Abstract
Our research focuses on demonstrating the existence of cryptic species named under Biblis aganisa Boisduval. We used COI sequences to delimit Biblis species for Mexico using species delimitation analyses and examined phylogenetic relationships with sequences from Mexico, Costa Rica, Argentina, USA, and Guana Island using a Bayesian inference tree. We performed a discriminant analysis with quantitative traits using female and male wing and genitalia, and a tree of maximum parsimony based on 39 qualitative characters of wings, head, and male genitalia. The results were congruent in the three analyses. Three groups were formed based on DNA, ECO 01 + DHJ02, ECO 02 + DHJ01, and ECO 03. The characters that contributed over 50% separation were for wings: wing length, anal margin length, and distance from the band to the outer margin; for male genitalia, angle of the integument, uncus, and the length of the hypandrium, while for females, it was the angle of the anteapophysis and the length of the abdomen. For the analysis of qualitative characters, a tree of maximum parsimony was obtained where 20 characters were informative. We confirmed the existence of three cryptic Biblis species in Mexico, two not yet described, and one corresponding to B. aganisa (ECO 02), which is sympatric in Oaxaca and Sinaloa (ECO 03) and in the Yucatan Peninsula (ECO 01).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Identifying species based on the DNA barcode has proven to help distinguish and discover new species (Blaxter 2003; Hallwachs et al. 2008; Janzen et al. 2017; Gaytán et al. 2020; D'ercole et al. 2021). The premise is that sequence diversity within a short, standardized segment of the mitochondrial genome (mtDNA) can provide a unique biological barcode, allowing species-level identifications; it presents a rapid mutation rate, a significant variation in sequences between different species, and a low variation within the same species (Hebert et al. 2003; Hajibabaei et al. 2006). A barcode system analyzes lineages whose species may present genetic divergence (generally greater than 2% in vertebrates and 3% in Lepidoptera: Hebert et al. 2003; Sperling 2003). However, this is not a substitute for a complete taxonomic study, and the use of barcodes must be combined with morphological and ecological data for the results to be decisive for the final recognition of the species (Hebert et al. 2004b; Sperling 2003).
Different studies have used the barcode in the Insecta class and have uncovered many cryptic species (e.g., Hallwachs et al. 2008; Seraphim et al. 2014; Nieukerken et al. 2012; Kim et al. 2020; Moraes et al. 2021). Cryptic species are morphologically similar taxa determined under a single name (Bickford et al. 2007; Gill et al. 2016). They may differ in biological attributes, such as host specificity, mating system, and susceptibility to different parasitoids, among others (Hebert et al. 2004a; Bickford et al. 2007; Pfenninger and Schwenk 2007; Trontelj and Fier 2009).
An excellent example of the usefulness of the COI, as a tool in molecular systematics, is in the genus Hermeuptychya Fabricius. The first publication with the use of this gene (Seraphim et al. 2014) revealed the unknown cryptic diversity within this genus and led to the discovery of new and corroborated species with morphological analysis of genitalia (Cong and Grishin 2014; Cong et al. 2021; Nakahara et al. 2017). This method has also been used to detect cryptic species of Astraptes fulgerator Walch (Hebert et al. 2004a) but not enough evidence has been presented to confirm this (Brower 2006). In another study, Janzen et al. (2017) confirmed the existence of cryptic species within the species previously known as Udranomia kikkawai Weeks (Hesperiidae), identifying two more species using complete genome sequencing and barcode (COI). These species presented subtle difference in adult color variation, size, ecological distribution, and natural history.
Jasso-Martínez et al. (2016) solved the taxonomic problem regarding the number of species within the Mexican species of the Enantia jethys (Boisduval) complex (Pieridae), using COI as a molecular character. They confirmed the three species reported in the literature and a fourth potential cryptic species not yet described. Later Jasso-Martínez et al. (2018) demonstrated hybridization events in three species of the genus Enantia that occur in partial sympatry using different genes. Burns et al. (2008) analyzed a complex of cryptic species of the genus Perichares Scudder (Hesperiidae: Hesperiinae) from northwestern Costa Rica based on genetic information and their relationship with host plant. They found that information on the host plants of this genus constitutes an ecological characteristic which helps distinguish female oviposition preferences of each species.
The genus Biblis Fabricius, 1807 (Lepidoptera: Nymphalidae), is monotypic and presents five described subspecies in the Americas and one putative subspecies from Peru (Lamas 2004). The type localities of these subspecies are the following: Biblis hyperia hyperia Cramer from St. Thomas, “Indies Occidentales”; Biblis hyperia laticlavia Thieme from Rio Napo in Ecuador; Biblis hyperia nectanabis Fruhstorfer from Rio Grande do Sul, Brazil; Biblis hyperia pacifica Hall from Huigra, Ecuador; Biblis hyperia aganisa Boisduval from “Java” (although Godman and Salvin (1879) mention that the locality of this species is an error and that the type specimen originates from Mexico, without stating the specific locality), and finally, Biblis hyperia nov. ssp., from Peru (Lamas 2004).
Initially, Biblis aganisa Boisduval, was described as a species, then changed to a subspecies of B. hyperia without sufficient evidence to justify this change (Lamas 2004). The same process occurred with B. laticlavia, described as a species easily recognized by its broad submarginal band of the posterior wing on the dorsal and ventral view and the marked difference in the size of each of the spots that make up this band, very different to other species of this group. There are also considerable differences in the male genitalia therefore could be considered a good species. Two more members, B. h. pacifica, and B. h. nectanabis, were described as subspecies and synonymized of B. hyperia, with very subtle differences in the width and coloration of the submarginal band.
Prado et al. (2011) examined the diversity of larvae and adults of the Nymphalidae (Papilionoidea) from the Yucatan Peninsula using Cytochrome C Oxidase I (COI), including B. hyperia, having shown divergence percentages above 3% in the sequences in individuals from the same geographic area, suggesting the presence of new species. B. hyperia was split into two separate groups with an average divergence of 4.6% and were named with a provisional OTU: Biblis hyperia ECO 01 and Biblis hyperia ECO 02.
However, no detailed studies include molecular characters combined with morphological data to verify the status of subspecies of B. hyperia. Recently, Zhang et al. (2021) proposed B. aganisa as a valid species and will classified as a distinct species from B. hyperia. However, morphological data are not included, and the only specimen used representing B. aganisa does not originate from Mexico (country where type specimen was collected). According to this study, the genus Biblis would be composed of two subgenera Biblis and Vila Kirby, 1871, with a separation percentage of 7% (46 bp) between both subgenera and 4.6% (30 bp) between species in the subgenera Biblis, which should be considered non-monotypic since it would be composed of the species of each of these subgenera. In the case of Vila, three valid species are known; two of these were considered in his study (Vila azeca E. Doubleday and Vila euidiformis Joicey & Talbot).
