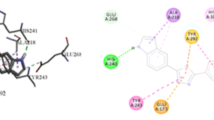
Abstract
A series of 1,2,4-triazole hydrazones 1–25 has been synthesized and characterized using different spectroscopic techniques including FT-IR, 1H-NMR, and ESI MS spectrometry. The synthetic derivatives were evaluated for their β-glucuronidase enzyme inhibition properties. Among them, 17 compounds demonstrated potential inhibitory activity towards β-glucuronidase with IC50 values ranging between 2.50 and 53.70 µM. Compounds 1 having IC50 = 2.50 ± 0.01 µM was found to be the most active compound of the series and showed remarkable activity and found to be far more potent than the standard d-saccharic acid 1,4-lactone (IC50 = 48.4 ± 1.25 µM). Furthermore, the possible binding interaction of active compounds was explored by in silico studies. These compounds can be used for anti-diabetic drug development process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterocyclic compounds have the top-most priority in drug discovery and development due to their unique ability to bind with various biomolecules, particularly the nitrogenous heterocyclic compounds gained more attention in this field. Among them, 1,2,4-triazole derivatives have been reported as very important class of heterocyclic compounds with diverse pharmacological, agricultural, as well as some industrial applications. Many of the commercial drugs such as triadimefon, triadimenol, flusilazole, and flupoxam have triazole scaffold [1] (Fig. 1).
Previously, 1,2,4-triazole derivatives have been reported for their antiparasitic [2], anti-inflammatory [3, 4], herbicidal [5], antifungal [6, 7], antimicrobial [8], brassinosteroid biosynthesis inhibitory [9], and cytostatic activities [10]. Beside this, the hydrazones (molecules containing –C=N– bonding) are also molecules of interest due to their unique application in many fields of chemistry [11]. Triazole hydrazones are also reported for their phosphodiesterase inhibitory activity [12].
Diabetes mellitus (DM) is a syndrome of irregular carbohydrate and protein metabolism that produces some acute chronic complications due to relative deficiency of insulin. Hyperglycaemia is the most common marker of diabetics. It has an adverse effect on vascular cells during the progression of diabetes and creates vascular complications. Moreover, individuals with diabetes may be more likely to develop periodontal and cardiovascular diseases than healthy individuals. These all functions may control by the lysosomal enzymes. Defects in the lysosomal enzymes provoke an accumulation of non-degraded molecules in the lysosomal system. The lysosomal enzyme, β-glucuronidase is found in spleen, lungs, biles, urine, etc. It is responsible for the catalytic degradation of glucuronosyl-O-bonds and glycosaminoglycans, including heparin, chondroitin, and dermatan sulphates. The high expression of β-glucuronidase enzyme was reported in many types of malignancies such as melanomas, bronchial tumour, and breast gastrointestinal tract carcinoma. The elevated levels of β-glucuronidase enzyme were found in borderline tuberculoid and lepromatous patients. Besides these, the low level of this enzyme may cause mucopolysaccharidosis type VII (MPS VII; Sly Syndrome). β-Glucuronidase and some other enzymes have been found to be up-regulated in many acute and chronic pathological conditions such as trauma, sepsis, and diabetes mellitus [13,14,15,16].
Various approaches were used to control diabetes through different modes of action such as gluconeogenesis inhibition, insulin release stimulation, increase of glucose transporters, and delaying glucose absorption in the intestine [17,18,19].
Rationale of the current study
For the exploration of new lead compounds as antidiabetic agents, earlier, we have reported many heterocyclic derivatives such as hydrazones A, oxadiazoles B, and flavones C with β-glucuronidase, antiglycation, and α-glucosidase activities, respectively [20,21,22,23,24]. In addition to that the standard d-saccharic acid 1,4-lactone, having one five-member heterocyclic ring with adjacent electron-donating substituents, i.e. hydroxyl and carboxylic groups. Both of these parts may interact with the enzyme efficiently, in the same manner our synthetic compounds also possess these two structural features, i.e. heterocyclic triazole ring and substituted aryl part (electron-donating/withdrawing substituents). So, it can be hypothesized that these compounds may also be bound to the protein as the standard. Therefore, in continuation of our previous work and resemblance with the standard, herein, we are reporting the β-glucuronidase inhibitory activities of 1,2,4-triazole hydrazone derivatives. The inhibitory activities were also supported by in silico studies (Fig. 2).
Results and discussion
Chemistry
The triazole derivatives 1–25 were synthesized by refluxing 2-amino-1,2,4-triazole with different substituted aryl carbonyl derivatives (aldehydes and ketones) (Scheme 1). The periodical TLC analysis is used to monitor the reaction. After refluxing and solvent evaporation, the crude product was crystallized from ethanol and pure triazole hydrazones 1–25 were obtained in excellent yield (Table 1 and Fig. 3).
β-glucuronidase inhibition activity
All the synthetic triazole hydrazone derivatives 1–25 with different substitution on aryl part obtained from aldehyde or ketone were evaluated for their β-glucuronidase inhibitory activities and results were compared with the standard d-saccharic acid 1,4-lactone (IC50 = 48.4 ± 1.25 µM) (Table 2).
