Abstract
Mutations in the SH3TC2 gene cause Charcot–Marie–Tooth disease type 4C (CMT4C), characterized by inherited demyelinating peripheral neuropathy. CMT4C is a common form of CMT4/autosomal recessive (AR) CMT1. This study examined the SH3TC2 variants, investigated genotype–phenotype correlations and explored the frequency of CMT4C in Chinese patients. A total of 206 unrelated patients of Chinese Han descent clinically diagnosed with CMT were recruited. All patients underwent detailed history-taking, neurological examination, laboratory workups, and electrophysiological studies. Genetic analysis was performed via high-throughput target sequencing (NGS). Three patients, one male and two females, were found to carry five SH3TC2 mutations: patient 1 (c.3154C > T, p.R1054X; c.929G > A, p.G310E); Patient 2 (c.2872_2872del, p.S958fs; c.3710C > T, p.A1237V) and Patient 3 (c.2782C > T, p.Q928X; c.929G > A, p.G310E). The c.2872_2872del, c.3710C > T and c.2782C > T variants were not reported before. CMT4C caused by SH3TC2 mutation is a very common type of CMT4/AR CMT1. Three novel mutations, c.2872_2872del, c.3710C > T and c.2782C > T, were found in this study. Combination of clinical phenotype, nerve conduction studies, genetic analysis and bioinformatics analysis are of vital importance in patients suspected as CMT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Charcot–Marie–Tooth disease (CMT), the most common hereditary motor and sensory neuropathy with a worldwide incidence of 1 in 2500, comprises a group of clinically and genetically heterogeneous peripheral neuropathies [1,2,3,4], and is roughly classified into Type 1 (CMT1; demyelinating) and Type 2 (CMT2; axonal) according to median nerve motor conduction velocity (MCV). More than 80 genes have been reported as being associated with CMT [5]. With the development of molecular genetics, the classification of CMT is refined.
All patterns of inheritance have been observed, with autosomal dominant (AD) being most common. Autosomal recessive (AR) demyelinating pattern is less common, which is referred to as CMT4 or AR-CMT1 [6,7,8]. CMT4 has been linked to 12 causative genes and referred to from CMT4A to CMT4K (see http://neuromuscular.wustl.edu/time/hmsn.html) [9]. AR-CMT is usually more severe than AD-CMT, characterized by earlier onset and more rapid clinical progression that results in more marked distal limb deformities and major spinal deformities [9].
Mutations in the SH3TC2 gene cause CMT type 4C (CMT4C) [8,9,10,11], which is characterized by variable onset, mostly in the first and second decade, demyelinating peripheral neuropathy, early progressive scoliosis and moderate severity (non-severe handicap) [12,13,14,15]. The incidence of CMT4C varies between countries [9, 14,15,16]. CMT4C is the most frequent form of AR-CMT1 in Italy (> 20%) [16], however, it is relatively rare in Japan (1.76%) [9] and the United States (0.8%) [17]. In this study, we examined the SH3TC2 mutations, investigated genotype–phenotype correlations and explored the frequency of CMT4C in a cohort of Chinese patients clinically diagnosed with CMT.
Materials and methods
Patients
A total of 206 unrelated Chinese Han patients clinically diagnosed with CMT were recruited from the Department of Neurology of the First Medical Center, Chinese PLA General Hospital (Beijing, China) from December 20th, 2015 to March 2nd, 2020. The patients were from 30 provinces, which covered most areas of China. The coverage was sufficient in all patients. The patients underwent detailed history-taking, neurological examination, laboratory examinations, electrophysiological studies, and genetic testing.
The study was approved by the Chinese PLA General Hospital Ethics Committee. Informed written consent was obtained from all of the patients enrolled in this study.