A similar case is found in the public database in GenBank/Bold System where Janzen and Hajibabaei (2009) registered sequences of Biblis specimens from Costa Rica, which were assigned to three BINs (BOLD: AAC6005, ABY4876, and AAC0692). BINs are a group of sequence assigned to an operational taxonomic unit (OTU), that often correspond to species (Ratnasingham and Hebert 2013). Janzen and Hajibabaei assigned a name to each group of sequences: Biblis sp. hyperiaDHJ01 (n = 5), Biblis sp. hyperiaDHJ02 (n = 4), and Biblis sp. hyperiaDHJ03 (n = 14). Out of a total of four BIN registrations in BoldSystem (http://www.boldsystems.org/index.php/Public_BarcodeIndexNumber_Home), two Biblis BINs share Mexico and Costa Rica as occurrence points.
According to the 4.6% separation percentage between the two groups of the Yucatan Peninsula in Mexico, there are two possible cryptic species (Prado et al. 2011) and maybe three cryptic species in Biblis from Costa Rica (according to the OTUS of Janzen and Hajibabaei on GenBank, not yet thoroughly analyzed). The groups from Mexico were determined using only a few specimens (11 specimens: Biblis hyperiaECO01 (1) and Biblis hyperiaECO 02 (10)) and by a single gene fragment (COI).
The study by Zhang et al. (2021) shows that the Mexican species differs from B. hyperia; therefore, continuing to consider the groups from Mexico as B. hyperia, as used in Prado et al. (2011), would be incorrect. The Mexican species from Oaxaca, B. aganisa, which was previously described as a species and now its species status has re-emerged. From here on, we will refer to the Biblis of Mexico groups as B. aganisa stat. rev.
There is no study that includes an analysis of morphological characters and sufficient samples from different sites in Mexico to verify if there are cryptic species named as B. aganisa. Therefore, the evidence is insufficient to confirm whether two or more cryptic Biblis species occur in Mexico. Consequently, it is essential that more detailed morphological and genetic studies are carried out in order to confirm the potential new cryptic species of the genus Biblis in the Americas, and specifically within the Yucatan Peninsula, Mexico.
The present study amplifies the molecular analyzes using the markers mtDNA (COI) and two nuclear genes (MDH and GAPDH). Furthermore, a morphological study verifies the existence of cryptic species in B. aganisa. Considering the following questions more specifically: (1) Are there adult morphological characters that support the presence of two or more cryptic species of Biblis? (2) Is there congruence among the groups formed by molecular analysis and by the morphometric analysis of adults? (3) Does the information from points 1 and 2 allow us to identify how many cryptic Biblis species occur? And finally (4) in which geographic areas are these species sympatric.
Methods
Biological Material
A total of 171 specimens of B. aganisa collected in the states of Campeche, Quintana Roo, Yucatan, Tabasco, Oaxaca, Queretaro, and Zacatecas and deposited in the Lepidoptera Collection of the Museo de Zoología of ECOSUR-Chetumal (ECOCH-L) and in the Lepidoptera Collection from the Museo de Zoología de la Facultad de Ciencias (MZFC) of the Universidad Nacional Autónoma de México (UNAM) were selected for morphological and genetic analyses. In addition, we downloaded COI gene sequences of the Biblis genus in GenBank (Benson et. al 2014): eleven sequences used by Prado et al. (2011), 22 sequences from Costa Rica, nine sequences from Argentina, four Biblis (two from the subgenera Biblis and two from the Vila) sequences used by Zhang et al. (2021), and finally one from V. azeca (GQ864818.1). Five sequences from Nymphalidae species of GenBank were taken as an outgroup (Colobura annulata Willmott, Constantino & J. Hall, Archimestra teleboas Ménétriés, Mestra amymone Ménétriés, Mestra hersilia Fabricius, and Mestra dorcas Fabricius) for molecular analyses, already used in other systematic studies (Chazot et al. 2020; Wahlberg et al. 2005a, b, 2009) (complete data in Supplementary Information, excel sheet). To obtain the sequences of COI of B. hyperia (NVG-19094E05), B. aganisa (NVG-17117F03), V. azeca (NVG-19095B04), and V. eueidiformis (NVG-19095B05) of the project (PRJNA731937) (Zhang et al. 2021), we conducted the following treatment: cleaning the adapters with the Trim using BBDuk tool in Geneious Prime 2020.2.3 (Kearse et al. 2012); subsequently, once the readings were cleaned, we created an assembly or COI mapping of these four species with the reference sequence used by Zhang et al. (2021); for later study, the consensus sequences were included in the alignment.
DNA Extraction, Sequencing, and Phylogenetic Analyses
The DNA extraction, amplification, and sequencing of the specimens of the Biblis from Mexico (N = 160), Costa Rica (N = 22), and Argentina (N = 9), and the five outgroups, were made considering the protocols from Hajibabaei et al. (2005) and Ivanova et al. (2006). Sequencing was carried out using COI primers with demonstrated efficacy in butterfly studies (Jasso-Martínez et al. 2016; Vodă et al. 2015), amplifying a ~ 657-bp fragment: LepR1 (5′-TAAACTTCTGGATGTCCAAAAAATCA-3′) and LepF1 (5′-ATTCAACCAATCATAAAGATATTGG-3′). Additionally, two nuclear genes were sequenced (for N = 17 specimens of Biblis from Mexico) that have been effective for butterfly phylogenetic studies (Wahlberg and Wheat 2008; Wahlberg et al. 2016) and cryptic species delimitation (Tóth et al. 2014): (1) GAPDH (HybFrigga 5′-TAATACGACTCACTATAGGGAARGCTGGRGCTGAATATG t-3′ and HybBurre 5′-ATTAACCCTCACTAAAGGGGWTTGAATGTACTTGATRAG RTC-3′) and (2) MDH (HybMDHF 5′-TAATACGACTCACTATAGGGGAYATNGCNCCNATGATGGGNGT-3′ and MDHmidR 5′-ATTAACCCTCACTAAAGGGAAYTGNGTRGATGARTGRTTNCC-3′).
A total of four alignments were realized consisting of one for each gene separately (COI, MDH, and GAPDH), and a concatenated alignment (COI + MDH + GAPDH) with 17 Mexican specimens for which we have the three genes sequenced plus one specimen of B. hyperia from GenBank (voucher number NW106-3) from Campinas, Sao Paulo, Brazil. All alignments were made in the Geneious Prime 2020.2.3 program (Kearse et al. 2012) using the Muscle 3.8.425 option and a refined alignment (Robert 2004; Edgar and Batzoglou 2006). The Mega X program (Kumar et al. 2018) was used for selecting the evolutionary model, which selected the GTR + G + I model with a lower BIC value (Huelsenbeck and Rannala 2004; Lecocq et al. 2013).