Compounds 1–25 showed remarkable β-glucuronidase inhibitory activity with IC50 values in the range of 2.50–33.50 µM. Out of 25, 17 triazole hydrazone derivatives 1 (IC50 = 2.50 ± 0.01 µM), 2 (9.40 ± 0.25 µM), 3 (4.60 ± 0.01 µM), 4 (3.60 ± 0.01 µM), 5 (24.30 ± 0.55 µM), 6 (22.10 ± 0.50 µM), 7 (23.510 ± 0.40 µM), 8 (29.60 ± 0.50 µM), 9 (9.40 ± 0.20 µM), 10 (13.50 ± 0.30 µM), 12 (43.10 ± 0.70), 13 (14.20 ± 0.35 µM), 14 (34.10 ± 0.65 µM), 15 (32.20 ± 0.21 µM), 16 (33.50 ± 0.63 µM), and 17 (19.40 ± 0.30 µM) showed superior activity than the standard d-saccharic acid 1,4-lactone (IC50 = 48.4 ± 1.25 µM) (Table 2).
Structure–activity relationship was evaluated in terms of substitution pattern on aryl part obtained from carbonyl derivatives. The most active compound of this series is compound 1 (IC50 = 2.50 ± 0.01 µM) and it was derived from isatin. The structural similarity with the standard having five-member ring with carbonyl at 2nd position with adjacent –NH moiety may be the reason for superb activity. In the same manner, compounds 15 (IC50 = 32.20 ± 0.21 µM) and 16 (IC50 = 33.50 ± 0.63 µM) derived from furfural and 5-methyl furfural also marked for their magnificent activity. These derivatives also have five-member ring system but these derivatives were not as active as compound 1. The suppression in activity might be due to aromatic nature of five-member ring or non-availability of carbonyl group at 2nd position, but still they showed higher activity than standard (Fig. 4).
Among chloro-substituted derivatives, the 2,4-dichloro-substituted compound 3 (IC50 = 4.60 ± 0.01 µM) was found to be most active compound. The 3-chloro-substituted compound 2 (IC50 = 9.40 ± 0.25 µM) also showed excellent inhibition potential. It is worth noting that mixed chloro substitution with electron-donating group, i.e. –OH as in compound 4 (IC50 = 3.60 ± 0.01 µM), showed an enhanced activity, while the addition of electron withdrawing such as –NO2 group with chloro substitution reduced the activity up to fivefolds as in compound 5 (IC50 = 24.30 ± 0.55 µM) (Fig. 5).
The 2,5-di-hydroxyl-substituted compound 9 (IC50 = 9.41 ± 0.20 µM) showed higher activity while mono –OH derivatives 6 (IC50 = 22.10 ± 0.50 µM) and 7 (IC50 = 23.510 ± 0.40 µM) showed lesser activity than compound 9. The decline in activity was observed in compound 8 (IC50 = 29.60 ± 0.50 µM) containing –OH group with additional –OCH3 group at 3rd position. However, all these derivatives showed higher activity than the standard (Fig. 6).
Among electron-withdrawing substituents nitro derivatives, compounds 10 (IC50 = 13.50 ± 0.30 µM) and 12 (IC50 = 43.10 ± 0.70 µM) containing 2 and 4 –NO2 group, respectively, showed better potential than the standard, whereas compound 11 with 3-NO2 (IC50 = 53.70 ± 0.80 µM) substitution was found to be a less active compound (Fig. 7).
The compounds 13 (IC50 = 14.20 ± 0.35 µM) and 14 (IC50 = 34.10 ± 0.65 µM) with substitutions N,N-di-CH3 and SCH3 at 4th position, respectively, showed significant β-glucuronidase inhibition potential. Compound 17 (IC50 = 19.40 ± 0.30 µM) with two triazole ring also showed better activity than the standard. All other derivatives were found to be inactive against β-glucuronidase (Fig. 8).
The above activity pattern suggested that substitution at 2nd position of aryl part may help to enhance the activity, as in most cases 2-substituted derivatives showed superior activity than the standard, but some other factors such as polarity, ability to form hydrogen bond, size, hydrophilic or hydrophobicity and electron-donating or -withdrawing nature of group may also be responsible for activity.
To understand the binding mode, some of the active compounds were subjected for in silico studies and results are discussed in following paragraphs.
Molecular docking studies
In silico studies were carried out to support the in vitro β-glucuronidase enzyme inhibitory potential studies. The docking of most active molecules 1–4, 9, and 11 with β-glucuronidase enzyme discloses that all the inhibitor ligands are displaying the bonding with different amino acids in the active site (Fig. 9).
Structure–activity relationship (SAR) was evaluated by three-dimensional human β-glucuronidase structure. For our study, X-ray crystal structure of human β-glucuronidase was downloaded from Protein Data Bank (http://www.rcsb.org/pdb) (PDB ID: 1BHG) (Fig. 9). Before docking the synthetic derivatives, the protein active site interaction was studied by docking the standard substrate molecule with it by Autodock Vina and Autodock Tools [25,26,27].
The modelled structure of d-saccharic acid 1,4-lactone-bound with human β-glucuronidase displayed the accurate orientation of substrate towards the catalytic residues Glu451 and Glu540 (Fig. 10a). During catalysis in human β-glucuronidase, it is proposed that Glu451 acts like acid/base catalyst, whereas Glu540 has nucleophilic characteristics [28]. As shown in Fig. 10a, the inhibitor precisely fits into the active site and interacts via hydrogen bonds with the receptor active site residues including Asp207, His385, Asn450, Glu540 and Lys606. The binding models most active compounds obtained after docking shown in Fig. 10.