Electrophysiological examination
The patients underwent nerve conduction study (NCS) in which their skin temperature was maintained at 32 °C or above during NCS [1, 2]. NCS were performed on the median, ulnar, tibial, peroneal and sural nerves using the Keypoint electromyography (EMG) system (Medoc Ltd, Israel) [1, 2]. The results were measured according to the normal reference values utilized by the EMG laboratory of Chinese PLA General Hospital (median motor nerve: amplitude ≥ 5.0 mV, velocity ≥ 50.0 m/s; median sensory nerve: amplitude ≥ 5.0 µV, velocity ≥ 50.0 m/s; ulnar motor nerve: amplitude ≥ 5.0 mV, velocity ≥ 50.0 m/s; ulnar sensory nerve: amplitude ≥ 5.0 µV, velocity ≥ 50.0 m/s; tibial motor nerve: amplitude ≥ 5.0 mV, velocity ≥ 40.0 m/s; peroneal motor nerve: amplitude ≥ 3.0 mV, velocity ≥ 45.0 m/s; and sural sensory nerve: amplitude ≥ 6.0 μV, velocity ≥ 50.0 m/s). NCS were considered abnormal if any of the parameters were found below the normal reference values [1, 2].
Genetic analysis
A total of 206 patients underwent genetic analysis using high-throughput target sequencing (NGS). The whole coding areas of CMT-associated genes (Table 1) were examined. Genomic DNA was extracted from the peripheral leukocytes of fresh blood samples obtained from the patients with clinical diagnosis of CMT [1, 2]. Target genes were captured by GenCap target region probe (MyGenostics Inc, Medford, MA, USA) and amplified by polymerase chain reaction. The eluted DNA was finally amplified for 15 cycles according to the following procedure: 98 °C for 30 s (1 cycle), 98 °C for 25 s, 65 °C for 30 s, 72 °C for 30 s (15 cycles), and 72 °C for 5 min (1 cycle) [1, 2]. The amplified products were purified using SPRI beads (Beckman Coulter, Brea, CA, USA) according to manufacturer’s protocol. Enriched libraries were sequenced using a HiSeq 2000 sequencer (Illumina, San Diego, CA, USA), which generated 100 bp paired reads [1, 2].
Depth reading of NGS identified PMP22 duplications/deletions, and multiplex ligation-dependent probe analysis (MLPA) was applied to confirm the results [1, 2]. Sanger direct sequencing was used to detect and confirm variants in the patients and their family members [1, 2].
The reference genome was UCSC hg19 (http://genome.ucsc.edu/). Read mapping was done using SOAP (Short Oligonucleotide Analysis Package) aligner (http://soap.genomics.org.cn/soapaligner.html) and Burrows–Wheeler Aligner (http://bio-bwa.sourceforge.net/bwa.shtml) software [1, 2]. Variant detection included the identification of single-nucleotide polymorphisms and indels with GATK and SOAPsnp (http://soap.genomics.org.cn/soapsnp.html) software [1, 2]. We searched the identified variants in 1000 normal Chinese population exome sequences (MyGenostics, Beijing, China). The genomic variants database included the 1000 Genomes Project (browser.1000genomes.org/index.html), the single-nucleotide polymorphism database (dbSNP) (http://www.ncbi.nlm.nih.gov/projects/SNP/) [1, 2], the Exome Aggregation Consortium (ExAC) and the East Asia ExAC.
Bioinformatics analysis
Polymorphism Phenotyping 2 (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph 2/), sorting intolerant from tolerant (SIFT) (http://sift.jcvi.org/), and Mutation Taster (http://www.mutationtaster.org/) were used to predict potential functional effects of SH3TC2 mutations [1, 2]. GeneSplicer and Human Splicing Finder were used to predict functional effects of splicing mutations. The pathogenicity was determined using the ACMG guideline.
Results
Twenty-six (26/206, 12.6%) patients were diagnosed with CMT1A, and three unrelated patients, 1 male and 2 females, were identified with 5 SH3TC2 variants from 206 patients of CMT (demyelinating/intermediate neuropathy: 146, axonal neuropathy: 60): Patient 1 (c.3154C > T, p.R1054X; c.929G > A, p.G310E); Patient 2 (c.2872_2872del, p.S958fs; c.3710C > T, p.A1237V) and Patient 3 (c.2782C > T, p.Q928X; c.929G > A, p.G310E). The frequency of CMT4C was 1.46% (3/206) in the whole cohort, and was 2.5% (3/120) in demyelinating/intermediate CMT negative for PMP22 duplication.