Before running the phylogenetic analysis, we applied two species delimitation methods to evaluate the informative potential of each gene and decide which ones to use for the phylogenetic tree: (1) ABGD (Automatic Barcode Gap Discovery) is an automatic procedure that classifies the sequences based on the width of the barcode, which is observed in the divergence between members within the same species and is always lower between different species (Puillandre et al. 2012). This analysis was performed with distance matrices according to the Kimura model (K80) and the default parameters; (2) Assemble Species by Automatic Partitioning (ASAP) is similar to ABGD and in both analyses the species hypothesis is indicated thanks to the partitions. In this analysis, distance matrices used the Kimura (K80) ts/tv 2.0 (Puillandre et al. 2021). Both analyses were ran on the online servers (ASAP: https://bioinfo.mnhn.fr/abi/public/asap/; ABGD: https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html).
A phylogenetic tree was constructed using the Bayesian inference (Huelsenbeck and Ronquist 2001; Holder and Lewis 2003). Trees under the Bayesian inference were constructed for the COI gene using MrBayes (Huelsenbeck and Ronquist 2001). Analyses considered a gamma distribution with five categories and a percentage of invariant sites; MCMC chains ran for 5 million generations, 25% of the trees were discarded (burned), and a consensus tree was calculated. The posterior probabilities summarized in the MrBayes consensus tree were drawn on the clade maximum credibility tree as recommended by García-Sandoval (2014) for better visualization. The mean intraspecific and interspecific sequence divergences were estimated using the Kimura two-parameter (K2P) distance model (Kimura 1980) in MEGA X (Tamura et al. 2011).
Finally, we constructed a Haplotype network with the alignment of 214 COI sequences based on statistical parsimony implemented in TCS version 1.23 (Clement et al. 2000). The haplotypes were classified with the tcsBU program (Múrias Dos Santos et al. 2016), and later the network was stylized in Photoshop 2020 to improve its presentation.
Morphological Analyses
Based on our results of the genetic analysis, we conducted morphological measurements on 148 specimens (85 males and 63 females) organized into three groups (ECO 01 (N = 4), ECO 02 (N = 137), and ECO 03 (N = 7)). For each specimen of both sexes, quantitative and qualitative characters were taken from the dorsal and the ventral view of the wings and male and female genitalia. For the genitalia analysis, abdomens were removed and placed in labeled glass jars with a 10% sodium hydroxide (KOH) solution for 1 or 2 days (depending on how dry the abdomens were). Genitalia was removed using a stereoscopic microscope, the fat and tissue using forceps, and a fine-tip dissecting needle, and scales were removed with a fine-bristle brush. Clean genitalia (including hypandrium) were placed on concave slides with 70% ethanol or glycerol. Lateral, dorsal, and ventral view photographs were taken using a millimeter scale to obtain later measurements and comparisons (Zubek et al. 2015). Image J software was used for the measurements, calibrated with the millimeter scale each time a new photograph was used (Abramoff et al. 2004; Collins 2007).
Quantitative Characters of Wings
The following twelve dorsal (D) and ventral (V) measurements were taken with a digital vernier, following the nomenclature of the wings according to Miller 1970. An alphanumeric key was assigned as described below: wing length (D1), distance from band to disc cell (D2), band distance to outer margin (D3), spot length in cell M3-CuA1 (D4), spot length in Rs-M1 (D5), spot length in cell CuA2-1A + 2A (D6), band to disc cell distance (V1), distance from band to outer margin (V2), spot length in cell M3-CuA1 (V3), length of the spot in cell Rs-M1 (V4), spot length in CuA2-1A + 2A (V5), and anal margin length (V6) (Supplementary Information: Fig. 1).
Quantitative Characters of Genitalia
The following ten male genitalia measurements were annotated: aedeagus length (EL), valva length (VL), tegument length (TL), uncus length (UL), tegumen angle (TA), tegumen to saccus distance (TSD), uncus angle (UA), saccus length (SL), valva width (VA), and androchondrial patch length (APL) (Supplementary Information: Fig. 2a). Additionally, measurements on the hypandrium were made: length (HL), width (HW), and angle (HA) (Supplementary Information: Fig. 2b). For the female genitalia: abdomen length (AL), width of ostium bursae (A), shortest distance between anteapophysis (B), longest distance between anteapophysis (C), length of anteapophysis (D), height of ostium bursae (E) (Supplementary Information: Fig. 2c), length of copus bursae (F), length of ductus bursae (G), width of papilla analis (H), and angle of anteapophysis (I) (Supplementary Information: Fig. 2d).
Four discriminant analyses were made based on the female and male wings and genitalia looking for groups within B. aganisa. Additionally, we performed a similarity percentage analysis (SIMPER) considering Euclidian distances as a dissimilarity measure for female and male wings and genitalia separately, to evaluate the contribution of the different measures. These analyses were processed with PAST version 4.03 (Hammer et al. 2001).
Qualitative Characters of Wings and Genitalia
A total of 23 characters were taken, five from dorsal view and 18 from ventral view of female and male (Supplementary Information: Figs: 3–5). A total of fifteen characters from male genitalia and one of the head (labial palps) were encoded (Supplementary Information: Figs. 5–9, and Tables 1 and 2). Using the matrix generated from the qualitative characters of wings of male and female, male genitalia, and head of male, a phylogenetic relationship based on maximum parsimony method was constructed using PAUP V4.0a (Swofford 2001).
Geographical Distribution
Using the coordinates of the 160 Mexican specimens belonging to the three groups (ECO 01 (4), ECO 02 (149), ECO 03 (7)), a distributional map was elaborate with the ArcGis program version 10.2.1
Results
Species Delimitation
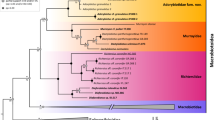
Analyses ran for each nuclear gene did not allow the delimitation of species inside the Biblis genus while regarding COI gene, we obtained an adequate species delimitation. Consequently, all further analyses were run using exclusively COI sequences. The best partitions generated by ASAP analysis with COI (including outgroup) consider five, nine, 11, and 12 species delimitation which correspond to one, three, four, and five possible species delimitation within the Biblis genus (Fig. 1). Additionally, other ASAP partitions were less probable (two, three, four, 13, and 60 with outgroups), and therefore were not considered. Examination of the ASAP score (the lower the score, the best partition) revealed that the optimal partition for species delimitation in Biblis is three species (Fig. 1; ASAP score 3). This implies that B. hyperia, B. nectanabis, and the group including ECO1 are only one species. We discarded this hypothesis because each of these species’ group presented morphological differences at the level of the extern red band (http://butterfliesofamerica.com/L/t/Biblis_hyperia_a.htm); thus, the next best partition is five internal species for the Bilbis genus (ASAP score = 5) which coincides with our phylogenetic analyses. With ABGD, four of the same groups were recovered with recursive partitioning (p = 0.004642), while ECO 01 + NW106-3 (Mexico-Brazil) separated from DHJ02 (Costa Rica), which also presents a difference at the level of the red band within the group, and that should be evaluated at a later stage at the genitalia level, as with the groups in Mexico (Fig. 1).
