Abstract
Concern over the prevalence of active pharmaceutical agents and subsequent occurrence of antimicrobial resistance in the environment is increasing. Incorruptible ability of Ganga water was evaluated using fresh, 8-year-old, and 16-year-old Ganga water samples spiked with pathogenic Escherichia coli serotype O157:H7. Survival of E. coli O157:H7 over the course of the experiment was 3, 7, and 15 days for fresh, 8-year-old, and 16-year-old Ganga waters, respectively. On the contrary, in Milli Q water the decline in viable count of E. coli O157:H7 up to 30 days was only 2 log units. Survival of E. coli O157:H7 was greater in boiled water compared with water after passage through a 0.2-μm-pore-size membrane filter, indicating involvement of heat-labile agents influencing survival of E. coli O157:H7 in Ganga water, which seems to indicate the role of antimicrobial peptides. Functional diversity of Ganga water’s native microbial community structure as assessed with Biolog Eco plates was not affected even in the presence of a 5-fold log units higher pathogenic load of E. coli O157:H7. These findings suggest that Ganga water has certain novel antimicrobial attributes, besides its remarkable fluidity, which may provide a much-needed basis for the development of new antimicrobial compounds.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The water of the River Ganga is frequently used for drinking, cooking, and bathing purposes due to ancient knowledge that Ganges water does not putrefy, even after long periods of storage. Water has been used from time immemorial for remedial purposes. Most religious beliefs involve some ceremonial use of “holy” water, and in India the water of the River Ganga is treated with such reverence. Under the continuous Saraswati-Indus civilization going back to ~7500 BC, the River Ganga is mentioned in Rigveda [16]. Hippocrates, going back to ~500 BC, wrote about the healing of disease with water. Bathing held a prominent place in the law that was prepared by Moses under divine instruction for the government of the Hebrew nation. The role of the bath in the treatment of leprosy also would lead one to believe that water was used for curative effects [11]. Outbreaks of acute diarrheal disease have been identified as causes of fatal disease dating back as far as the Sanskrit literature and during Hippocratic times [15]. Ernest Hankin, a British bacteriologist, reported in 1896 on the presence of marked antibacterial activity against Vibrio cholerae, which he observed in the water of the River Ganga river India, and he suggested that it might help to decrease the incidence of cholera in people using water from the Ganges. Though invisible, it was possible to show that this principle was particulate and D’Herelle called it “bacteriophage” [6]. Thus in a way the world owes the discovery of bacteriophages to the Ganges water.
Overuse in human medicine and for agricultural purposes has become a recognized medical problem, and scientists have become increasingly concerned about the occurrence of antibacterial resistance in the environment. To curtail the development and spread of antimicrobial resistance will require both the preservation of current antimicrobials through their appropriate use and the discovery and development of new agents. Technologies for accessing and screening new sources of badly needed and novel antibiotics have improved dramatically during the past decade [10, 12, 14, 22]. This study was conducted to validate our ancient knowledge about the antimicrobial effect of Ganga water and to evaluate the potential of Ganga water in our endeavor to explore the possibility of using it as a novel source of antimicrobial compounds. Enterohemorrhagic Escherichia coli is a worldwide cause of infection in humans and animals. E. coli O157:H7 is a major enteropathogen responsible for causing outbreaks of hemorrhagic colitis and hemolytic uremic syndrome [1]. The human infectious dose is very low, and ingestion of as few as 10 cells is thought to be sufficient to cause illness [3]. The objective of this study was to evaluate the incorruptible self-purificatory characteristic and microbial community structure of Ganga water when spiked with E. coli O157:H7.