First, most active derivative 1 (IC50 = 2.50 ± 0.01 µM) was predicted for its binding conformations to the straight binding groove of β-glucuronidase. Graphical investigation of the top-most graded pose of compound 1 showed that the amine group of indolin-2-one mediated interaction with the side chain carboxyl oxygen (Oε1) of Glu540 (2.43 Å) and hydroxyl oxygen (Oη) of Tyr504 (2.4 Å) via strong hydrogen bonding. These two hydrogen bonds could be the main reason of the high inhibition potential of compound 1. The phenyl ring of indolin-2-one is involved in π–anion and π–π interaction with the Glu451 (3.57 Å) carbonyl residue and phenyl ring of Tyr504 (5.0 Å), respectively. Additionally, carbon–hydrogen bond interaction is found between =CH– of triazole group and the carbonyl oxygen of Tyr205 (3.71 Å) along with π-donor hydrogen bond between triazole ring and amine group of Asp207 (2.66 Å) (Fig. 10e). Other residues such as Phe208, His385, Val410, Asn450, Asn484, Tyr508, Trp587, and Lys606 stabilize the most active compound 1.
Binding mode of compound 4 which is the second-most active derivative of the series (IC50 = 3.60 ± 0.01 µM) showed that one nitrogen atom of the triazole ring is interacting through strong hydrogen bonding with the amino residue (Hζ3) of Lys606 (2.45 Å). Other van der Waals, π–π, π–anion and π–cation interactions of the phenyl and triazole rings of 4 with the side residues especially Glu451 and Glu540, can be visualized in Fig. 10c. These interactions stabilize the second-most potent compound 4 in the active site of enzyme.
The binding mode of analogue 3 (IC50 = 4.60 ± 0.01 µM) showed that the amine group of triazole ring interacts with the carboxyl oxygen (Oδ2) counterpart of Asp207 (2.52 Å) via hydrogen bonding, while one of the imine nitrogen of triazole ring is involved in forming two hydrogen bonds with two side residues, i.e. hydroxyl group of Tyr508 (2.18 Å) and amine hydrogen (Hζ2) of Lys606 (3.09 Å), respectively (Fig. 10b). π–Anion interaction was also found between the triazole ring and the carboxyl oxygen (Oε2) of Glu540 (3.63 Å).
Docking result of another active compound 2 (IC50 = 9.41 ± 0.25 µM) revealed that the imine nitrogen is engaged in hydrogen bonding with the hydroxyl residue of Tyr508 (2.45 Å), while one of the imine nitrogen of triazole group was also interacting with the amine hydrogen (Hζ3) of Lys606 (2.66 Å). Carbon–hydrogen bond interaction was spotted between hydrogen of imine group and carboxyl oxygen (Oε1) of Glu451 (3.69 Å). Carboxyl oxygen (Oε2) of Glu540 was also spotted in π–anion interaction (3.49 Å) with the triazole ring of 2 (Fig. 10d).
Binding interaction of 9 (IC50 = 9.40 ± 0.20 µM) showed that the imine nitrogen of triazole ring is engaged with hydroxyl group of Tyr508 (2.24 Å) by hydrogen bonding, while the triazole ring was found involved in π–anion interaction with the carbonyl oxygen (Oε2) of Glu540 (3.50 Å) (Fig. 10f).
Compound 11 (IC50 = 53.70 ± 0.80 µM) showed lesser activity but still it showed good interaction with the binding site as its IC50 value is negligibly higher than the standard compound, i.e. d-saccharic acid 1,4-lactone (Fig. 10a)
Hydrogen bond surface of predicted docked poses of all the selected compounds inside the active site of β-d-glucuronidase was analyzed. The results show that the ligand exactly fits into the active site of the enzyme (Fig. 11).
The line with black dashes indicates different interactions and the distances are in Angstrom. Amino acid residues involved in interaction are shown in element coloured stick (Figs. 11, 12).
Conclusion
Triazole hydrazones 1–25 with different functional groups have been synthesized and their β-glucuronidase activity was explored. Among them, 17 compounds showed remarkable β-glucuronidase inhibition activity. The results were also supported by molecular docking studies which identified various structural features that interact with the active site of enzyme. This work will enable us to identify new inhibitors and structural features contributing towards β-glucuronidase inhibition activity.
Supplementary information
The H1NMR spectra of most active compounds are available in supplementary information.
Experimental
Materials and methods
The β-glucuronidase was purchased from Sigma-Aldrich, USA. 3-Amino-4(H)-1,2,4-triazole, and various substituted aryl aldehydes/ketone were purchased from Merck/TCI, Germany/Japan. Dried ethanol was used in all experiments and Avance Bruker AM 300, 500 MHz machines were used for 1H- and 13C-NMR experiments, respectively. Agilent 6330 Ion Trap instrument using positive or negative mode was used for ESI MS determination. CHNS analyses were performed on a Perkin Elmer, Milano, Italy. Kieselgel 60, 254, E. Silica gel-coated aluminium plates, Merck, Germany, were used for thin-layer chromatography. UV light source with wavelengths 254 and 365 nm were used for TLC visualization.
Typical method for synthesis of compounds 1–25
Variously substituted aryl aldehydes (0.03 moles) were dissolved in 25 mL of ethanol with catalytic amount of acetic acid, reaction mixture was stirred and equimolar amount of 3-amino-1,2,4-triazole was added into it and refluxed for 6 h. After refluxing, solvent was evaporated under rotary evaporator; the obtained precipitates were washed with water, dried, and characterized through various spectroscopic techniques.