Bayesian phylogenetic relationship based on COI gene for the species of Biblis Fabricius (Nymphalidae: Biblidinae). The numbers in the clades correspond to the posterior probability values supporting the clade. The colors of the branches indicate species, and the same color on the right side of the bars present the results of the two species delimitation methods. The numbers below the bars indicate the number of species or groups recovered
Phylogenetic Analyses
The Bayesian phylogeny constructed using COI gene dataset (Fig. 1; 214 sequences: 22 from Costa Rica, nine from Argentina, 171 from Mexico, one from EU, and one from the British Virgin Islands: Guana Island) revealed two well-supported sister clades: (1) the first clade is confirmed by the majority of Costa Rica samples (DHJ03, DHJ02), samples from Argentina (B. nectanabis), and four samples from Mexico belonging to the ECO 01. The only sample of B. hyperia was combined with DHJ03. Remarkably, these four Mexican samples are pooled with some Costa Rica samples (DHJ02) and they are sister to Argentina samples; (2) a second clade consists of the remaining Mexican samples, with very good separation between the ECO 03 and ECO 02 samples. Some Costa Rica samples (DHJ01) and B. aganisa are pooled with ECO 02 samples from Mexico in this clade.
The results of percentages of identity among the species confirm the results of phylogeny (Table 1). The samples from Mexico (ECO 01), Costa Rica, and Brazil present very high values (from 99.22 to 99.81%); in contrast, ECO 01 present lower values with Costa Rica (DHJ03; 97.48%), with Argentina (B. nectanabis) 97.67%, and with B. hyperia from Guana Island (95.54%). The clade including the two other groups from Mexico (ECO 02 and ECO 03) presents a percentage of identity of 96.12%. Our samples identified as ECO 02 (Mexico) present maximum values with samples from Costa Rica (DHJ01), the sample from Texas, and B. aganisa.
The genetic divergence values (Table 2) were obtained in the groups of the first clade, comprised Costa Rica, Argentina, Guana Island, and Mexico. In the group composed by ECO 01, Costa Rica (DHJ02), and Brazil (NW106-3), the mean intraspecific distance was 0.42%. However, between this group and Costa Rica (DHJ03), the interspecific distance was 2.80%, and 2.44%. with Argentina (B. nectanabis). The second clade, composed primarily of samples from Mexico (ECO 02), some from Costa Rica (DHJ01), and a sample of B. aganisa from Hidalgo (Texas, USA), presents an intraspecific divergence of 0.22%. This clade presented 4.12% of divergence with ECO 03. The most considerable divergence between the groups from Mexico (ECO 01 and ECO 03) was 5.85%.
The haplotype network analysis of 214 specimens including outgroups and all specimens from Biblis resulted in the formation of five haplogroups (Fig. 2). In Fig. 2, the blue dotted circle is composed of specimens from Mexico (ECO 01), Costa Rica (DHJ02), and Brazil (NW106-3). Green haplotypes regroup specimens from Mexico (ECO 02), Costa Rica (DHJ01), and USA (B. aganisa). The specimens from Mexico (ECO 03) are shown in pink, while specimens from Argentina (B. nectanabis) are in yellow, and finally the haplogroup formed by individuals from Costa Rica (DHJ03) and the British Virgin Islands (B. hyperia) are in pistachio green.
Haplotype network based on COI barcodes of Biblis and outgroup that provide evidence of the 5 haplogroups of Biblis that correspond to the phylogenetic tree. The size of the colored circle is proportional to the number of samples represented. Small uncolored circles represent unsampled haplotypes or mutational step difference
Morphological Analyses
Quantitative Characters
The discriminant analysis based on male wings’ characters separated the samples into three groups (Fig. 3A; ECO 01 in blue, ECO 02 in green, and ECO 03 in fuchsia). ECO 01 is completely separated from the two remaining groups, while a slight overlap is noted between ECO 02 and ECO 03. The SIMPER analysis based on male wing measurements (Fig. 10A, Supplementary Information) indicates that three variables could explain 50% of the separation between groups: wing length (D1), anal margin length (V6), and band to outer margin (V2).
Discriminant analysis with morphometric measurements of the wings and genitalia of the male and female. A Male wings, B female wings, C genitalia in male, and D genitalia in female. Filled blue squares: Biblis aganisa ECO 01, filled green triangles: Biblis aganisa, ECO 02, and pink filled diamonds: Biblis aganisa ECO 03
The discriminant analysis, considering the characters of females’ wings (Fig. 3B), also resulted in the separation of three groups. In this case, ECO 03 (fuchsia) is separate from the two other groups, while a slight overlap is observed between ECO 02 (green) and ECO 01 (blue). The SIMPER analysis (Fig. 10B Supplementary Information) revealed that the same characters of wing females explain 50% of the separation between groups: wing length (D1), anal margin length (V6), and band to outer margin (V2).
The discriminant analysis based on male genitalia characters (Fig. 3C) results in the formation of three groups (ECO 01, ECO 02, and ECO 03) without any overlap. The SIMPER analysis (Fig. 10C, Supplementary Information) indicates that only two characters explain 90% of the separation of the three groups: tegumen angle (TA) and uncus angle (UA).
The discriminant analysis based on female genitalia characters (Fig. 3D) also results in the separation of three groups (ECO 01, ECO 02, and ECO 03) without any overlap. The SIMPER analysis (Fig. 3D, Supplementary Information) indicates that only two characters explain 90% of the separation of the three groups: angle of anteapophysis (I) and length of abdomen (AL).