Materials and Methods
Sampling Stations
Water samples for the present study were collected form the upper stretch of River Ganga (hilly region) at Rishikesh, having the geographical coordinates of longitude 78°42′E and latitude 30°7′N. The water samples were collected from the same site in March 2000 and 2007. A 16-year-old sample was collected in 1991 from Gomukh in the snout of the Gangotri Glacier, a vast expanse of ice 5 × 15 miles in the higher Himalaya, at an altitude of 3920 m, having the geographical coordinates of longitude 78°54′E and latitude 30°54′N. The samples thus collected were stored at Lucknow in clean glass bottles fitted with screw caps indoors in a cool dark place. Lucknow is located in mid-Ganges plains at an altitude of 123 m, having geographical coordinates of longitude 80°59′E and latitude 26°55′N and a mean ambient temperature in the range of 20 to 35°C. A fresh Ganga water sample was filtered with a sterilized filtration unit, through a 0.22-μm-pore-size membrane filter (diameter, 47 mm; Millipore Inc., Billerica, MA, USA). The purpose of this filtration was to allow viruses to pass through the filter while removing organic and inorganic chemicals, natural organic matter, protozoans, algae, zooplankton, and free-living aquatic bacteria. Boiled water was obtained by boiling a fresh Ganga water sample for 20 min to kill micro-organisms [13].
Bacterial Analysis
Heterogeneous bacterial counts in the water were elucidated by serial dilution plating directly on nutrient agar (from HI-MEDIA Laboratories Pvt. Ltd., Mumbai, India) plates. After 72 h of incubation at 28°C, the colonies that developed on the plate were counted [17]. Strains of E. coli were not detected in Ganga water. E. coli O157:H7 (ATCC 43895) was obtained from King George Medical University, Lucknow. Rainbow agar O157:H7 was used for enhanced detection of E. coli O157:H7 (Biolog, Inc., Hayward, CA, USA).
Survival of E. coli O157:H7 in Water
Inoculation of E. coli in Ganga water and Milli-Q water was carried out using overnight grown culture to assess the impact of water on survival of E. coli O157:H7. Overnight-grown culture was centrifuged and the pellet was washed three times using 0.85% sterile saline (w/v NaCl), then inoculated in Ganga water and Milli-Q water to a starting concentration of about 5 × 107 CFU/mL, and the mixture placed in three replicate (sterile) polypropylene tubes at 30°C under static conditions. E. coli O157:H7 surviving in the water was quantified at the designated time up to 30 days, diluted, and plated on Hi-Crome ECC agar plates (from HI-MEDIA Laboratories Pvt. Ltd.). All experiments were performed independently at least three times.
Microbial Diversity Using the Carbon Source Utilization Pattern
Biolog Eco plates (Biolog, Inc.) were used to determine the effect of E. coli on carbon source utilization pattern after different times of incubation in Ganga water and Milli-Q water. Inoculation of E. coli in Ganga water and Milli-Q water was carried out as described earlier [16]. Data were recorded for days 1–7 at 590 nm. Microbial activity in each microplate, expressed as average well color development (AWCD), was determined as described by Garland [8]. Diversity and evenness indexes were calculated as described by Staddon et al. [21]. Principal component analysis (PCA) was performed on data divided by the AWCD as described by Garland and Mills [9]. Formulas used for diversity calculations were described by Staddon et al. [21]; data collected after day 5 were used. At least three independent experiments were conducted for each treatment. Statistical analyses were performed using SPSS 16.0 and Windowstat 7.5.
Results and Discussion
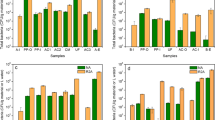
Studies of factors affecting the survival of E. coli in Ganges water is of great interest due to its importance as an indicator of fecal pollution in natural waters. It is ancient knowledge that Ganges water does not putrefy, even after long periods of storage, thus water from the Ganges has for millennia been regarded as incorruptible [4, 6, 19]. To facilitate a fair assessment of the potential of its self-purificatory and incorruptible abilities, Ganga water having a resident bacterial population of 4.0 × 102 CFU/mL was spiked with about 5.0 × 107 CFU/mL E. coli O157:H7. The incorruptible nature of the water was studied in fresh, 8-year-old, and 16-year-old Ganga water samples spiked with E. coli O157:H7. Figure 1 shows the decline in viable counts of E. coli O157:H7 in fresh, 8-year-old, and 16-year-old Ganga water during the course of the experiment. In general, the number of culturable E. coli O157:H7 declined over time but tended to be greater in fresh water than in 8- and 16-year-old water. Survival of E. coli O157:H7 over the course of the experiment was 3, 7, and 15 days for fresh, 8-year-old, and 16-year-old Ganga waters, respectively. On the contrary, in Milli-Q water the decrease in the viable count of E. coli O157:H7 up to 30 days was 2.0 × 104 CFU/mL (Fig. 1).
