Zhenyao Shen.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baruah, R., Mishra, S.K., Kalita, D.J. et al. Assessment of bacterial diversity associated with crude oil-contaminated soil samples from Assam. Int. J. Environ. Sci. Technol. 14, 2155–2172 (2017). https://doi.org/10.1007/s13762-017-1294-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13762-017-1294-2