3-(1H-[1, 2, 4]-Triazol-3-ylimino)-1,3-dihydro-indol-2-one (1)
Yield: 81%; 1H-NMR (DMSO-d6): δ 8.40 (s, 1H), 8.07 (m, 2H), 7.48 (m, 2H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.1, 161.3, 146.4, 146.3, 138.5, 131,3, 129.1, 124.0, 123.2, 120.2: ESI MS (m/z): 214.1 [M + 1]+; FTIR (νcm− 1): –NH– 3189.54, –C=O– 1724.12, –C=N– 1614.10, –C=C– 1482.73, –C–N– 1330.61; anal. calcd. for C10H7N5O, C = 56.32, H = 3.30, N = 32.84, found, C = 55.31, H = 3.29, N = 32.87.
(3-Chloro-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (2)
Yield: 90%; 1H-NMR (DMSO-d6): δ 9.43 (s, 1H), 8.57 (s, 1H), 7.92 (d, 1H, J = 8.3 Hz), 7.49 (dd, 1H, J = 7.9 Hz), 7.04 (d, 1H, J = 8.3 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.5, 146.4, 146.3, 133.6, 132.4, 131.1, 130.1, 129.2, 127.3: ESI MS (m/z): 219.1 [M + 3]+, 207 [M + 1]+; FTIR (νcm− 1): –NH– 3233.02, –N=C– 1586.27, –C–N– 1329.00, –C=C– 1477.10, –C–Cl– 835.74; anal. calcd. for C9H7ClN4, C = 51.29, H = 3.40, N = 26.12, found, C = 51.28, H = 3.40, N = 26.09.
(2,4-Dichloro-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (3)
Yield: 60%; 1H-NMR (DMSO-d6): δ 9.02 (s, 1H), 8.27 (s, 1H) 7.81 (d, 1H, J = 8.4 Hz), 7.34 (d, 1H, J = 8.2 Hz), 6.92 (s, 1H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.2, 146.5, 146.4, 137.3, 135.5, 131.3, 129.2, 129.1, 126.9; ESI MS (m/z): 245.0 [M + 5]+, 243 [M + 3]+, 243 [M + 1]+; FTIR (νcm− 1): NH– 3123.27, –C=N– 1605.13, –C=C– 1440.63, –N–C– 1319.34, –C–Cl– 799.70; anal. calcd. for C9H6Cl2N4, C = 44.82, H = 2.50, N = 23.23, found, C = 44.79, H = 2.45, N = 23.19.
5-Chloro-2-[(1H-[1,2,4]-triazol-3-ylimino)-methyl]-phenol (4)
Yield: 75%; 1H-NMR (DMSO-d6): δ 7.46 (d, 1H, J = 8.2 Hz), 7.42 (d, 1H, J = 8.1 Hz), 7.40 (s, 1H), 7.06 (s, 1H), 7.04 (s, 1H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.1, 159.5, 146.4, 146.6, 137.3, 131.7, 121.3, 116.4, 116.1; ESI MS (m/z): 225.0 [M + 3]+, 223 [M + 1]+; FTIR (νcm− 1): –OH/NH– 3111.30, –N=C– 1605.87, –C=C– 1569.32, –C–N– 1355.70, –C–Cl– 844.03; anal. calcd. for C9H7ClN4, C = 51.30, H = 3.40, N = 26.10, found, C = 51.27, H = 3.37, N = 26.08.
2-Chloro-5-nitro-benzylidene-(1H-[1, 2, 4]-triazol-3-yl)-amine (5)
Yield: 57%; 1H-NMR (DMSO-d6): δ 9.13 (s, 1H), 8.51(s, 1H), 7.60–7.12 (m, 3H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.3, 146.4, 146.3, 146.5, 140.2, 132.4, 129.6, 127.2, 125.1 ESI MS (m/z): 254.0 [M + 3]+, 252 [M + 1]+; FTIR (νcm− 1): –NH– 3244.91, –N–C– 1342.28, –N=C– 1574.50, –NO2 1523.34, –C=C– 1487.34, –C–Cl– 875.15; anal. calcd. for C9H6ClN5O2, C = 42.96, H = 2.40, N = 27.83, found, C = 42.93, H = 2.38, N = 27.80.
2-[(1H-[1,2,4]-Triazol-3-ylimino)-methyl]-phenol (6)
Yield: 70%; 1H-NMR (DMSO-d6): δ 9.92 (s, 1H), 7.99 (d, 1H, J = 8.5 Hz), 7.59 (m, 2H), 7.08 (d, 1H, J = 8.4 Hz), 6.92 (s, 1H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 157.6, 146.5, 146.4, 132.1, 130.2, 121.1, 118.0, 115.3; ESI MS (m/z): 189.0 [M + 1]+; FTIR (νcm− 1):–OH– 3251.93, –NH– 3123.01, –C=N– 1612.18, –N–C– 1353.24; anal. calcd. for C9H8N4O, C = 56.44, H = 4.28, N = 29.75, found, C = 56.41, H = 4.26, N = 28.75.
3-[(1H-[1,2,4]-Triazol-3-ylimino)-methyl]-phenol (7)
Yield: 98%; 1H-NMR (DMSO-d6): δ 9.17 (s, 1H), 8.52 (s, 1H), 7.93 (d, 1H, J = 9.1 Hz), 7.72 (m, 1H), 7.40 (d, 1H, J = 9.4 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.2, 157.6, 146.5, 146.4, 132.2, 130.1, 121.6, 118.5, 116.1; ESI MS (m/z): 189.0 [M + 1]+; FTIR (νcm− 1): OH/NH– 3239.31, –C=N– 1582.11, C=C– 1493.92, –C–N– 1338.33; λmax (nm): 310. anal. calcd. for C9H8N4O, C = 57.43, H = 4.27, N = 29.78, found, C = 57.41, H = 4.25, N = 28.76.