Survival of E. coli O157:H7 in Milli-Q water and Ganga water. Overnight-grown culture was inoculated into Ganga water and Milli-Q water to an initial concentration of about 5 × 107 CFU mL−1 in polypropylene tubes before the experiment was carried out, incubated under static conditions at 30°C during the course of the experiment, and plated on Hi-Crome ECC agar plates (HI-MEDIA Laboratories Pvt. Ltd., Mumbai, India). E. coli O157:H7 surviving in the water was quantified at the designated times up to 30 days. The plotted data are averages from three independent experiments
Age of the water seems to influence survival of E. coli O157:H7, thus its fate was further studied in boiled water and after passage through a 0.2-μm-pore-size membrane filter. To elucidate the involvement of active principals and their sensitivity to high temperatures, the water was boiled. Water samples thus prepared were spiked with E. coli O157:H7 to evaluate the antibacterial ability of the water. Boiling water at 100°C kills microbes; filtration is becoming increasingly the method of choice for sterilization of biologicals, especially when the product is heat labile, because the filtration process is inherently nondestructive. In general, 0.2 μm will remove algae, protozoa, and most bacteria, while a 0.01-μm filter is needed to remove viruses [13]. Overall, survival was higher in boiled water (3.5 × 102 CFU/mL for up to 25 days; Fig. 1) than in water after passage through a 0.2-μm-pore-size membrane filter (3.9 × 102 CFU/mL for up to 15 days; Fig. 1), indicating that heat-labile agents influence the survival of E. coli O157:H7 in Ganga water. An interesting observation was the ability of the 8- and 16-year-old Ganga water to influence survival of E. coli O157:H7. Eight-year-old water had a better ability to kill E. coli O157:H7 compared with boiled water and water passed through a 0.2-μm-pore-size membrane filter. While antibacterial activity of 16-year-old water was better than that of boiled water and almost comparable to that of water passed through a 0.2-μm filter, indicating that a combination of factors controls the rate of decline and does not let the water putrefy, even after long periods of storage. Further studies should be undertaken to establish which factors are the key regulators influencing the death of E. coli O157:H7 in Ganga water.
To investigate the well-known self-purificatory characteristic of Ganga water, the impact of the addition of E. coli O157:H7 on the microbial community structure in Milli-Q water and Ganga water after incubation for 0, 3, 5, and 7 days was assessed, using Biolog Eco plates. Eco plates are intended for environmental samples; they contain 31 different carbon sources in a triplicate pattern. Patterns of carbon source utilization were used to calculate diversity indexes of Shannon, Simpson, and McIntosh and related evenness indexes. Analysis of carbon source utilization patterns by microbial samples using Biolog plates shows promise as a means of assessing microbial community structure, which examines the functional capabilities of the microbial population, and the resulting data can be analyzed using multivariate techniques to compare the metabolic capability of communities. Biolog plates have found application in the assessment of microbial metabolic diversity in water [2]. We used the Biolog plates as a way to estimate the metabolic diversity of the native heterotrophic bacteria in Ganga water in the absence and presence of E. coli O157:H7. Such information allows examination of the natural variation and diversity of microbial communities. Most importantly, this technique offers the potential to monitor changes in microbial diversity caused by environmental fluctuations, management practices, and pollution. Variable significant differences among treatments were noted for water samples collected at 0, 3, 5, and 7 days using the McIntosh, Shannon, and Simpson indexes (Table 1). Evenness calculated from both the Shannon and the McIntosh indexes, however, was not significantly different among treatments (Table 1). The principal components (PC) score plots describe the characteristics of the samples and help to clarify their distribution and clustering. The PC score plot (PC-I and PC-II) shows the spatial distribution of the samples. PCA of the carbon source utilization pattern on Biolog Eco plates showed clustering among the Ganga water samples incubated with E. coli for 0, 3, 5, and 7 days, while the samples from Milli-Q water samples incubated with E. coli for 0, 3, 5, and 7 days distributed separately from each other, at 34 and 77% on the PCA vector 1 and 2 axis (Fig. 2). There was distinct resolution of Milli-Q water microbial communities from Ganga water microbial communities spiked with E. coli O157:H7 (Fig. 2). PCA thus indicated no impact of the addition of E. coli O157:H7 on microbial community structure in Ganga water, while the addition of E. coli O157:H7 in Milli-Q water resulted in significant differences in the microbial community structure.