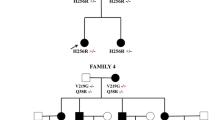
Patient 1 was a 26-year-old female who carried the c.3154C > T and the c.929G > A variants. The above two variants were reported to be associated with CMT4C [17, 18]. She presented with gait abnormality at the age of 5 and with bilateral distal lower limbs weakness and atrophy at the age of nine. The symptoms progressed slowly and the patient had difficulty climbing stairs and standing up when squatting down at the age of 26. Her parents did not have the same presentation as the patient, and she had no siblings. Sanger test confirmed that the patient’s mother carried the c.3154C > T variant and her father carried the c.929G > A variant (Fig. 1a–f). Physical examination showed scoliosis and muscle atrophy in bilateral thenar eminences, interosseous muscles and tibialis anterior muscles. Weakness of distal upper and lower limbs, more severe in lower limbs with gait abnormality, was found. Superficial and vibration perception were lost in distal lower extremities. Deep tendon reflexes were decreased, and bilateral Babinski signs were negative. NCS indicated demyelinating neuropathy and MCV of the median nerve was 28.0 m/s (Table 2).
a Patient 1, c.3154C > T, p.R1054X heterozygous mutation; b patient 1′s father, no c.3154C > T, p.R1054X mutation; c patient 1′s mother, c.3154C > T, p.R1054X heterozygous mutation; d patient 1, c.929G > A, p.G310E heterozygous mutation; e patient 1′s father, c.929G > A, p.G310E heterozygous mutation; f: patient 1′s mother, no c.929G > A, p.G310E mutation
Patient 2 was a 66-year-old female who carried the c.2872_2872del and the c.3710C > T variants (Fig. 2a, b). The two variants were not reported before. She presented with bilateral distal lower limbs weakness and atrophy in her thirties. The symptoms progressed slowly and the patient had difficulty climbing stairs in her sixties. Her parents were dead and they did not have the same presentation as the patient. She had no children. She has an elder sister without neuropathy symptoms who refused to provide DNA sample. Sanger test was not conducted on the patient’s family members due to the unavailability of their genomic DNA. Bioinformatic analysis indicated “pathogenic” of the two variants. Physical examination showed pes cavus and muscle atrophy of bilateral tibialis anterior muscles. Weakness of distal upper and lower limbs, more severe in distal lower limbs, was found. Gait abnormality was observed. Superficial and vibration perception were lost in distal lower extremities. Deep tendon reflexes were decreased, and bilateral Babinski signs were negative. NCS indicated demyelinating neuropathy and MCV of the median nerve was 32.4 m/s. (Table 2).
Patient 3 was a 49-year-old male who carried the c.2782C > T and the c.929G > A variants. The c.929G > A variant was reported in 2016 [18] and the c.2782C > T variant was not reported. He presented with gait abnormality and bilateral distal lower limbs weakness and atrophy in his 20 s. The symptoms progressed slowly. His parents and two children did not have the same presentation as the patient. He had no siblings. Sanger test confirmed that the patient’s mother carried the c.2782C > T variant, his daughter carried the c.929G > A variant and his son carried the c.2782C > T variant (Fig. 3a–h). His father died years ago without a Sanger test result. Bioinformatic analysis indicated “pathogenic” of the c.2782C > T variant. Physical examination showed scoliosis and muscle atrophy in bilateral thenar eminences, interosseous muscles and tibialis anterior muscles. Weakness of distal upper and lower limbs was found. Gait abnormality was observed. Superficial and vibration perception were lost in distal upper and lower extremities. Deep tendon reflexes were decreased, and bilateral Babinski signs were negative. NCS indicated demyelinating neuropathy and MCV of the median nerve was 30.0 m/s (Table 2).
