Abstract
A combination of the smoothly clipped absolute deviation (SCAD) method and the artificial neural network (ANN) was utilized as a novel methodology (SCAD-ANN) in the quantitative structure-retention indices relationship (QSRR). The proposed SCAD method reduces the dimension of data before using the robust ANN modeling method. The efficiency of the SCAD-ANN methods was evaluated by the construction of a QSRR model between the most relevant molecular descriptors (MDs) and RIs for two sets of volatile organic compounds. The SCAD method was applied to training data, and effective MDs were selected in a λ with the lowest cross-validation error (λmin) and were defined as the inputs to the ANN modeling method. All ANN parameters were optimized simultaneously. Some statistical parameters were computed, and the obtained results indicate that the constructed QSRR models have acceptable values. Also, the applicability domain analysis reveals that more than 95% of the data are in the confidence range, indicating that the prediction results of the SCAD-ANN models are reliable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It is well acknowledged that the properties of compounds are critically dependent on the structural features. Therefore, in computational chemistry, finding the relationship between the structural characteristics of compounds and their properties is of interest to researchers. Among the various computational chemistry approaches, the quantitative structure–property relationship (QSPR) study is a helpful tool to find such a relationship and has been widely used in various fields such as chemometrics, biological chemistry, and medicinal chemistry. QSPR has been established to be a potent tool for the analysis of chromatographic properties. Recently the calculation of the retention indices (RIs) has been reported in the literature [1,2,3,4,5,6,7,8,9,10]. As a kind of QSPR study, the study of chromatographic retention is defined by the general name of quantitative structure retention indices relationship (QSRR). The retention indices obtained using chromatographic phases can serve as physicochemical properties correlated to the structural features. RI is a quantitative criterion that indicates the relative retention of each sample component relative to the normal alkanes (hydrocarbons) on a stationary phase at a given temperature. RIs are mainly independent of the chromatographic parameters (such as column length, diameter, column gas flow) and allow comparing values measured by different analytical laboratories under different conditions. In general, RIs can be calculated using the GC technique. However, it should be noted that RI and mass spectra are not always suitable for providing structural profiles of compounds, and the measurements with GC suffer from the following limitations. The compounds identification is often carried out by matching GC peaks of the analyte with the peaks of standards, while the pure standard samples are sometimes not available. Also, it is always challenging to determine the RI of chemicals due to the complexity of quantitative analysis methods. The GC technique also requires sample and column preparations and parameter optimization, which are time-consuming and costly [2, 11]. Therefore, the introduction of efficient methods for predicting RI values for unknown compounds without any measurement or experiments is of interest [1]. The QSRR models can be used to predict the retention indices of chemical structures and have recently received much attention from researchers [1,2,3]. The QSRR model establishes a mathematical relationship between RI as a response of a chromatographic system and the molecular properties that describe the structure of the analyte to predict the RI parameter.
The main goal of the QSPR study is to find an accurate relationship between the structural characteristics and properties of the studied compounds. In this context, the performed QSRR modeling consisted of the following steps: the definition of endpoints and coding the molecular descriptors, selecting the optimal molecular descriptors using an unambiguous algorithm, appropriate measures of goodness-of-fit, robustness, and predictability, a defined domain of applicability, the evaluation of models [12, 13]. Coding the structural properties and descriptors generation is crucial in QSPR studies, in which the empirical and computational information of structures is converted into experimental and theoretical descriptors [14]. Empirical descriptors can be calculated using the experimental results in the specific conditions for each compound. The theoretical descriptors are calculated according to the structural features of the studied compounds using available software and online computational web tools [15,16,17]. With recent progress in computational chemistry, a large number of MDs could be calculated for a given chemical compound; therefore, the QSRR studies are encountered with high-dimensional data (HDD) [18]. Generally, HDD contains redundant and irrelative MDs without vital information. Correlated and redundant descriptors do not add useful and relevant information to the QSRR studies and are the source of miss modeling and the complexity of the QSRR models. Therefore, to construct QSRR models with the smallest MDs, the highest predictability power and appropriate interpretability are desirable. For the construction of QSRR models with the characteristics mentioned above, the irrelevant, redundant MDs should be omitted from the data set. Therefore, introducing new and efficient variable selection techniques is required to increase the modeling performance, improve the interpretability of the model, and decrease the computation complexity and storage