Principal component analysis (PCA) of carbon source utilization pattern on Biolog Eco plates (Biolog, Inc., Hayward, CA, USA) of E. coli O157:H7 samples in Milli-Q water (nos. 1, 3, 5, and 7) and Ganga water (nos. 2, 4, 6, and 8) incubated for 0, 3, 5, and 7 days, respectively, was carried out. Biolog Eco plates were used to determine the effect of E. coli on carbon source utilization pattern at different times of incubation in Ganga water and Milli-Q water. Data on Biolog Eco plates were recorded every 24 h at 590 nm with an automated microplate reader (BioTek Instruments Inc., USA). At 5th day PCA was performed on blank subtracted data divided by the average well color development (AWCD) as described by Garland and Mills [9]. The plotted data are averages of three independent experiments. PCA was performed using Windowstat 7.5
The survival of fecal bacteria in aquatic environments is in general dependent on their ability to tolerate a range of biological, physical, and chemical stresses. These include the influence of temperature, UV radiation, predation, and nutrient availability [7]. For boiled and membrane-filtered Ganga water in which survival of E. coli O157:H7 was monitored, it appeared that biotic factors had a strong influence on survival. These results attain further importance when one considers the fact that the resident bacterial population of 4 × 102 CFU/mL Ganga water was spiked with about 5 × 107 CFU/mL E. coli O157:H7. Thus even a 5-fold log units higher pathogenic load of E. coli O157:H7 did not affect the Ganga water’s native microbial community structure in the studied environment. To the best of our knowledge this is the first report demonstrating that the functional diversity of Ganga water as assessed with Biolog Eco plates was not affected even in the presence of a high pathogenic load of E. coli O157:H7. The studies clearly demonstrate that Ganga water indeed has certain novel antimicrobial attributes, besides its remarkable fluidity and adaptability in the presence of a heavy load of E. coli O157:H7, thereby validating the river’s remarkable “magical” self-cleansing properties. Although it is not possible to extrapolate the behavior of a single strain of E. coli O157:H7 under laboratory conditions to that of all strains under environmental conditions, this study has provided an insight into the impact of Ganga water on E. coli O157:H7 survival. Technologies for accessing and screening new sources of badly needed and novel antibiotics have improved dramatically during the past decade [5, 18, 20]. Where combinatorial chemistry and genomics have failed, could an exploration of untapped sources usher in a second golden age of antibiotic discovery? The involvement of heat-labile agents influencing survival of E. coli O157:H7 in Ganga water seems to indicate a role of antimicrobial peptides (AMPs). AMPs are part of the innate immune system, and an important component of immune defense. They are produced by plants, animals, insects, and single-celled organisms, and possess antimicrobial properties. As such, they are an ideal target for future antibiotic production [5, 18, 20]. The encouraging results of these experiments demonstrating the Ganga’s antimicrobial capacity, indicating involvement of heat-labile agents, if carefully developed, could eventually provide a much-needed basis for the development of new antimicrobial compounds.