a Patient 3, c.2782C > T, p.Q928X heterozygous mutation; b patient 3′s daughter, no c.2782C > T, p.Q928X mutation; c patient 3′s son, c.2782C > T, p.Q928X heterozygous mutation; d patient 3′s mother, c.2782C > T, p.Q928X heterozygous mutation; e patient 3, c.929G > A, p.G310E heterozygous mutation; f patient 3′s daughter, c.929G > A, p.G310E heterozygous mutation; g patient 3′s son, no c.929G > A, p.G310E mutation; h patient 3′s mother, no c.929G > A, p.G310E mutation
Discussion
The SH3TC2, the causative gene of CMT4C, codes for the SH3 domain and tetratricopeptide repeat domain 2 localized in the plasma membrane of the Schwann cells of peripheral nerves [8,9,10]. To date, more than 70 variants of SH3TC2 have been reported, and scattered along the whole protein (HGMD database, professional release, June 2017). In this study, we identified 3 patients with 5 SH3TC2 variants from 206 patients with a clinical diagnosis of CMT. Among the five mutations, the c.3154C > T and the c.929G > A variants have been reported [17, 18], while the c.2872_2872del, the c.3710C > T and the c.2782C > T variants have not been reported before. No homozygous variants were found, which may be partly due to legal prohibition of consanguineous marriage in China.
The c.2872_2872del variant caused frameshift deletion and was very strong (PVS1) evidence of pathogenicity. This variant was absent in either the 1000 genomes project, ExAC or East Asia ExAC. Bioinformatics analysis indicated pathogenic. The variant was likely pathogenic according to the ACMG guideline. The c.3710C > T variant was absented in the 1000 genomes project and present as a rare mutation in ExAC (minor allele frequency, MAF = 0.00003301) and East Asia ExAC (MAF = 0.0003). Bioinformatics analysis indicated pathogenic, and PROVEAN indicated that the variant affected the tetratricopeptide-like helical domain of the protein. The clinical presentation of Patient 2 was in accordance with CMT4C. It was of uncertain significance according to the ACMG guideline. The ACMG guideline may underestimate the pathogenicity of the above two variants owing to the unavailability of the family members’ genomic DNA [19]. In Patient 3, compound heterozygous variants, the c.929G > A and the c.2782C > T, were identified, which were cosegregated among his unaffected mother and children. The c.2782C > T variant was a novel null mutation which was PSV1 evidence of pathogenicity according to the ACMG guideline and was absent in the above public databases. Bioinformatics analysis indicated the variant was pathogenic, and PROVEAN indicated the variant affected the tetratricopeptide-like helical domain of the protein. The c.2782C > T variant was pathogenic according to ACMG guideline.
The c.929G > A variant was found in both Patient 1 and 3. In a Korean study, 4 CMT4C families were found in 504 CMT patients, and each family carried the c.929G > A variant [15]. In another Korean study, the c.929G > A variant was also identified in one family [18]. It indicates that the c.929G > A variant may be common in CMT4C, especially common in Asian CMT4C patients, which requires further investigations to confirm.
Patients with CMT4C usually present with remarkably broad spectrum of clinical phenotypes. The age of onset may vary greatly [11,12,13,14,15,16]. Spine deformities and cranial nerve involvement were reported in previous studies [20, 21]. Motor and sensory deficits were common manifestations of CMT4C [9, 22, 23]. Patients with the c.3154C > T and the c.929G > A variants usually present with foot deformities, mild scoliosis, distal weakness, areflexia, cranial nerve involvement, and hearing loss [17, 18]. In this study, the onset age ranged from childhood to middle age. Spine deformities were observed in Patient 1 and 3. Motor and sensory deficits were observed in the three patients. However, cranial nerve involvement was not found in the patients in our study. VEP and BAEP should be conducted to further assess subclinical cranial nerve involvement.