3-Methoxy-4-[(1H-[1,2,4]-triazol-3-ylimino)-methyl]-phenol (8)
Yield: 81%; 1H-NMR (DMSO-d6): δ 8.43 (s, 1H), 8.30 (s, 1H), 8.29 (s, 1H), 8.27 (d, 1H, J = 7.6 Hz), 7.47 (d, 1H, J = 7.3 Hz), 1.32 (s, 3H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.7, 162.5, 160.1, 146.3, 146.2, 131.1, 109.4, 108.7, 101.2, 56.2; ESI MS (m/z): 219 [M + 1]+; FTIR (νcm− 1): –OH/NH– 3104.36; –C=C– 1538.80, –C=N– 1579.53, –C–N– 1330.07; anal. calcd. for C10H10N4O2, C = 55.05, H = 4.63, N = 25.69, found, C = 55.03, H = 3.58, N = 24.64.
2-[(1H-[1,2,4]-Triazol-3-ylimino)-methyl]-benzene-1,4-diol (9)
Yield: 68%; 1H-NMR (DMSO-d6): δ 8.48 (s, 1H), 7.73 (s, 1H), 8.43 (s, 1H), 7.08 (d, 1H, J = 7.3 Hz), 7.01 (d, 1H, J = 7.8 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.2, 150.1, 150.3, 146.5, 146.4, 119.7, 119.2, 117.5, 117.1; ESI MS (m/z): 205 [M + 1]+; FTIR (νcm− 1): –OH/NH– 3131.04, –C=N– 1621.17, –C=C– 1517.52, –C–N– 1313.69; anal. calcd. for C9H8N4O2, C = 52.95, H = 3.94, N = 27.45, found, C = 51.91, H = 2.92, N = 26.42.
(2-Nitro-benzylidene)-(1H-[1, 2, 4] triazol-3-yl)-amine (10)
Yield: 79%; 1H-NMR (DMSO-d6): δ 8.45 (s, 1H), 8.25 (s, 1H), 8,21 (d, 2H, J = 9.2 Hz), 7.64 (d, 1H, J = 9.3 Hz), 7.47 (m, 2H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.5, 148.7, 146.5, 146.4, 134.5, 131.4, 129.7, 126.2, 123.4; ESI MS (m/z): 218.0 [M + 1]+; FTIR (νcm− 1): –NH– 3152.34, –C=N– 1520.96, –C=C– 1445.74, NO2- 1343.31, –C–N– 1309.86; anal. calcd. for C9H7N5O2, C = 49.78, H = 3.27, N = 32.26, found, C = 48.74, H = 2.59, N = 31.21.
(3-Nitro-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (11)
Yield: 60%; 1H-NMR (DMSO-d6): δ 7.60 (s, 1H), 7.29 (s, 1H), 8.63 (s, 1H), 8.02 (bd, 2H), 7.75 (d, 1H, J = 6.8 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 148.5, 146.5, 146.4, 135.2, 132.4, 129.3, 125.7, 124.1; ESI MS (m/z): 218.0 [M + 1]+; FTIR (νcm− 1): –NH– 3233.78 –N=C– 1583.43, –C=C– 1524.85, –NO2– 1478.41, –N–C– 1344.16; anal. calcd. for C9H7N5O2, C = 49.79, H = 3.26, N = 32.27, found, C = 48.75, H = 2.23, N = 31.23.
(4-Nitro-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (12)
Yield: 73%; 1 H-NMR (DMSO-d6): δ 8.28 (s, 1H), 7.29 (s, 1H), 7.83 (d, 2H, J = 8.3 Hz), 7.54 (d, 2H, J = 8.2 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 150.4, 146.5, 146.4, 137.2, 129.5, 129.5, 123.3, 123.3; ESI MS (m/z): 218.0 [M + 1]+; FTIR (νcm− 1): –NH– 3100.35, –N=C– 1596.27, –C=C– 1516.14, –NO2– 1470.29, –N–C– 1344.89; anal. calcd. for C9H7N5O2, C = 49.79, H = 3.27, N = 32.27, found, C = 48.74, H = 2.22, N = 31.21.
(4-Dimethylamino-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (13)
Yield: 75%; 1H-NMR (DMSO-d6): δ 9.02 (s, 1H), 8.51 (s, 1H), 7.81 (d, 2H, J = 7.6 Hz), 6.79 (d, 2H, J = 7.8 Hz), 3.39 (s, 6H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.7, 146.5, 146.4, 129.7, 129.7, 120.5, 113.1, 113.1, 43.3, 43.3; ESI MS (m/z): 216.0 [M + 1]+; FTIR (νcm− 1): –NH– 3093.36, –C=N– 1581.58, –C=C– 1526.20, –C–N– 1327.57, anal. calcd. for C11H13N5, C = 61.37, H = 6.08, N = 32.55, found, C = 60.34, H = 5.06, N = 31.52.