needed [18, 19]. Variable selection approaches are generally distributed into different classes, including filter, wrapper, and embedded procedures [20]. The filter-based variable selection method ranks descriptors using a simple algorithm; this approach reduces descriptors based on a specific function: correlation coefficient, chi-square test, Fisher score, variance threshold, and other statistical parameters [21]. The wrapper-based variable selection approach attempts to find the optimal subset by iteratively selecting descriptors to improve the model performance. Some widely used methods, such as forward selection, backward elimination, and stepwise regression, belong to the wrapper category. However, these classical approaches suffer from high bias, instability, and false determination coefficient [20]. The embedded method complements the filter and wrapper techniques and benefits from the advantages of both complemented techniques. These advantages are high computational speed and the existence of suitable learning algorithms. One of the most efficient embedded variable selection methods is the penalized regression approach [20, 21]. The least absolute shrinkage and selection operator (LASSO) is the first and widely used penalized method presented by Tibshirani [22]. LASSO minimizes the regression error same as the ordinary least square (OLS), except that a penalty parameter has been added to the squared error term of OLS, which constrains the coefficients of the irrelevant variables to zero according to the following equation [23]:
where Y and X are the vector of the response variable and the design matrix of independent variables, respectively. β parameter represents regression coefficients, and λ is the tuning parameter and has a non-negative value (λ ≥ 0) that controls the quantity of penalty related to the magnitude of estimated coefficients. LASSO has well-known inherent advantages over OLS; therefore, it has been used to construct successful linear QSPR models [24,25,26,27,28,29,30]. An exciting and useful aspect of some penalized methods, such as LASSO, is their ability to select the most significant variables via shrinking the coefficients to zero. In this context, the important variables are selected according to the λ with the minimum cross-validation (CV) error value (λmin) [22]. Recently, LASSO as a first penalized method has been used as a variable selection technique coupled with MLR (LASSO-MLR) [31] and with stepwise regression (LASSO-SR) [32] for the prediction of RT and retention factor at 5% acetonitrile (log kACN) with satisfactory results. However, LASSO suffers from drawbacks. It is biased and shrinks the coefficients more than needed; the parameters are severely underestimated [33]. Also, for HDD cases, where the number of variables (p) is higher than the number of sample points (n), LASSO only can select at most the number of n variables due to its convex optimization nature [33]. Therefore, the LASSO ability is reduced due to these limitations, mainly when used as a variable selection method. To overcome the limitations of LASSO, a new and concave penalized technique called smoothly clipped absolute deviation (SCAD) has been proposed [33], in which two tuning parameters were added to the OLS function. SCAD estimates the regression coefficients using the following equation:
which \({{\rm P}}{{(.; {\rm a},\lambda )}}\) is the penalty function of SCAD and has the following three arguments form:
where \(a > 0\) and λ ≥ 0 are the penalty parameters and control the magnitude of the coefficients. Fan and Li proved that the optimal value of \(a\) is 3.7, using a Bayesian interpretation [33]. The SCAD method is an oracle, unbiased, and predictive model with fewer variables, preventing over-fitting. Therefore, as a powerful modeling method, SCAD has been used to construct a few linear and nonlinear QSRR models with satisfactory results [26, 30, 34]. Due to the exciting aspects of SCAD, such as sparsity and consistency, could be a good alternative for LASSO in the stable variable selection. Nevertheless, there is no report on using a SCAD-ANN in QSRR studies based on our best knowledge. Therefore, we decided to use the SCAD method as the variable reduction technique in the QSRR study of the RI values for some VOCs.
For the first time in this study, the SCAD method as a new variable selection method was coupled with the ANN method as powerful nonlinear modeling (SCAD-ANN) to predict the GC retention indices (RI) values of Volatile Organic Compounds (VOCs). VOCs are aliphatic and aromatic chemical compounds with low molecular weights and boiling points [35]. VOC sources include solvents, fuels, dyes, detergents, cigarettes, and foodstuffs. Most of the VOCs produced by the industries often contain benzene, toluene, xylene, furan, and chloroform, which are harmful to human health [36]. After calculating MDs for different sets of VOCs and screening the variables, tenfold cross-validation SCAD was performed, and the significant descriptors corresponded to the λmin (a λ with the minimum CV error) were extracted. Due to the complex and nonlinear relationship between the dependent and independent variables, the ANN was used to establish a QSRR model. The performance of the developed SCAD-ANN models was evaluated using statistical parameters for the prediction of RIs of the test and validation sets and whole dataset through the leave-one-out (LOO) technique. Additionally, the suggested SCAD-ANN was evaluated using an applicability domain technique based on a leverage matrix computation. It was found that the nonlinear ANN modeling method combined with the penalized variable selection technique produced accurate QSRR models for the prediction of VOC retention indices (RIs).