In this study, the patients were from 30 provinces, which covered most areas of China. Thus, the patients recruited can represent the prevalence of CMT in China. In our study, the frequency of CMT4C was 1.46% (3/206) of the whole cohort, and was 2.5% (3/120) in demyelinating/intermediate CMT negative for PMP22 duplication. The frequency of CMT4C was 1.76% of demyelinating/intermediate CMT negative for PMP22 duplication in Japan [9]. The frequency was 2.7% of demyelinating/intermediate CMT including PMP22 duplication in Germany [14]. The frequency was 0.79% of demyelinating/intermediate CMT (including PMP22 duplication) and 2.02% of demyelinating/intermediate CMT (negative for PMP22 duplication) in Korea [15]. The frequency was over 20% of AR demyelinating CMT (negative for PMP22 duplication, GJB1 and MPZ) in Italy [16]. The frequency of CMT4C was 0.8% of the CMT cohort (both demyelinating and axonal, including PMP22 duplication) in the USA [17]. The reasons for the different frequencies of gene mutations in different studies may include different genetic background and gene distribution among different areas, different methods of gene screening and different criteria for patient inclusion. Thus, standard gene screening strategy and inclusion criteria may help identify a more accurate frequency. Furthermore, studies with larger sample size are also helpful to reveal a more reliable frequency of CMT4C.
In general, CMT4 caused by SH3TC2 mutation is a common type of CMT4. With the advance of next-generation sequencing technologies including disease-specific gene panels, whole-exome sequencing, whole-genome sequencing, etc., novel likely pathogenic genes and mutations would be found increasingly [1, 2]. Our study suggests the importance of a comprehensive assessment with clinical phenotype, NCS and genetic analysis in patients with suspected CMT.
References
Sun B, Chen Z, Ling L et al (2017) Clinical and genetic spectra of Charcot-Marie-Tooth disease in Chinese Han patients [J]. J Peripher Nerv Syst 22(1):13–18. https://doi.org/10.1111/jns.12195 (PMID:27862672)
Sun B, Chen ZH, Ling L et al (2016) Mutation analysis of gap junction protein Beta 1 and genotype-phenotype correlation in X-linked Charcot-Marie-Tooth disease in Chinese patients [J]. Chin Med J 129(9):1011–1016. https://doi.org/10.4103/0366-6999.180511 (PMID:27098783)
Szigeti K, Lupski JR (2009) Charcot-Marie-Tooth disease. Eur J Hum Genet 17(6):703–710. https://doi.org/10.1038/ejhg.2009.31
Pareyson D (1999) Charcot-Marie-Tooth disease and related neuropathies: molecular basis for distinction and diagnosis. Muscle Nerve 22(11):1498–1509. https://doi.org/10.1002/(sici)1097-4598(199911)22:11%3c1498::aid-mus4%3e3.0.co;2-9
Gentile L, Russo M, Fabrizi GM et al (2020) Charcot-Marie-Tooth disease: experience from a large Italian tertiary neuromuscular center. Neurol Sci 41(5):1239–1243. https://doi.org/10.1007/s10072-019-04219-1
Lupo V, Galindo MI, Martínez-Rubio D et al (2009) Missense mutations in the SH3TC2 protein causing Charcot-Marie-Tooth disease type 4C affect its localization in the plasma membrane and endocytic pathway. Hum Mol Genet 18(23):4603–4614. https://doi.org/10.1093/hmg/ddp427
Reilly MM, Shy ME (2009) Diagnosis and new treatments in genetic neuropathies. J Neurol Neurosurg Psychiatry 80(12):1304–1314. https://doi.org/10.1136/jnnp.2008.158295
Laššuthová P, Mazanec R, Vondráček P et al (2011) High frequency of SH3TC2 mutations in Czech HMSN I patients. Clin Genet 80(4):334–345. https://doi.org/10.1111/j.1399-0004.2011.01640.x
Yuan JH, Hashiguchi A, Okamoto Y et al (2018) Clinical and mutational spectrum of Japanese patients with recessive variants in SH3TC2. J Hum Genet 63(3):281–287. https://doi.org/10.1038/s10038-017-0388-5