(4-Methylsulfanyl-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (14)
Yield: 90%; 1H-NMR (DMSO-d6): δ 9.17 (s, 1H), 8.52 (s, 1H), 7.93 (d, 2H, J = 7.3 Hz), 7.40 (d, 2H, J = 7.1 Hz), 2,53 (s, 1H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4, 137.7, 129.1, 129.1, 126.3, 126.3, 18.7; ESI MS (m/z): 219.0 [M + 1]+; FTIR (νcm− 1): –NH– 3140.76, –N=C– 1614.20, –C=C– 1590.21, –N–C– 1329.10, –C–S– 705.86; anal. calcd. for C10H10N4S, C = 55.03, H = 4.61, N = 25.68; S = 14.67, found, C = 54.00, H = 3.59, N = 24.65, S = 14.64.
Furan-2-ylmethylene-(1H-[1, 2, 4]-triazol-3-yl)-amine (15)
Yield: 36%; 1H-NMR (DMSO-d6): δ 8.39 (s, 1H), 8.23 (s, 1H), 7.97 (d, 1H, J = 6.4 Hz), 7.28 (dd, 1H), 7.84 (d, 1H, J = 6.7 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.3, 146.5, 146.4, 143.1, 143.1, 110.3, 110.3; ESI MS (m/z): 163.0 [M + 1]+; FTIR (νcm− 1): –NH– 3404.45, –C=N– 1622.50, –C=C– 1582.85, –C–N– 1295.75, –C–O– 1250.00; anal. calcd. for C7H6N4O, C = 51.86, H = 3.74, N = 34.56, C, 54.54; H, 4.58; N, 31.80 °C = 50.83, H = 2.71, N = 33.53.
(5-Methyl-furan-2-ylmethylene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (16)
Yield: 42%; 1H-NMR (DMSO-d6): δ 8.41 (s, 1H), 8.01 (s, 1H), 7.78 (d, 1H, J = 6.3 Hz), 6.78 (d, 1H, J = 6.4 Hz), 1.49 (s, 3H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.3, 153.4, 146.5, 146.4, 141.2, 111.2, 106.7,14.5; ESI MS (m/z): 177.0 [M + 1]+ ; FTIR (νcm− 1): –NH– 3130.14, C = N– 1621.48, –C=C– 1581.44, –C–N– 1249.88, –C–O– 1249.88, –CH– 2934.86; anal. calcd. for C8H8N4O, C = 54.55, H = 4.58, N = 31.81, C = 53.50, H = 3.53, N = 30.74.
N-{2-[(1H-1,2,4-triazol-3-ylimino)methyl]phenyl}methylidene-1H-1,2,4-triazol-3 amine (17)
Yield: 90%; 1H-NMR (DMSO-d6): δ (s, 9.77, 2H), 7.71 (dd, 2H), 7.48 (d, 2H, J = 8.1 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 162.5, 163.3, 146.4, 146.4, 146.3, 146.3, 131.6, 131.5, 130.8, 130.6, 129.4, 129.3; ESI MS (m/z): 267.0 [M + 1]+; FTIR (νcm− 1): –NH– 3119.65, –C=N– 1647.49, –C–N– 1340.28, –C=C– 1557.76; anal. calcd. for C12H10N8 C = 53.11, H = 2.71, N = 41.01, Found, C = 53.08, H = 2.72, N = 42.06.
(2,4-Dimethyl-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (18)
Yield: 80%; 1H-NMR (DMSO-d6): δ 9.42 (s, 1H), 8.43 (s, 1H), 7.97 (d, 1H, J = 8.4 Hz), 7.87 (s, 1H), 7.17 (dd, 1H), 2.33 (s, 6H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4, 139.5, 138.3, 130.1, 128.7, 128.7, 126.2, 21.1, 14.7; ESI MS (m/z): 201.0 [M + 1]+; FTIR (νcm− 1): –NH– 3149.08, –N=C– 1600.47, –C=C– 1437.95, –N–C– 1322.23, –CH– 2965.42; anal. calcd. for C11H12N4, C = 65.97, H = 6.05, N = 27.97, found, C = 64.95, H = 5.01, N = 26.97.
(3,4-Dimethyl-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (19)
Yield: 60%; 1H-NMR (DMSO-d6): δ 9.13 (s, 1H), 8.51 (s, 1H), 7.55 (d, 1H, J = 7.8 Hz), 7.48 (s, 1H), 7.13 (d, 1H, J = 7.6 Hz), 1.90 (s, 6H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4, 140.5, 138.4, 129.4, 129.2, 128.3, 125.7, 14.6, 14.6 ; EI ESI MS (m/z): 201.0 [M + 1]+; FTIR (νcm− 1): –NH– 3198.28, –N=C– 1619.06, –C=C– 1605.59, –N–C– 1344.23; anal. calcd. for C11H12N4, C = 64.96, H = 5.02, N = 27.96, found, C = 64.94, H = 6.01, N = 27.92.
(3-Methoxy-benzylidene)-(1H-[1, 2, 4]-triazol-3-yl)-amine (20)
Yield: 90%; 1H-NMR (DMSO-d6): δ 8.85 (s, 2H), 8.34 (s, 1H), 8.18 (s, 1H), 7.83 (d, 1H, J = 7.2 Hz), 7.53 (dd, 1H), 7.15 (d, 1H, J = 7.5 Hz), 3.88 (s, 3H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 162.3, 146.5, 146.4, 132.1, 129.7, 121.2, 116.2, 114.5, 56.1; ESI MS (m/z): 203.0 [M + 1]+; FTIR (νcm− 1): –NH– 3236.29, –N=C– 1584.34, –C=C– 1491.33, –N–C– 1328.64; anal. calcd. for C10H10N4O, C = 59.41, H = 4.97, N = 27.72, found, C = 59.39, H = 3.97, N = 26.68.