Qualitative Characters
Maximum parsimony analysis of the qualitative characters of the wings, head (labial palps), and the male genitalia produced a tree with three terminal branches: ECO 01, ECO 02, and ECO 03, and two terminal branches in a different clade: B. hyperia and B. h. laticlavia (Fig. 11, Supplementary information). We use a matrix of 39 characters (23 from the wings, 1 from the head, and 15 from male genitalia; Table 1 in the Supplementary information). This character matrix resulted in a single, more parsimonious tree of 55 steps with a consistency index (CI) equal to 0.8182 and a retention index (RI) equal to 0.6154. The informative characters were band shape, spot color in cell Sc + R1, spines on the distal part of the hypandrium, proximal portion of the uncus in lateral view, distal portion of the uncus bifurcated in dorsal view, proximal portion of the uncus in dorsal view, tegumen in dorsal view, saccus in lateral view (size and shape), and valvae in ventral view (size) (Table 1 and Figs. 4–9, Supplementary Information). Of the characters with the best CI fit and apomorphies, ECO 03 presents a postdiscal red dot in cell CuA1 of the anterior wing in dorsal view. The point size in cell Sc + R1 is not vestigial, and the color of this point is not totally red (it contains white scales). The basal points do not present white scales. The proximal portion of the hypandrium is slightly narrow in the center. The tegumen in the lateral view is slightly curved, and the proximal portion of the valvae in the lateral view is flattened. ECO 02, the thin red band pattern, is present in ventral view of the posterior wing (as in the type specimen of B. aganisa), and the spot size in the cell Sc + R1 can be medium and white. The saccus in lateral view can be straight or curved. ECO 01 does not present a clear broadband pattern, the cilia on the labial palps in the middle segment in the inner part only with few white cilia. The distal portion of the hypandrium is semi-rectangular, the valvae in ventral view are slender, and the distal portion of the valvae in lateral view is wider than the width of the valvae. A comparison of these diagnostic characters of the groups can be seen in more detail in Table 3 (Supplementary Information).
Geographical Distribution
The map shows the records of the three groups, i.e., ECO 01, ECO 02, and ECO 03, in México (Fig. 4) highlighting a broad distribution for B. aganisa ECO 02 (green circles), predominantly in southern and southeastern Mexico with records in the Yucatán Peninsula (Quintana Roo, Yucatán and Campeche states), Chiapas, and Oaxaca and only two record from Queretaro and Zacatecas. Meanwhile, the records for the B. aganisa ECO 01 (blue circles) have a restricted distribution in the southern region of the Yucatan Peninsula (Quintana Roo and Campeche states), near the border with Belize and Guatemala. Finally, B. aganisa ECO 03 (pink circles) presents a restricted distribution in the north of Oaxaca and Sinaloa in northern Mexico.
The map indicates several sympatric areas between groups. The ECO 01 and ECO 02 groups present a sympatric distribution in the southern part of the Yucatan Peninsula, more precisely in the Calakmul region of Campeche. Furthermore, ECO 02 and ECO 03 are in sympatry in the north of Oaxaca (Fig. 4).
Discussion
Previous studies with molecular traits using COI in B. hyperia have documented the separation of B. hyperia, establishing the possibility that it is a complex of cryptic species. Prado et al. (2011) suggested the division into two species when analyzing the specimens in the Yucatan Peninsula. Later, Zhang et al. (2021) recognized that the typical subspecies is different at a specific level from the taxon of north and central America, renaming it B. aganisa. Our study confirms the result of Zhang et al. (2021) when comparing all our samples with the B. aganisa and B. hyperia sequences, which are grouped into different clades. Moreover, all the samples of the ECO 02 group are the same as B. aganisa. Our research confirms previous findings by Prado et al. (2011) by studying a more significant number of samples; in addition, a third lineage (ECO 03) is recognized (Fig. 1). Furthermore, in GenBank/BoldSystem, Janzen and Hajibabaei (2009) recognize three lineages from Costa Rica (DHJ01, DHJ02, and DHJ03), two of which are genetically close to the lineages of Mexico (ECO 01 with DHJ02 and ECO 02 with DHJ01) (Fig. 1). The three lineages discovered with specimens from Mexico are strongly supported by the Bayesian analysis using COI. The genetic distances according to the Kimura model (Kimura 1980) between groups or species, found both by Prado et al. (2011) and by Zhang et al. (2021), established that there is a valid difference at a specific level, having a threshold above 4% with COI for Biblis and above 3% for Lepidoptera as suggested by Hebert et al. (2003). In our study, genetic distances between ECO 01 and ECO 02 were similar to Prado et al. (2011). In addition, the new group (ECO 03) presents a distance of 5.87% from ECO 01 and 4.12% from ECO 02; this is not surprising since their biogeographic areas (Fig. 4) are different from their evolutionary history (Morrone 2019). Additionally, the results of the haplotype network, ASAP, and ABGD analysis corroborated the groups formed based on the genetic distances and tree topology.
The use of discriminant analysis in other groups of Lepidoptera using measurements of parts of the wings and structures of the male genitalia has been previously used to delimit species (Kolev 2005; Hernández-Roldán and Munguira 2008; Prieto et al. 2009; Núñez et al. 2021) and even to confirm cryptic species (Dincâ et al. 2011). According to the discriminant analyses in which linear measurements were used in wing characters (Fig. 3A, B), the results were consistent with the Bayesian tree. This method was effective for species discrimination, as there was no overlap between the three groups. The male genitalia of the three groups are similar, but when applying this analysis, the difference is clear, as there is no overlap between the groups (Fig. 3C, D). Hypandrium examination has been helpful for reliable diagnosis in the delimitation of species within Biblidinae, despite only shape and proportions being used (Jenkins 1990; Leite et al. 2017; Zubek et al. 2015). In our study, the length and angle of the hypandrium were two fundamental measures to support the separation. The impossibility of differentiation by a simple observation means that these three species must be considerate cryptic species.
Phylogenetic studies where molecular and morphological characters are included are crucial to resolve relationships between species, particularly in unresolved nodes that become more robust when more characters are added to the analysis (Wahlberg and Nylin 2003). Moreover, if there is a good selection of characters, the results may be consistent with genetic analyzes (Shi et al. 2015). The cladistics analysis where we included 39 morphological characters (qualitative characteristics) of wings and genitalia of a single more parsimonious tree produced results that were consistent with the molecular phylogeny, since the same relationships were recovered between groups (ECO 01, ECO 02 + ECO 03).
Some of the characters that contribute to the formation of these groups, for example in the ECO 02 group, which is the most variable in its coloring pattern, present an exclusive characteristic that coincides with that presented in the type specimen of B. aganisa described by Boisduval of “Java” in 1836, specifically that in the ventral view, the submarginal band is thin and red scales predominate. However, this unique feature is rare since it was present in only 8% of all individuals. In addition, this group presents another variant of the submarginal band, which is wide clear, with a few red scales, which resembles the type specimen of Didonis pasira Doubleday, which is currently considered synonymous with B. aganisa. Therefore, we suggest that ECO 02 is the group that represents B. aganisa. However, this form of the submarginal band is not exclusive to the group since it is also observed in individuals of ECO 03.
The ECO 01 group shares the pattern of the intermediate submarginal band with both groups (Fig. 3, Supplementary Information); however, the pinkish scales are absent in the submarginal area of the anterior wing in ventral view, a characteristic that is also shared with the other two groups. We also observed a red spot in cell CuA1 of the forewings in dorsal view within the ECO 03 group; however, this is not present in all individuals. When examining the qualitative characteristics of the three groups individually, we consider that it is difficult to identify any group since they share several characteristics with the other groups; therefore, it is not very reliable to use only qualitative characters to discriminate species.