Materials and methods
Dataset
In order to develop QSPR models, several datasets were used. The first dataset consisted of Kovats RI of 132 VOCs consisting of alkanes, alkenes, amines, ethers, alcohols, alkyl-benzenes alkyl-halides. The RIs of studied compounds of dataset 1 were measured using a modified Packard Becker (Delft, the Netherlands) Model 439 GC equipped with two thermal conductivity detectors. The stationary phase was C67 with the chemical name of 19,19-Diethyl-14,24 – ditridecylheptatricontane, and GC data for about 132 VOCs were measured on C67 at 130 °C [37]. The second dataset consists of the RI values of 52 VOCs, including pyrazines, pyridines, furans, etc. The RIs of studied compounds of dataset 2 were extracted using automatic solid-phase microextraction (SPME) onto the headspace coated fiber. The SPME fiber was inserted into the GC injection port to desorb the VOCs for 5 min at 270 °C. The Varian CP-3800 GC column is fitted with DB-5 ms (30 m × 0.25 mm × 0.25 mm). The oven temperature was adjusted in the range of 40 to 280 °C with an increment rate of 4 °C/min. Helium was used as a carrier gas with a flow rate of 1.0 ml min−1. Mass spectrometer (MS) analysis was performed using the ion trap mass spectrometer (Varian Inc., Walnut Creek, CA) [38]. The structures of compounds were converted to a simplified molecular-input line-entry system (SMILES) format file. Tables S1 and S2 summarize the chemical names and SMILES format of studied structures with the corresponding RI values. RI values for all compounds in their own category were measured under the same conditions. It should be noted that the two data sets did not have homogeneity to be combined in one dataset, so due to the homogeneity in their own category, they were used as two separate datasets for further studies. Therefore, each dataset has enough homogeneity for a QSRR model with low experimental outliers. RI values were considered as the dependent variable in the whole study. The dataset was divided into the train set (60 percent of the entire dataset), validation set (20 percent of the entire dataset), and test set (20 percent of the entire dataset) with the Kennard‐Stone (KS) algorithm based on Euclidean distance using R-package [39].
Optimization of three-dimensional structures
The three-dimensional structures of all VOCs were sketched using HyperChem software [40]. The structures were optimized and reached a minimum state of energy. The optimization process was performed using the semi-empirical AM1 method. The optimization process was continued until the root mean square (RMS) value of the energy gradient reached 0.001 kcal/mol. Optimal structures were used as DRAGON inputs for the descriptor computation [16].
Screening and descriptor generation
The MDs were calculated from the optimized structures. The optimum structures of compounds were subjected to the DRAGON 5.5 software [16], and 3224 MDs were calculated. The calculated descriptors belong to 22 different classes, which are: constitutional indices, topological indices, walk and path counts, connectivity indices, information indices, 2D autocorrelations, edge adjacency indices, Burden eigenvalues, topological charge indices, eigenvalues based indices, Randic molecular profiles, geometrical descriptors, RDF descriptors, 3D-MoRSE descriptors, WHIM descriptors, GETAWAY descriptors, functional group counts, atom-centered fragments, charge descriptors, molecular properties, 2D binary fingerprints, and 2D frequency fingerprints [16].
Selection of the most significant descriptors
In order to reduce the instability of the developed model, the number of descriptors should be reduced. For this purpose, a two-step strategy of variable screening and variable selection using SCAD was implemented. MDs with constant and near-constant values (variables with variance lower than 0.001) were eliminated in the screening step. Also, among two descriptors with a correlation above 0.9, the descriptor with the lowest relevance to the response was eliminated. After the screening, the remaining descriptors were arranged in a data matrix and used in the subsequent variable selection method. At initially, MDs and RI values were considered as the independent and dependent variables, respectively. The test set data were uninvolved from the dataset; then, the new matrix was subjected to the SCAD method. The SCAD method was implemented using the tenfold cross-validation method, and CV errors were computed for all possible values of λ. The best λ value minimum CV error (λmin) was identified, and the MDs corresponding to such λ were considered as the most effective descriptors. In all SCAD calculations, the α parameter was adjusted to 3.7 according to the bayesian study of Fan and Li [33]. The programs were written in R software [39] using the caret and ncvreg packages [41, 42].
ANN modeling
The relationship of the MDs and the RIs was established using a feed-forward ANN trained with the back-propagation algorithm using train set data. To evaluate the efficiency of the variable selection method combined with the ANN, two different training functions were used for ANN modeling. The ANN models were constructed using the Levenberg- Mardquat (LM) training function for dataset 1 and the Bayesian regularization (BR) training function for dataset 2. After optimization and developing QSRR models, the ANN model with the LM training function (trainlm in MATLAB nnet toolbox) and logarithmic sigmoid as the transfer function (logsig in MATLAB nnet toolbox) was adopted as the best ANN model for dataset 1 (the SCAD-LM-ANN). Additionally, for dataset 2, the optimal ANN model was developed with the BR algorithm as a training function (trainbr in MATLAB nnet toolbox) and logarithmic sigmoid as the transfer function (logsig in MATLAB nnet toolbox), respectively. The architectures of the optimal ANN models are 10-2-1 and 7-4-1 for datase1 and dataset 2, respectively. The corresponding SCAD-ANN models were optimized using SCAD selected descriptors to minimize the root mean square error (RMSE) of the validation set. The superior models were applied to predict RI values of VOCs in the external test set.