(1H-[1, 2, 4]-Triazol-3-yl)-(3,4,5-trimethoxy-benzylidene)-amine (21)
Yield: 64%; 1H-NMR (DMSO-d6): δ 9.87 (s, 1H), 8.50 (s, 1H), 7.49 (s, 1H), 7.40 (s, 1H), 3.76 (s, 9H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 148.7, 148.7, 146.5, 146.4, 135.6, 125.2, 107.1, 56.2, 56.2, 56.2; ESI MS (m/z): 263.0 [M + 1]+; FTIR (νcm− 1): –NH– 3204.87, –N=C– 1682.54, –C=C– 1622.23, –N–C– 1354.88; anal. calcd. for C12H14N4O3, C = 53.96, H = 5.38, N = 20.36, found, C = 53.93, H = 5.35, N = 21.34.
Naphthalen-2-ylmethylene-(1H-[1, 2, 4]-triazol-3-yl)-amine (22)
Yield: 88%; 1H-NMR (DMSO-d6): δ 9.77 (s, 1H), 8.81 (s, 1H), 8.23 (s, 1H), 7.71 (d, 1H, J = 8.1 Hz), 7.70 (d, 1H, J = 7.9 Hz), 7.49 (d, 2H, J = 7.6 Hz), 7.48 (m, 2H), 6.98 (d,1H, J = 7.8 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4, 135.8, 133.4. 128.5, 128.3, 128.2, 128.2, 128.1, 126.3, 125.7, 125.7; ESI MS (m/z): 223.0 [M + 1]+; FTIR (νcm− 1): –NH– 3235.54, –C=N– 1689.92, –C=C– 1586.64, –C–N– 1326.26; anal. calcd. for C13H10N4, C = 70.26, H = 3.54, N = 24.21, found, C = 70.24, H = 3.51, N = 24.17.
Anthracen-9-ylmethylene-(1H-[1, 2, 4]-triazol-3-yl)-amine (23)
Yield: 71%; 1H-NMR (DMSO-d6): δ 10.47 (s, 1H), 9.03 (s, 1H), 8.98 (m, 4H), 7.74 (m, 4H), 8.20 (s, 1H), 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4, 132.2, 132.2, 131.7, 131.7, 128.7, 128.4, 128.1, 128.1, 128.0, 128.0, 125.2, 125.2, 125.2, 125.2; ESI MS (m/z): 273.0 [M + 1]+; FTIR (νcm− 1): –NH– 3048.61, –N=C– 1621.87, –C=C– 1475.13, –N–C– 1342.41; anal. calcd. for C17H12N4, C = 74.97, H = 4.44, N = 20.59, found, C = 73.96, H = 3.42, N = 21.55.
Pyren-1-ylmethylene-(1H-[1, 2, 4] triazol-3-yl)-amine (24)
Yield: 74%; 1H-NMR (DMSO-d6): δ 8.66 (s, 1H), 8.46 (s, 1H), 7.72 (m, 3H), 7.52 (d, 1H, J = 8.1 Hz), 7.49 (d, 1H, J = 8.3 Hz), 7.48 (d, 2H, J = 8.5 Hz), 6.90 (d, 2H, J = 8.1 Hz); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 146.5, 146.4 132.3, 132.1, 131.8, 130.2, 129.3, 128.3, 128.2, 126.7, 126.5, 126.5, 126.5, 126.5, 126.1, 126.1, 122.7, 122.2; ESI MS (m/z): 297.0 [M + 1]+; FTIR (νcm− 1): –NH– 3036.92, –C=N– 1676.60, –C–N– 1647.48; anal. calcd. for C19H12N4, C = 77.02, H = 4.09, N = 18.90, found, C = 77.01, H = 3.05, N = 17.88.
2-[1-(1H-[1,2,4]Triazol-3-ylimino)-ethyl]-phenol (25)
Yield: 48%; 1H-NMR (DMSO-d6): δ 8.25 (s, 1H), 8.45 (d, 1H, J = 7.4 Hz), 7.64 (m, 2H), 7.40 (d, 1H, 7.6), 1.91 (s, 3H); 13C-NMR (DMSO-d6, 125 MHz): δ 163.4, 157.7, 146.5, 146.4, 132.1, 130.2, 121.1, 118.1, 115.7, 14.3; ESI MS (m/z): 287.0 [M + 1]+; FTIR (νcm− 1): –OH– 3403.16, –NH– 3325.70, –C=N– 1636.31, –C=C– 1592.85, –C–N– 1268.54; anal. calcd. for C10H10N4O, C = 59.42, H = 4.97, N = 27.72, found, C = 58.37, H = 3.96, N = 26.69.
β-Glucuronidase Inhibition Activity
β-Glucuronidase (E.C. 3.2.1.31 from bovine liver, G-0251) inhibition activity of reported compounds was evaluated by previously reported method. In this study, d-saccharic acid 1,4-lactone was used as the standard drug [23, 24].
Molecular docking studies
Autodock vina was used to study the positioning of inhibitors bound in the active site of β-glucuronidase enzyme. PDBQT files of ligands and receptor to be used for docking were generated from their PDB files using Autodock Tools. Protein structure of human β-glucuronidase (PDB ID: 1BHG) as shown in Fig. 8 was taken from PDB (http://www.rcsb.org/pdb). Missing atoms, contacts and bonds were checked in the structure. B-chain and the hetero-atoms were removed from the original file. Calculation of partial atomic charges and addition of hydrogen atoms to the protein molecule were done using Autodock tools. Configuration file containing parameters for a grid box of size 16 × 16 × 16 Å cantered at the coordinates of X: 85.22, Y: 86.62, Z: 87.37 was used to cover the active site. Docking results were analyzed using Discovery Studio Visualizer [28].