The data on the distributions of the sequenced specimens was necessary to detect sympatry areas and delimitation of the groups. One specimen from Campinas, Sao Paulo, Brazil (NW106-3), is related to ECO 01 from Mexico. Nevertheless, the shape, size, and position of the red band on the hindwing is different in Mexico, Costa Rica, and Brazil. So, we suggest that there are three subspecies with the following distribution range: one subspecies is widely distributed from Mexico to Nicaragua (ECO 01), the other represented by DHJ02 from Nicaragua to Brazil, and the last one represented by NW106-3 in Brazil. Our results on the actual distributions supported ECO 01 as a different group and more distant from ECO 03, particularly as it did not present areas of sympatry and was in a different historic biogeographical area (Fig. 4). Similarly, there was no overlap of the groups using quantitative characteristics in both wings and genitalia (Fig. 3). There are other groups of Lepidoptera that do not extend their distribution beyond the Yucatan Peninsula (Llorente-Bousquets et al. 2006), such as Heraclides rogeri rogeri (Boisduval) and Hamadryas julitta Fruhstorfer, while ECO 02 and ECO 03 have wider distributions in Mexico. The distribution of ECO 02 is probably towards the Gulf slope. ECO 03 exists towards the Mexican Pacific slope, with the zone of sympatry for both groups in northern Oaxaca, as demonstrated in this study and for other groups of Lepidoptera (e.g., Lasaia, Arellano-Covarrubias et al. 2019).
This paper provides evidence that that the genus Biblis is more diverse in Mexico than was supposed in recent times. We also confirm the proposal of Zhang et al. (2021) to reinstall the Boisduval nomination of Biblis aganisa and report the existence of two new species in Mexico in sympatry with B. aganisa.
To conclude, we consider these species as cryptic species.
References
Abramoff M, Magalhães PJ, Ram SJ (2004) Image processing with Image. J Biophotonics Int 11(7):36–42
Arellano-Covarrubias A, Trujano-Ortega M, Luis-Martínez A, Luna-Reyes M, Llorente-Bousquets J (2019) Geographical distribution of Lasaia Bates, 1868 (Lepidoptera: Riodinidae) across the biogeographical provinces of Mexico. Zootaxa 4656(2):243–273. https://doi.org/10.11646/zootaxa.4656.2.3
Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW (2014) GenBank. Nucleic Acids Res 42(D1):32–37. https://doi.org/10.1093/nar/gkt1030
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22(3):148–155. https://doi.org/10.1016/J.TREE.2006.11.004
Blaxter M (2003) Counting angels with DNA. Nature 421(6919):122–123. https://doi.org/10.1038/421122a
Brower AVZ (2006) Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae). Syst Biodivers 4(2):127–132. https://doi.org/10.1017/S147720000500191X
Burns JM, Janzen DH, Hajibabaei M, Hallwachs W, Hebert PDN (2008) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proc Natl Acad Sci U S A 105(17):6350–6355. https://doi.org/10.1073/pnas.0712181105
Chazot N, Condamine FL, Dudas G, Peña C, Matos-Maraví P, Freitas AVL, Willmott KR, Elias M, Warren A, Aduse-Poku K et al (2020) The latitudinal diversity gradient in brush-footed butterflies (Nymphalidae): conserved ancestral tropical niche but different continental histories. bioRxiv:2020.04.16.045575. https://doi.org/10.1101/2020.04.16.045575
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9(10):1657–1659. https://doi.org/10.1046/J.1365-294X.2000.01020.X
Collins TJ (2007) ImageJ for microscopy. Biotechniques 43(1S):S25–S30. https://doi.org/10.2144/000112517
Cong Q, Grishin NV (2014) A new Hermeuptychia (Lepidoptera, Nymphalidae, Satyrinae) is sympatric and synchronic with H. sosybius in southeast US coastal plains, while another new Hermeuptychia species - not hermes - inhabits south Texas and Northeast Mexico. Zookeys 379:43–91. https://doi.org/10.3897/zookeys.379.6394
Cong Q, Barbosa EP, Marín MA, Freitas AVL, Lamas G, Grishin NV (2021) Two new species of Hermeuptychia from North America and three neotype designations (Nymphalidae: Satyrinae). Taxon Rep 9(7):1–21. https://doi.org/10.5281/zenodo.5622602
D’ercole J, Dincă V, Opler PA, Kondla N, Schmidt C, Phillips JD, Robbins R, Burns JM, Miller SE, Grishin N et al (2021) A DNA Barcode Library for the Butterflies of North America. Peer J. https://doi.org/10.7717/peerj.11157
Dincâ V, Dincâ D, Lukhtanov VA, Talavera G, Vila R (2011) Unexpected layers of cryptic diversity in wood white Leptidea butterflies. Nat Commun.https://doi.org/10.1038/ncomms1329; http://www.nature.com/naturecommunications. Accessed 30 May 2021
Edgar RC, Batzoglou S (2006) Multiple sequence alignment. Curr Opin Struct Biol 16(3):368–373. https://doi.org/10.1016/j.sbi.2006.04.004
García-Sandoval R (2014) Why some clades have low bootstrap frequencies and high Bayesian posterior probabilities. Isr J Ecol Evol 60(1):41–44. https://doi.org/10.1080/15659801.2014.937900
Gaytán Á, Bergsten J, Canelo T, Pérez-Izquierdo C, Santoro M, Bonal R (2020) DNA Barcoding and geographical scale effect: the problems of undersampling genetic diversity hotspots. Ecol Evol 10:10754–10772. https://doi.org/10.1002/ece3.6733
Gill BA, Kondratieff BC, Casner KL, Encalada AC, Flecker AS, Gannon DG, Ghalambor CK, Guayasamin JM, Poff NL, Simmons MP et al (2016) Cryptic species diversity reveals biogeographic support for the ‘mountain passes are higher in the tropics’ hypothesis. Proc R Soc B Biol Sci 283(1832):7–12. https://doi.org/10.1098/rspb.2016.0553
Godman FD, Salvin O (1879–1901) Biologia Centrali Americana. Insecta. Lepidoptera: Rhopalocera, Volumes I-III. London: Published for the editors by Porter RH. 782 p. Available on-line at http://www.sil.si.edu/digitalcollections/bca/. Accessed 5 Nov 2021
Hajibabaei M, DeWaard JR, Ivanova NV, Ratnasingham S, Dooh RT, Kirk SL, Mackie PM, Hebert PDN (2005) Critical factors for assembling a high volume of DNA barcodes. Philos Trans R Soc B Biol Sci 360(1462):1959–1967. https://doi.org/10.1098/rstb.2005.1727
Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci 103(4):968–971. https://doi.org/10.1073/pnas.0510466103
Hallwachs W, Janzen DH, Burns JM, Hebert PDN, Hajibabaei M (2008) DNA barcodes and cryptic species of skipper butterflies in the genus Perichares in Area de Conservacion Guanacaste, Costa Rica. Proc Natl Acad Sci 105(17):6350–6355. https://doi.org/10.1073/pnas.0712181105
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electron 4(1):1–9
Hebert PDN, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc Lond Ser B Biol Sci 270(1512):313–321. https://doi.org/10.1098/rspb.2002.2218
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004a) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci 101(41):14812–14817. https://doi.org/10.1073/PNAS.0406166101
Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004b) Identification of birds through DNA barcodes Charles Godfray editor. PLoS Biol 2(10):e312. https://doi.org/10.1371/journal.pbio.0020312
Hernández-Roldán JL, Munguira ML (2008) Multivariate analysis techniques in the study of the male genitalia of Pyrgus bellieri (Oberthür 1910) and P alveus (Hübner 1803) (Lepidoptera: Hesperiidae): species discrimination and distribution in the Iberian Peninsula. Ann La Soc Entomol Fr 44(2):145–155. https://doi.org/10.1080/00379271.2008.10697551
Holder M, Lewis PO (2003) Phylogeny estimation: traditional and Bayesian approaches. Nat Rev Genet 4(4):275–284. https://doi.org/10.1038/nrg1044
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Huelsenbeck JP, Rannala B (2004) Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol 53(6):904–913. https://doi.org/10.1080/10635150490522629
Ivanova NV, Dewaard JR, Hebert PDN (2006) An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecol Notes 6(4):998–1002. https://doi.org/10.1111/j.1471-8286.2006.01428.x
Janzen DH, Hajibabaei M (2009) Biblis sp. hyperiaDHJ01, Biblis sp. hyperiaDHJ02 and Biblis sp. hyperiaDHJ03. Genbank. https://www.ncbi.nlm.nih.gov/genbank/. Accessed 12 May 2021
Janzen DH, Burns JM, Cong Q, Hallwachs W, Dapkey T, Manjunath R, Hajibabaei M, Hebert PDN, Grishin NV (2017) Nuclear genomes distinguish cryptic species suggested by their DNA barcodes and ecology. Proc Natl Acad Sci U S A 114(31):8313–8318. https://doi.org/10.1073/pnas.1621504114
Jasso-Martínez JM, Castañeda-Sortibrán AN, Pozo C, García-Sandoval R, Prado BR, Luis Martínez A, Llorente-Bousquets J, Rodríguez-Arnaiz R (2016) The Enantia jethys Complex1: Insights from COI confirm the species complex and reveal a new potential cryptic species. Southwestern Entomologist 41(4):1005–1020. https://doi.org/10.3958/059.041.0401
Jasso-Martínez JM, Machkour-M’Rabet S, Vila R, Rodríguez-Arnaiz R, Castañeda-Sortibrán AN (2018) Molecular evidence of hybridization in sympatric populations of the Enantia jethys complex (Lepidoptera: Pieridae). PLoS ONE 13(5):1–23. https://doi.org/10.1371/journal.pone.0197116
Jenkins DW (1990) Neotropical Nymphalidae VIII. Revision of Eunica. Bull Allyn Museum 131:1–177
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Kim S, Lee Y, Mutanen M, Seung J, Lee S (2020) High functionality of DNA barcodes and revealed cases of cryptic diversity in Korean curved-horn moths (Lepidoptera: Gelechioidea). Sci Rep 10(1). https://doi.org/10.1038/s41598-020-63385-x
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative Studies of Nucleotide Sequences. J Mol Evol 16(2):111–120. https://doi.org/10.1007/BF01731581
Kolev Z (2005) New data on the taxonomic status and distribution of Polyommatus andronicus Coutsis & Ghavalas, 1995 (Lycaenidae). Nota Lepidopterol 28(1):35–48
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Lamas G (2004) Hesperioidea-Papilionoidea. In: Heppner JB (ed) Atlas of Neotropical Lepidoptera. Checklist, Part 4a. Association for Tropical Lepidoptera, Gainesville, p 439
Lecocq T, Vereecken NJ, Michez D, Dellicour S, Lhomme P, Valterová I, Rasplus J-Y, Rasmont P (2013) Patterns of genetic and reproductive traits differentiation in mainland vs. Corsican populations of bumblebees. Plos One 8(6):e65642. https://doi.org/10.1371/journal.pone.0065642
Leite LAR, Bonfantti D, Lidke AL, Casagrande MM, Mielke OHH (2017) Comparative study on the hypandrium of the Neotropical Biblidinae (Lepidoptera: Nymphalidae). Shil Rev Lepidopterol 45(178):263–282. https://www.redalyc.org/articulo.oa?id=45551614013. Accessed 28 Nov 2019
Llorente-Bousquets J, Luis-Martínez A, Vargas-Fernández I (2006) Apéndice general de Papilionoidea: Lista sistemática, distribución estatal y provincias biogeográficas. In: Morrone JJ, Llorente-Bousquets J (eds) Componentes bióticos principales de la entomofauna mexicana. vol II. Las Prensas de Ciencias UNAM, México, D. F., pp 945–1009
Miller LD (1970) Nomenclature of wing veins and cells. J Res Lepid 8:37–48
Moraes SS, Montebello Y, Stanton MA, Yamaguchi LF, Kato MJ, Freitas AVL (2021) Description of three new species of Geometridae (Lepidoptera) using species delimitation in an integrative taxonomy approach for a cryptic species complex. PeerJ 9:e11304. https://doi.org/10.7717/peerj.11304
Morrone JJ (2019) Regionalización biogeográfica y evolución biótica de México: encrucijada de la biodiversidad del Nuevo Mundo. Rev Mexicana de Biodiversidad 90:e902980. https://doi.org/10.22201/IB.20078706E.2019.90.2980
Múrias Dos Santos A, Cabezas MP, Tavares AI, Xavier R, Branco M (2016) tcsBU: a tool to extend TCS network layout and visualization. Bioinformatics 32(4):627–628. https://doi.org/10.1093/BIOINFORMATICS/BTV636
Nakahara S, Tan D, Lamas G, Parus A, Willmott KR (2017) A distinctive new species of Hermeuptychia Forster, 1964 from the eastern tropical Andes (Lepidoptera: Nymphalidae: Satyrinae). Trop Lepid Res 26(2):77–84
Nieukerken EJV, Doorenweerd C, Stokvis FR, Groenenberg DSJ (2012) DNA barcoding of the leaf-mining moth subgenus Ectoedemia s. str. (Lepidoptera: Nepticulidae) with COI and EF1-α: two are better than one in recognising cryptic species. Contrib Zool 81(1):1–24. https://doi.org/10.1163/18759866-08101001