Arnaud E, Zenker J, de Preux Charles AS et al (2009) SH3TC2/KIAA1985 protein is required for proper myelination and the integrity of the node of Ranvier in the peripheral nervous system. Proc Natl Acad Sci USA 106(41):17528–17533. https://doi.org/10.1073/pnas.0905523106
Roberts RC, Peden AA, Buss F et al (2010) Mistargeting of SH3TC2 away from the recycling endosome causes Charcot-Marie-Tooth disease type 4C. Hum Mol Genet 19(6):1009–1018. https://doi.org/10.1093/hmg/ddp565
Yger M, Stojkovic T, Tardieu S et al (2012) Characteristics of clinical and electrophysiological pattern of Charcot-Marie-Tooth 4C. J Peripher Nerv Syst 17(1):112–122. https://doi.org/10.1111/j.1529-8027.2012.00382.x
Manganelli F, Tozza S, Pisciotta C et al (2014) Charcot-Marie-Tooth disease: frequency of genetic subtypes in a Southern Italy population. J Peripher Nerv Syst 19(4):292–298. https://doi.org/10.1111/jns.12092
Rudnik-Schöneborn S, Tölle D, Senderek J et al (2016) Diagnostic algorithms in Charcot-Marie-Tooth neuropathies: experiences from a German genetic laboratory on the basis of 1206 index patients. Clin Genet 89(1):34–43. https://doi.org/10.1111/cge.12594
Lee AJ, Nam SH, Park J-M et al (2019) Compound heterozygous mutations of SH3TC2 in Charcot–Marie–Tooth disease type 4C patients. J Hum Genet 64:961–965. https://doi.org/10.1038/s10038-019-0636-y
Piscosquito G, Saveri P, Magri S et al (2016) Screening for SH3TC2 gene mutations in a series of demyelinating recessive Charcot-Marie-Tooth disease (CMT4). J Peripher Nerv Syst 21(3):142–149. https://doi.org/10.1111/jns.12175
DiVincenzo C, Elzinga CD, Medeiros AC et al (2014) The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy. Mol Genet Genomic Med 2(6):522–529. https://doi.org/10.1002/mgg3.106
Nam SH, Hong YB, Hyun YS et al (2016) Identification of genetic causes of inherited peripheral neuropathies by targeted gene panel sequencing. Mol Cells 39(5):382–388. https://doi.org/10.14348/molcells.2016.2288
Murphy SM, Herrmann DN, McDermott MP et al (2011) Reliability of the CMT neuropathy score (second version) in Charcot-Marie-Tooth disease. J Peripher Nerv Syst 16(3):191–198. https://doi.org/10.1111/j.1529-8027.2011.00350.x
Lee AJ, Nam SH, Park JM et al (2019) Compound heterozygous mutations of SH3TC2 in Charcot-Marie-Tooth disease type 4C patients. J Hum Genet 64(9):961–965. https://doi.org/10.1038/s10038-019-0636-y
Forrester N, Rattihalli R, Horvath R et al (2020) Clinical and genetic features in a series of eight unrelated patients with neuropathy due to Glycyl-tRNA synthetase (GARS) variants. J Neuromuscul Dis 7(2):137–143. https://doi.org/10.3233/JND-200472
Senderek J, Bergmann C, Stendel C et al (2003) Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. Am J Hum Genet 73(5):1106–1119. https://doi.org/10.1086/379525
Azzedine H, Ravisé N, Verny C et al (2006) Spine deformities in Charcot-Marie-Tooth 4C caused by SH3TC2 gene mutations. Neurology 67(4):602–606. https://doi.org/10.1212/01.wnl.0000230225.19797.93
Funding
The study is financially supported by National Natural Science Foundation of China: The study of long non-coding RNA in the pathogenesis of CMT1A (81870989) and the study of mitochondrial unfolded protein response in the pathogenesis of CMT1B (81901274).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest.
Ethical approval
The study was approved by the Chinese PLA General Hospital Ethics Committee. Informed written consent was obtained from all the patients enrolled in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, B., He, ZQ., Li, YR. et al. Screening for SH3TC2 variants in Charcot–Marie–Tooth disease in a cohort of Chinese patients. Acta Neurol Belg 122, 1169–1175 (2022). https://doi.org/10.1007/s13760-021-01605-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13760-021-01605-5