References
Aitken MD, Sobsey MD, Van Abel NA et al (2007) Inactivation of Escherichia coli O157:H7 during thermophilic anaerobic digestion of manure from dairy cattle. Water Res 41:1659–666
Beauchamp CJ, Simao-Beaunoir AM, Beaulieu C et al (2006) Confirmation of E. coli among other thermotolerant coliform bacteria in paper mill effluents, wood chips screening rejects and paper sludges. Water Res 40:2452–2462
Chart H (2000) VTEC enteropathogeicity. J Appl Microbiol 88:12S–23S [Symposium Supplement]
Darian SG (1978) The Ganges in myth and history. University Press of Hawaii, Honolulu
DasGupta SM, Chauhan PS, Nautiyal CS (2007) Search for an ever elusive “guardian-angel” novel antibiotic: myth or reality. In: Chauhan AK, Kharkwal H, Verma A (eds) Microbes for human life. IK International, New Delhi, pp 311–335
D’Herelle F (translated to English by Smith GH) (1922) The bacteriophage: its role in immunity. Williams and Wilkins/Waverly Press, Baltimore
Flint KP (1987) The long-term survival of Escherichia coli in river water. J Appl Bacteriol 63:261–270
Garland JL (1996) Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem 28:213–221
Garland JL, Mills AL (1991) Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359
Häusler T (2006) Viruses vs. superbugs: a solution to the antibiotics crisis? Macmillan Science, New York
Kloss J (1939) Back to Eden. Back to Eden books, Loma Linda
Lerner CG, Hajduk PJ, Wagner R et al (2007) From bacterial genomes to novel antibacterial agents: discovery, characterization, and antibacterial activity of compounds that bind to HI0065 (YjeE) from Haemophilus influenza. Chem Biol Drug Des 69:395–404
Liberman D, Berman T (2006) Analysis and monitoring: MSC—a biologically oriented approach. Filtrat Sep 43:439–440
Lock RL, Harry EJ (1994) Cell-division inhibitors: new insights for future antibiotics. Nature Rev Drug Dis 7:324–338
McMahan ZH, Dupont HL (2007) The history of acute infectious diarrhea management—from poorly focused empiricism to fluid therapy and modern pharmacotherapy. Aliment Pharmacol Ther 25:759–769
Nautiyal CS, Chauhan PS, Nene YL (2007) Medicinal smoke reduces airborne bacteria. J Ethnopharmacol 114:446–451
Nautiyal CS, Govindarajan R, Lavania M et al (2008) Novel mechanism of modulating natural antioxidants in functional foods: involvement of plant growth promoting rhizobacteria NRRL B–30488. J Agr Food Chem 56:4474–4481
Rossi LM, Rangaswamy P, Zhang J et al (2008) Research advances in the development of peptide antibiotics. J Pharm Sci 97:1060–1070
Sharma Y (1997) The Ganga, India. In: Helmer R, Ivanildo H (eds) Water pollution control—a guide to the use of water quality management principles. WHO/UNEP, Geneva
Sheridan C (2006) Antibiotics au naturel. Nat Biotechnol 24:1494–1496
Staddon WJ, Duchesne LC, Trevors JT (1997) Microbial diversity and community structure of post disturbance forest soils as determined by sole-carbon-source utilization patterns. Microb Ecol 34:125–130
Vicente M, Hodgson J, Massidda O et al (2006) The fallacies of hope: will we discover new antibiotics to combat pathogenic bacteria in time? FEMS Microbiol Rev 30:841–852
Acknowledgments
The author is deeply indebted to R.K. Trivedi, former Governor of the State of Gujarat, India, for providing the 16-year-old Ganga water sample; V. Sharma, King George Medical University, Lucknow, for providing the strain of E. coli O157:H7; and B. Staddon, Eastern Kentucky University, Richmond, USA for, his generous help in the Diversity/Evenness Index analysis. Thanks are due to the Director, National Botanical Research Institute, Lucknow, for the necessary support. This work was supported by the New Millennium Indian Technology Leadership Initiative (NMITLI) program and Task Force Grant NWP-0019 from the Council of Scientific and Industrial Research, New Delhi.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nautiyal, C.S. Self-Purificatory Ganga Water Facilitates Death of Pathogenic Escherichia coli O157:H7. Curr Microbiol 58, 25–29 (2009). https://doi.org/10.1007/s00284-008-9260-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-008-9260-3