Núñez R, Willmott KR, Álvarez Y, Genaro JA, Pérez-Asso AR, Quejereta M, Turner T, Miller JY, Brévignon C, Lamas G et al (2021) Integrative taxonomy clarifies species limits in the hitherto monotypic passion-vine butterfly genera Agraulis and Dryas (Lepidoptera, Nymphalidae, Heliconiinae). Syst Entomol n/a(n/a). https://doi.org/10.1111/syen.12523
Pfenninger M, Schwenk K (2007) Cryptic animal species are homogeneously distributed among taxa and biogeographical regions. BMC Evol Biol 7(1):121. https://doi.org/10.1186/1471-2148-7-121
Prado BR, Pozo C, Valdez-Moreno M, Hebert PDN (2011) Beyond the Colours: Discovering Hidden Diversity in the Nymphalidae of the Yucatan Peninsula in Mexico through DNA Barcoding. Smith MA, editor. PLoS One 6(11):e27776. https://doi.org/10.1371/journal.pone.0027776
Prieto CG, Munguira ML, Romo H (2009) Morphometric analysis of genitalia and wing pattern elements in the genus Cupido (Lepidoptera, Lycaenidae): are Cupido minimus and C. carswelli different species? Dtsch Entomol Zeitschrift 56(1):137–147. https://doi.org/10.1002/mmnd.200900012
Puillandre N, Lambert A, Brouillet S, Achaz G (2012) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21(8):1864–1877. https://doi.org/10.1111/j.1365-294x.2011.05239.x
Puillandre N, Brouillet S, Achaz G (2021) ASAP: assemble species by automatic partitioning. Mol Ecol 21:609–620. https://doi.org/10.1111/1755-0998.13281
Ratnasingham S, Hebert PDN (2013) A DNA-based registry for all animal species: the Barcode Index Number (BIN) System. PLoS ONE 8(8):e66213. https://doi.org/10.1371/journal.pone.0066213
Robert C (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797. https://doi.org/10.1093/nar/gkh340
Sperling F (2003) Butterfly molecular systematics: from species definitions to higher-level phylogenies. In: Butterflies: Ecology and Evolution Taking Flight (eds Boggs CL, Watt WB and Ehrlich PR), pp. 431–458. University of Chicago Press, Chicago, Illinois
Seraphim N, Marín MA, Freitas AVL, Silva-Brandão KL (2014) Morphological and molecular marker contributions to disentangling the cryptic Hermeuptychia hermes species complex (Nymphalidae: Satyrinae: Euptychiina). Mol Ecol Resour 14(1):39–49. https://doi.org/10.1111/1755-0998.12161[consultado2019mar28]
Swofford DL (2001) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sinauer Associates, Sunderland
Shi Q-H, Sun X-Y, Wang Y-L, Hao J-S, Yang Q (2015) Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS ONE 10(4):e0124349. https://doi.org/10.1371/journal.pone.0124349
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. https://doi.org/10.1093/molbev/msr121
Tóth JP, Bereczki J, Varga Z, Rota J, Sramkó G, Wahlberg N (2014) Relationships within the Melitaea phoebe species group (Lepidoptera: Nymphalidae): new insights from molecular and morphometric information. Syst Entomol 39:749–757. https://doi.org/10.1111/syen.12083
Trontelj P, Fier C (2009) Cryptic species diversity should not be trivialised. Syst Biodivers 7(1):1–3. https://doi.org/10.1017/S1477200008002909
Vodă R, Dapporto L, Dincă V, Vila R (2015) Why do cryptic species tend not to co-occur? A case study on two cryptic pairs of butterflies. PLoS ONE 10(2):e0117802
Wahlberg N, Nylin S (2003) Morphology versus molecules: resolution of the positions of Nymphalis, Polygonia, and related genera (Lepidoptera: Nymphalidae). Cladistics 19(3):213–223. https://doi.org/10.1111/j.1096-0031.2003.tb00364.x
Wahlberg N, Wheat CW (2008) Genomic outposts serve the phylogenomic pioneers: designing novel nuclear markers for genomic DNA extractions of Lepidoptera. Knowles LL, editor. Syst Biol 57(2):231–242. https://doi.org/10.1080/10635150802033006
Wahlberg N, Braby MF, Brower AVZ, de Jong R, Lee MM, Nylin S, Pierce NE, Sperling FA, Vila R, Warren AD, Zakharov E (2005a) Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc Biol Sci 272(1572):1577–86. https://doi.org/10.1098/rspb.2005.3124
Wahlberg N, Brower AVZ, Nylin S (2005b) Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae). Biol J Linn Soc 86(2):227–251. https://doi.org/10.1111/j.1095-8312.2005.00531.x
Wahlberg N, Leneveu J, Kodandaramaiah U, Peña C, Nylin S, Freitas AVL, y Brower AVZ (2009) Las mariposas ninfálidas se diversifican tras su casi desaparición en el límite Cretácico / Terciario. Proc R Soc B 276:4295–4302. https://doi.org/10.1098/rspb.2009.1303
Wahlberg N, Peña C, Ahola M, Wheat CW, Rota J (2016) PCR primers for 30 novel gene regions in the nuclear genomes of lepidoptera. Zookeys 2016(596):129–141. https://doi.org/10.3897/zookeys.596.8399
Zhang J, Cong Q, Shen J, Opler PA, Grishin NV (2021) Genomics-guided refinement of butterfly taxonomy. Taxon Rep Int Lepid Surv 9(3):1–55
Zubek A, Lorenc-Brudecka J, Pyrcz TW (2015) Hypandrium as a key character in resolving species-level taxonomy on the example of Perisama oppelii (Latreille) (lepidoptera: Nymphalidae, biblidinae). Zootaxa 3990(1):32–40. https://doi.org/10.11646/zootaxa.3990.1.2
Acknowledgements
We thank Alma Estrella García Morales and Manuel Elías Gutiérrez from Node of MEXBOL from ECOSUR for their help in processing and sending the samples for sequencing; to Humberto Bahena Basave for his help in taking photographs to make part of the figures; to Holger Weissenberger from ECOSUR Department of Observation and Study of the Earth, the Atmosphere and the Ocean for his help in making the map; and to the Consejo Nacional de Ciencia y Tecnología (CONACYT) for the scholarship awarded to the first author.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Hugo Álvarez García and Carmen Pozo. The first draft of the manuscript was written by Hugo Álvarez García, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Edited by André VL Freitas
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Álvarez-García, H., Machkour-M’Rabet, S., Martínez, A.L. et al. New Complex of Cryptic Species Discovered in Genus Biblis (Papilionoidea: Nymphalidae: Biblidinae) in Mexico. Neotrop Entomol 51, 557–569 (2022). https://doi.org/10.1007/s13744-022-00969-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13744-022-00969-4