References
N. Sun, J. Fu, J. Weng, J. Jin, C. Tan, X. Liu, Molecules 18, 12725–12739 (2013)
T.N. Franklim, L. Freire-de-Lima, J.D.S. Diniz, J.O. Previato, R.N. Castro, L. Mendonca-Previato, M.E.F. de Lima, Molecules 18, 6366–6382 (2013)
W.S. El-Serwy, N.A. Mohamed, E.M. Abbas, R.F. Abdel-Rahman, Res. Chem. Intermed. 39, 2543–2554 (2013)
B. Kocyigit-Kaymakcioglu, A.O. Celen, N. Tabanca, A. Ali, S.I. Khan, I.A. Khan, D.E. Wedge, Molecules 18, 3562–3576 (2013)
W. Ke, N.B. Sun, H.K. Wu, J. Chem. Soc. Pak. 35, 1239–1244 (2013)
X.H. Liu, C.X. Tan, J.Q. Weng, Phosphorus, Sulfur Silicon Relat. Elem. 186, 558–564 (2011)
J.Y. Tong, H.K. Wu, N.B. Sun, X.H. Liu, Chin. J. Struct. Chem. 32, 607–611 (2013)
M.A. Al-Omar, E.S. Al-Abdullah, I.A. Shehata, E.E. Habib, T.M. Ibrahim, A.A. El-Emam, Molecules 15, 2526–2550 (2010)
K. Yamada, Y. Yoshizawa, K. Oh, Molecules 17, 4460–4473 (2012)
K. Benci, L. Mandic, T. Suhina, M. Sedic, M. Klobucar, S.K. Pavelic, K. Pavelic, K. Wittine, M. Mintas. Molecules 17, 11010–11025 (2012)
A. Kajal, S. Bala, S. Kamboj, N. Sharma, V. Saini, J. Catal., 1–14 (2013)
K.M. Khan, S. Siddiqui, M. Saleem, M. Taha, S.M. Saad, S. Perveen, M.I. Choudhary, Bioorg. Med. Chem. 2, 6509–6514 (2014)
H.H. Jajarm, N. Mohtasham, A. Rangiani, J. Oral. Sci. 50, 335–340 (2008)
S. Roy, K. Trudeau, S. Behl, Y. Roy, S. Dhar, A. Chronopoulos, J. Dent. Res. 89, 116–127 (2010)
L. Santacroce, R.G. Carlaio, L. Bottalico, Endoc. Metab. Immune. Disord. Drug Targets 10, 57–70 (2010)
F. Tanwir, M. Altamash, A. Gustafsson, Acta Odontol. Scand. 67, 129–133 (2009)
Y. Yao, W. Sang, M. Zhou, G. Ren, J. Agric. Food Chem. 58, 770–774 (2009)
M. Yoshikawa, T. Murakami, K. Yashiro, H. Matsuda, Kotalanol. Chem. Pharm. Bull. 46, 1339–1340 (1998)
T. Nishioka, J. Kawabata, Y. Aoyama, Baicalein, an α-glucosidase inhibitor from Scutellaria baicalensis. J. Nat. Prod. 61, 1413–1415 (1998)
W. Jamil, S. Perveen, S.A.A. Shah, M. Taha, N.H. Ismail, S. Perveen, N. Ambreen, K.M. Khan, M.I. Choudhary, Molecules 19, 8788–8802 (2014)
M. Taha, N.H. Ismail, W. Jamil, S. Imran, F. Rahim, S.M. Kashif, M. Zulkefeli, Med. Chem. Res. 25, 225–234 (2016)
S. Imran, M. Taha, N.H. Ismail, S.M. Kashif, F. Rahim, W. Jamil, H. Wahab, K.M. Khan, Chem. Biol. Drug Des. 87, 361–373 (2016)
K.M. Khan, F. Rahim, S.A. Halim, M. Taha, M. Khan, S. Perveen, Z. -ul-Haq, M.A. Mesaik, M.I. Choudhary, Bioorg. Med. Chem. 19, 4286 (2013)
N.K.N.Z. Abdullah, M. Taha, N. Ahmat, A. Wadood, N.H. Ismail, F. Rahim, M. Ali, N. Abdullah, K.M. Khan, Bioorg. Med. Chem. 23, 3119 (2015)
O. Trott, A.J. Olson, J. Comput. Chem. 31, 455–461 (2010)
G.M. Morris, R. Huey, W. Lindstrom, M.F. Sanner, R.K. Belew, D.S. Goodsell, J. Comput. Chem. 30, 2785–2791 (2009)
J. Sanjeev, Nat. Struct. Mol. Biol. 4, 375–381 (1996)
Dassault Systèmes BIOVIA, Discovery Studio Modeling Environment, Release 4.5 (Dassault Systèmes, San Diego, 2015)
Acknowledgements
The authors are thankful to University of Sindh, Jamshoro, and Higher Education Commission (HEC), Pakistan, for their financial support under “National Research Support Program for Universities”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jamil, W., Kumari, D., Taha, M. et al. Synthesis, β-Glucuronidase Inhibition, and Molecular Docking Studies of 1,2,4-Triazole Hydrazones. J IRAN CHEM SOC 15, 2441–2454 (2018). https://doi.org/10.1007/s13738-018-1433-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-018-1433-9