Diversity analysis
The diversity analysis refers to the chemical space of the proposed model and shows the degree of similarity of the studied compounds [43, 44]. Therefore, the diversity analysis was performed to investigate how the training, validation, and test set compounds are distributed and examine whether the test and validation sets are the suitable representation of the chemical space of train set data. In this context, the distance between the two compounds was calculated using the Euclidean distance norm (rij) according to the following equation [43]:
where pij and qij are MD values for two compounds i and j. The Euclidean distance between 2 variables (rij) is equal to the summation from the first variable (k = 1) to the last variable (n). Euclidean distance is found by finding the differences for each value of an individually variable (i value for the kth variable being pki and j value being qkj). Then, the mean Euclidean distances of each compound (\({{{\bar r}}_{{\text{ij}}}}\)) was calculated and normalized (\(0 < \overline{r}_{\rm ij} < 1\)) [23].
Applicability domain
The applicability domain (AD) is recognized as an approach to QSRR model evaluation. AD validates part of the response or chemical space of structures of the compounds in which the model provides reliable predictions. The AD calculation aims to prove the applicability of the recommended ANN model for the compounds of the datasets, similarly confirm the presence of the predicted RI values in the chemical space range, and prove the accuracy of the results. AD was calculated using the Mahalanobis distance to the center of the train set distribution through the leverage method. The leverage function was computed using the projection of RI on the predicted response. The leverage denotes the diagonal of the Hat matrix. The H values were obtained according to the equation as follows [45, 46]:
where X corresponds to the MDs matrix of the training set compounds, xi is the row vector of each MDs for each compound, and T represents the transpose of matrix or vector. The threshold of leverage is defined as the following equation [44, 47]:
where p refers to the number of the descriptors that participated in the SCAD-ANN models, and N is the number of train set observations. AD approach can be illustrated using William's plot, which is obtained by plotting the H values versus the standardized residuals. If the H value is greater than the threshold of h*, it means that the studied compound is structurally very different from the training set compounds. Furthermore, if a compound exceeds the range of σ < -3 or σ > 3, it is not in the reliable range, and such a compound is known as an outlier.
Results and discussion
Selection of relevant descriptors
Due to a large number of MDs, the presence of redundant descriptors is high; hence, a combination of a pre-processing and variable selection method was implemented on the total descriptors to reduce the data dimensions before the construction of a predictive and straightforward ANN model. Therefore, in the pre-processing (screening) step, descriptors with constant and near-constant values (variance less than 0.001) were removed. The descriptor with the lowest correlation with response was omitted among two descriptors with a pair-wise correlation above 0.9. Before applying a variable selection technique, the dataset was splitted into the train (60 percent of whole datasets), validation (20 percent of whole datasets), and test set (20 percent of whole datasets), using the Kennard‐Stone (KS) algorithm and based on the Euclidean distance. In all variable selection and modeling steps, the test set was discarded, and the variable selection was performed using the training and validation sets data. Due to the intrinsic limitations of OLS-based variable selection methods, SCAD as a novel penalized method with well-known advantages, such as sparsity and consistency, was used to reduce dataset dimension further. For this purpose, the SCAD method was implemented to the remained descriptors of training and validation sets. At first, the α tuning parameter was adjusted to 3.7 [33], and the SCAD was performed using a tenfold cross-validation technique. The cross-validation errors for 100 different values of λ were computed. The tuning parameter of λ was selected based on a grid search approach between 0 and 100, and the λ value corresponded to 8 and 15 variables, with the minimum CV error was selected as the λmin for each dataset. Therefore, the most effective MDs with the highest relevance with RI were chosen in such λmin and used as inputs of the ANN models.
The selected MDs of the SCAD method were investigated for the existence of correlation and multicollinearity. In statistical contexts, the jth variance inflation factor (VIF) can be computed using \(VIF_{j} = \frac{1}{{1 - R_{j}^{2} }}\), where \(R_{j}^{2}\) is coefficient of determination for the regression of each descriptor on other descriptors. According to the recently published papers, VIF > 10 is a sign of severe multicollinearity. According to Table 1, all the VIF values are lower than 10. A comprehensive explanation of VIF and significant values for detecting multicollinearity can be referred to the recently published papers [48,49,50,51,52].
Construction of SCAD—ANN-based QSRR Model
The artificial neural network procedure does not involve any initial information of the mathematical architecture of the data. Therefore, the use of ANN is ideal for data analysis, in which there is a hidden nonlinear relationship or a complex interdependence between independent and dependent variables. Given that nonlinear models are more suitable for simulating molecular properties in real situations [53], ANN modeling was adapted to create a nonlinear relationship between RI and SCAD-selected MDs. In this context, a feed-forward neural network model learning with a back-propagation error technique was considered for QSRR modeling. To obtain an optimal ANN model, the simultaneous optimization of all significant parameters such as the number of inputs, number of nodes in the hidden layer, training epochs, and the training and transfer functions is necessary. In most computational chemistry researches, the ANN model with a single hidden layer is satisfactory [53]. Hence, ANN models containing one input layer, a single hidden layer, and one output layer were used to optimize the ANN parameters. In the simultaneous optimization of ANN parameters, ANN models with different architectures were designated by the use of two diverse training algorithms (Levenberg–Marquardt (LM) for dataset 1 and Bayesian Regularization (BR)for dataset 2) and logarithmic sigmoid as the transfer function. The linear purlin function is used as the output transfer function in the ANN training procedure. To optimize the number of ANN inputs, the number of input layer neurons was first defined from 2 to the whole number of SCAD selected descriptors in each dataset. Given that the number of subgroups that can be formed from this number of inputs is very large, not all of these subgroups can be used to train the ANN because it is very time-consuming. Thus, the ANN method was utilized to find the optimal combination of SCAD selected descriptors. The generated ANN models were optimized with respect to the total number of descriptors in the case of the examined datasets. The optimal node and the training epochs were obtained. Following that, all of the descriptors were randomized within their respective ranges of variation. Each time, a new set of descriptors concluding a manipulated descriptor in the presence of other actual descriptors was used to train the ANN constructed models. Therefore, the development of the ANN models was performed as the above procedure for all SCAD selected descriptors. The RI values were predicted using the ANN models. The RMSEi value of the validation set was calculated in the presence of all selected descriptors with a manipulated ith descriptor. This process was repeated for the number of SCAD selected descriptions so that 15 and 8 RMSE values were finally obtained for datasets 1 and 2, respectively. The higher RMSEi rate indicates that the ANN model suffers more errors in the absence of the descriptor with the actual values. So ith descriptor with more error is more important than other descriptors. The descriptors were arranged decreasingly in terms of RMSE values and defined as the ANN inputs. So that the ANN was first trained with the first two descriptors with more importance until the ANN was optimized with the total number of important descriptors. Subsequently, in optimizing the number of inputs of ANN models, arranged subsets with the different number of descriptors in the range of 2 to the maximum number of SCAD selected descriptors (15 and 8 MDs for dataset 1 and dataset 2, respectively) were designed and used as the ANNs inputs. Therefore, in the optimization of ANN conditions, the number of 14 subsets for dataset 1 and 7 subsets for dataset 2 were used as inputs according to the importance of descriptors, while the number of neurons in the hidden layer was changed in the ranges of 2 to 10 with a step size of 1. The designed ANN models were trained with training epochs of 5 to 50 (with step 5) using the training set data. Consequently, the trained model was applied to the prediction of RI values in the validation set. The optimization goal was used to minimize the RMSE for the validation set. The best ANN model was selected according to the lowest RMSE value of the validation set. The RMSE value was calculated using the following equation:
where N is the number of validation compounds, and \(\hat{y}_{i}\) and \(y_{i}\) parameters are the model predicted and actual values of RI, respectively. The best architectures of ANN models with different training and transfer functions according to the minimum RMSE values are summarized in Table 2. The obtained results showed that the ANN models with the mentioned optimum conditions have low RMSE values and were introduced as the superior model for predicting RI values for compounds in the external test set and further evaluations. The process of modeling steps is summarized in Fig. 1 for more clarity. The descriptor arrangement in order of their importance was summarized in Table S5 in the supplementary material file.
Validation of SCAD-ANN modes
Predictableness of the superior SCAD-ANN model
The predictability of the proposed models was estimated using different approaches. The SCAD-LM_ANN model with 10–2-1 architecture and the SCAD-BR-ANN model with the architecture of 7–4-1 were practical to predict RI of the test sets of dataset 1 and dataset 2, respectively. Using the obtained results, the predicted RI values were plotted against their corresponding actual values. The LOO technique was also used to predict the RI values of all compounds. Each compound was excluded once as test data in this technique, and the optimal models were trained using the remaining data. After running the corresponding models, the RI values of all compounds were predicted, and the predicted values were plotted in terms of actual RI values (Fig. 2). Q2LOO for datasets 1 and 2 was equal to 0.94 and 0.88, respectively. The prediction results of the model were also evaluated using standardized residual diagrams. For this purpose, the standardized residuals (ri) were calculated for the LOO predicted values according to the following equation:
where ei is the difference between the observed and predicted responses for each observation i = 1, …, n, and sei represents the standard deviation of residual values. The standard residuals are plotted in terms of actual RI values in Fig. 2c, f. The obtained standardized residual graph (Fig. 2c, f) shows a reasonably random pattern. Thus, it implies that there was no systemic error in the developed ANN models as the spread of residuals was pragmatic on both sides of zero [54]. Different statistical parameters were also calculated for further evaluation of the prediction ability of the proposed SCAD-ANN models. As displayed in Table 3, the results specify the good generalizability and predictability of the developed model in predicting response values for test set data and entire data using the LOO technique. The formulas and values of the estimated statistical parameters are summarized in Table 3. The values of statistical parameters and comparison with their acceptable values or ranges confirm the satisfactory prediction ability of the proposed SCAD-ANN models.
Additionally, the SCAD efficiency as the variable selection for the ANN modeling was compared with different traditional and penalized methods. For this purpose, the prediction power of the constructed SCAD-ANN models was compared with QSAR models derived from the combinations of stepwise regression (SR) and LASSO, as a penalized variable selection method, with the ANN modeling method. In this regard, selected variables by SR and LASSO methods were individually entered into the corresponding optimal ANN models (named as (SR-LM-ANN and LASSO-LM-ANN for dataset 1, and SR-BR-ANN and LASSO-BR-ANN for dataset 2, respectively) as the inputs. After the training and optimization, the RI values of the test set data and the whole data (using the LOO technique) were predicted using the corresponding ANN models with the architectures mentioned in Table 3. The calculated statistical parameters for the predicted values are summarized in Table 3. The obtained results confirm that the developed SCAD-ANN models have good predictability compared to other models, indicating the high performance of SCAD in selecting significant variables for ANN-based QSAR studies. Additionally, the data distribution was illustrated using the plot of the normalized \({{\overline{\rm r} }}_{{{\text{ij}}}}\) values versus the response values (Fig. 3). The results in Fig. 3 demonstrate the proper distribution of test and validation sets in the chemical space of the training set data. Also, in this study, the applicability domain (AD) of created models was investigated based on the calculation of leverage matrix (h), and the William plot was drawn for all developed models. According to Fig. 4, the values of h* for both datasets were equal to 0.31 and 0.77, respectively. The William plot indicates that more than 95% of the data are in the confidence range, meaning that the prediction results of the SCAD-ANN models are reliable.
For further investigation, an additional dataset (concluding 206 chemical structures of VOCs) [54] was employed to validate the developed SCAD-ANN model. After the pre-processing step, 317 variables were defined as the input of the SCAD method. Fourteen significant SCAD selected descriptors were arranged based on the ANN procedure and then defined as the ANN input. After simultaneous optimization of the parameters, the optimum LM-ANN with 5-2-1 architecture was obtained. RI values of the test set data were predicted using the optimum LM-ANN model. Several statistical parameters such as R2test, CCC2 test, R2adj, MAEtest, and RMSEtest were equal to 0.97, 0.97, 0.96, 24.66, and 31.41. The findings of the suggested SCAD-ANN model reveal that it has satisfactory predictability and is comparable to the previously presented results.
Y-scrambling test
The validity of the established relationship by the superior SCAD-ANN models was also evaluated using the Y-scrambled data in which the manipulated RI values are used [55, 56]. This approach aims to prove that the relationship between independent and dependent variables is not by chance. In this regard, about 1000 sets of RI as scrambled responses for the data set were randomly generated in their own range of variation. The proposed SCAD-ANN models were constructed using manipulated responses. The statistical parameters of the SCAD-ANN models using manipulated responses (\(R_{manipulated}^{2}\)) were calculated. The values of \(R_{{{\text{manipulated}}}}^{2}\) values were plotted versus the times of runs. The obtained results in Fig. 5 show that the \(R_{{{\text{manipulated}}}}^{2}\) values are significantly smaller than that of the test data set (R2test = 0.89) and smaller than the acceptable value of 0.6 [57]. Therefore, the results obtained prove that the established QSRR relationship between the significant MDs and RI values is not random and created based on an accurate and reasonable relationship between the molecular descriptors and RI values.
Contribution of MDs in the ANN model
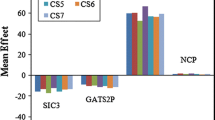
Due to the nonlinear model developed, the contribution of MDs in the final model was investigated [23] using the optimal proposed to correspond SCAD-ANN models with 10–2-1 and 7–4-1 architectures. To calculate the contribution percentage of each MDs, the values of each descriptor were randomized in the range of its own variations. The optimal SCAD-ANN models were developed each time in the presence of one MD with randomized values and other MDs with their actual values. The developed models were used to predict RI values in the validation set, and the RMSEi value of the validation set was obtained when ith MD had randomized values. This process was repeated until all RMSEi values were obtained for MDs that appeared in the final SCAD-ANN model. Finally, the contribution percentage of ith MD (Ci) was calculated using the following equation:
Figure 6 shows the calculated contribution percentage (%Ci) for all selected MDs. The negative or positive influences of MDs on the responses were determined using the sign of the standardized coefficients of the corresponding SCAD models for dataset 1 (Eq. 10) and dataset 2 (Eq. 11). The best fitted SCAD model was estimated as follows:
RI = 0.97 X1sol—0.002 F01CO—0.22 BAC—0.01 H047 -0.03 Mor25m + 0.03 H050 + 0.03 Hy -0.25 BLI -0.02 RDF060p -0.01 GGI5 + 0.015 F03CN + 0.016 C025 + 0.02 HATS5V + 0.03 TPSATot + 0.001 E1m (10).
RI = 1.01 X2sol -0.005G2e—0.13 Mor27u + 0.027 TIC5 + 0.006 TIC1 + 0.005 AMR -0.005 Mor07e (11).
Conclusion
For the first time in this study, the SCAD as an efficient penalized method was used to select important MDs, and the selected MDs were used to predict the RI values of VOCs using the nonlinear ANN model to construct the QSRR model. The proposed corresponding SCAD-ANN models simultaneously benefit from the advantage of SCAD, such as sparsity and extremely high prediction ability of the ANN method due to the nonlinearity of the relationships between the MDs. The evaluation of the developed SCAD-ANN models using test set data, LOO techniques, and applicability domain revealed the high prediction power of the model. The results (Table 3) showed that all statistical parameters of the model for the test data and the whole data are acceptable. The value of \({\text{Q}}_{{{\text{LOO}}}}^{{2}}\) is greater than the acceptable value of 0.5, and the value of MAELOO is smaller than the allowable value (0.1 × RangeTrain), which indicates the accurate prediction of RI values.
Availability of data and material
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Code for data cleaning and analysis is provided as part of the replication package. It is available at "r-project.org" for review.
References
R. Kaliszan, Chem. Rev. 107, 3212 (2007)
Y. Marrero-Ponce, S.J. Barigye, M.E. Jorge-Rodríguez, T. Tran-Thi-Thu, Chem. Papers 72, 57 (2018)
L. Wu, P. Gong, Y. Wu, K. Liao, H. Shen, Q. Qi, H. Liu, G. Wang, H. Hao, J. Chromatogr. A 1303, 39 (2013)
M. Aćimović, L. Pezo, V. Tešević, I. Čabarkapa, M. Todosijević, Ind. Crops Prod. 154, 112752 (2020)
B.C. Naylor, J.L. Catrow, J.A. Maschek, J.E. Cox, Metabolites 10, 237 (2020)
M. Acimovic, L. Pezo, J. S. Jeremic, M. Cvetkovic, M. Rat, I. Cabarkapa, V. Tesevic, J. Essential Oil Bearing Plants 23, 464 (2020)
B. Pavlić, N. Teslić, P. Kojić, L. Pezo, J. Serb. Chem. Soc. 85, 9 (2020)
D.D. Matyushin, A.Y. Sholokhova, A.E. Karnaeva, A.K. Buryak, Chemometrics Intell. Lab. Syst. 202, 104042 (2020)
S. Đurović, Green Sustainable Process for Chemical and Environmental Engineering and Science, Elsevier (2021)
R. Kaliszan, Handbook of Analytical Separations 8, 587 (2020)
P. Kalhor, O. Yarivand, Anal. Chem. Lett. 6, 371 (2016)
J. Dearden, M.T. Cronin, K.L. Kaiser, SAR QSAR Environ. Res. 20, 241 (2009)
M. Vračko, V. Bandelj, P. Barbieri, E. Benfenati, Q. Chaudhry, M. Cronin, J. Devillers, A. Gallegos, G. Gini, P. Gramatica, SAR QSAR Environ. Res. 17, 265 (2006)
C. Zisi, I. Sampsonidis, S. Fasoula, K. Papachristos, M. Witting, H.G. Gika, P. Nikitas, A. Pappa-Louisi, Metabolites 7 (2017)
I. Sushko, S. Novotarskyi, R. Körner, A.K. Pandey, M. Rupp, W. Teetz, S. Brandmaier, A. Abdelaziz, V.V. Prokopenko, V.Y. Tanchuk, J. Comput. Aided Mol. Des. 25, 533 (2011)
T. Srl, Italy (2007)
O. Soufan, W. Ba-alawi, A. Magana-Mora, M. Essack, V. B. Bajic, Sci. Rep. 8, 1 (2018)
J.P.M. Andries, M. Goodarzi, Y.V. Heyden, Talanta 219, 121266 (2020)
W. Zheng, M. Jin, Digital Scholarship in the Humanities (2019).
Z. M. Hira, D. F. Gillies, Adv Bioinfo. 2015, 198363 (2015)
A. Jović, K. Brkić, N. Bogunović, in: 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), IEEE (2015)
R. Tibshirani, J.R. Stat, Soc. Series. B. Stat. Methodol. 58, 267 (1996)
Z. Mozafari, M.A. Chamjangali, M. Arashi, Chemometrics Intell. Lab. Syst., p. 103998 (2020)
O. Farkas, I.G. Zenkevich, F. Stout, J.H. Kalivas, K. Héberger, J. Chromatogr. A 1198, 188 (2008)
E. Daghir-Wojtkowiak, P. Wiczling, S. Bocian, Ł. Kubik, P. Kośliński, B. Buszewski, R. Kaliszan, M.J. Markuszewski, J. Chromatogr. A 1403, 54 (2015)
A.M. Al-Fakih, Z.Y. Algamal, M.H. Lee, M. Aziz, SAR QSAR Environ. Res. 28, 691 (2017)
A. Al-Fakih, Z. Algamal, M. Lee, M. Aziz, SAR QSAR Environ. Res. 29, 339 (2018)
Z.T. Al‐Dabbagh, Z.Y. Algamal, J. Chemom. 33, e3139 (2019)
J. Krmar, M. Vukićević, A. Kovačević, A. Protić, M. Zečević, B. Otašević, J. Chromatogr. A, p. 461146 (2020).
Z.Y. Algamal, M.H. Lee, A.M. Al-Fakih, M. Aziz, SAR QSAR Environ. Res. 27, 703 (2016)
L. Kubik, P. Wiczling, J. Pharm. Biomed. Anal. 127, 176 (2016)
E. Daghir-Wojtkowiak, P. Wiczling, S. Bocian, L. Kubik, P. Koslinski, B. Buszewski, R. Kaliszan, M.J. Markuszewski, J. Chromatogr. A 1403, 54 (2015)
J. Fan, R. Li, J. Am. Stat. Assoc. 96, 1348 (2001)
X.-L. Peng, H. Yin, R. Li, K.-T. Fang, Anal. Chim. Acta 578, 178 (2006)
M.E. Fleming-Jones, R.E. Smith, J. Agric. Food. Chem. 51, 8120 (2003)
R. M. Vinci, L. Jacxsens, B. De Meulenaer, E. Deconink, E. Matsiko, C. Lachat, T. de Schaetzen, M. Canfyn, I. Van Overmeire, P. Kolsteren, Food Control 52, 1 (2015)
R. Ghavami, S. Faham, Chromatographia 72, 893 (2010)
J. Xu, W. Zhang, K. Adhikari, Y.-C. Shi, J. Cereal Sci. 75, 77 (2017)
R. C. Team, Vienna, Austria (2013)
K. Wolinski, J. Hinton, D. Wishart, B. Sykes, F. Richards, A. Pastone, V. Saudek, P. Ellis, G. Maciel, J. McIver, Inc., Gainsville (2007)
P. Breheny, M.P. Breheny, R Foundation for Statistical Computing, Vienna, Austria. URL https://cran.r-project.org/package=ncvreg (2020)
M. Kuhn, R Foundation for Statistical Computing, Vienna, Austria. URL https://cran.r-project.org/package=caret (2012)
A. G. Maldonado, J. Doucet, M. Petitjean, B.-T. Fan, Mol. Divers. 10, 39 (2006)
S.M. Behgozin, M.H. Fatemi, J. Iran. Chem. Soc. 16, 2159 (2019)
A. Tropsha, P. Gramatica, V. K. Gombar, QSAR and Combinatorial Science 22, 69 (2003)
B. Sepehri, Z. Hassanzadeh, R. Ghavami, J. Iran. Chem. Soc. 13, 1525 (2016)
Z. Mozafari, M. Arab Chamjangali, M. Beglari, R. Doosti, Chem. Biol. Drug Des. (2020)
D. C. Montgomery, E. A. Peck, G. G. Vining, Introduction to linear regression analysis, John Wiley & Sons, London (2021)
A. M. E. Saleh, M. Arashi, B. G. Kibria, Theory of ridge regression estimation with applications, John Wiley & Sons, London (2019)
K. Gholivand, A.A.E. Valmoozi, M. Salahi, F. Taghipour, E. Torabi, S. Ghadimi, M. Sharifi, M. Ghadamyari, J. Iran. Chem. Soc. 14, 427 (2017)
L. Asadi, K. Gholivand, K. Zare, J. Iran. Chem. Soc. 13, 1213 (2016)
F. Sadeghi, A. Afkhami, T. Madrakian, R. Ghavami, J. Iran. Chem. Soc., p. 1 (2021)
J. Zupan, J. Gasteiger, Neural networks for chemists: an introduction, John Wiley & Sons, Inc., London (1993)
B. Sepehri, R. Ghavami, S. Farahbakhsh, R. Ahmadi, Int. J. Environ. Sci. Technol. (Tehran), p. 1 (2021)
C. Rücker, G. Rücker, M. Meringer, J. Chem. Inf. Model. 47, 2345 (2007)
R. Ghavami, B. Sepehri, J. Iran. Chem. Soc. 13, 519 (2016)
A. Golbraikh, A. Tropsha, J. Mol. Graphics Modell. 20, 269 (2002)
Acknowledgements
The authors are thankful to the Shahrood University of Technology Research Council for supporting this work.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
ZM: Methodology, Software, Writing—original draft, Investigation, Writing—review and editing. MAC: Supervision, Writing—review and editing, Data curation. MA: Methodology, Software, Validation, Writing—review and editing. NG: Review and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Rights and permissions
About this article
Cite this article
Mozafari, Z., Arab Chamjangali, M., Arashi, M. et al. QSRR models for predicting the retention indices of VOCs in different datasets using an efficient variable selection method coupled with artificial neural network modeling: ANN-based QSPR modeling. J IRAN CHEM SOC 19, 2617–2630 (2022). https://doi.org/10.1007/s13738-021-02488-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-021-02488-2