Abstract
Two new precious metal coordination complexes [Pd(HL)2] (1) and [PtL2] (2) (where H2L is 1-(carboxymethyl)-1H-pyrazole-3-carboxylic acid) were synthesized by one-pot in situ hydrolysis. The complexes are assembled into a 3D structure by hydrogen bonding interactions and intermolecular contacts. The structures have been established by single-crystal X-ray diffraction, and characterized by FT-IR and liquid state fluorescent spectroscopy. Hirshfeld surface analysis reveals that the O···H contacts outnumber the other contacts in both structures (49.4 % of the total interactions for 1 and 41.6 % for 2).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pyrazolyl ligands are among the important building units used in ligand designing [1] and crystal engineering [2]. The pyrazole carboxylate are known as multidentate bridging ligands [3]. Some of their metal complexes have been used as precursors for chemical vapor phase deposition and found to show luminescent properties [4]. The pyrazole–carboxylate segments can be integrated as part of many polydentate donors [5]. The polypyrazolylborates are among the most useful and widely employed ligands [6].
Coordination chemistry of palladium(II) and platinum(II) complexes with heterocyclic ligands with two potential donor atoms is one of the research areas that has undergone a fast development during the last years [7]. Among all the examples reported so far, those containing pyrazole derivatives are especially attractive because they have multiple applications. For example, the complex of Pt(II) with some pyridylpyrazole ligand has shown greater antitumor activity and lower toxicity than the common cis-[PtCl2(NH3)2] complex [8]. Applications of palladium and platinum complexes including pyrazolic ligands extend also to macromolecular chemistry [9] and homogeneous catalysis [10].
More recently, our group has strong interests in introducing a carboxylic group to the pyrazole rings, several amazing MOFs have been successfully synthesized [11]. Our findings show that the pyrazole carboxylate ligands bearing bulky conjugate groups can indicate strong coordination ability and various coordination modes. This prompted us to prepare more novel ligands to investigate their coordination features. Herein, we adopt ester group to modify the pyrazole unit to prepare one novel organic ligand, 1-(2-ethoxy-2-oxoethyl)-1H-pyrazole-3-carboxylate (Eopzc). To the best of our knowledge, this ligand has been never reported so far. However, in order to further enhance the ability of this ligand, we use one-pot in situ hydrolysis to obtain two new complexes [Pd(HL)2] (1) and [PtL2] (2) (where H2L is 1-(carboxymethyl)-1H-pyrazole-3-carboxylic acid). The photoluminescence property of complexes 1 and 2 has been investigated.
Experimental
General
All chemicals were commercially available and used as received without further purification. The C, H, N microanalyses were carried out with an Elemental Vario-EL CHNS elemental analyzer. The FT-IR spectra were recorded from KBr pellets in the range of 4000–400 cm−1 on a Bio-Rad FTS-7 spectrometer. The crystal structure was determined by single-crystal X-ray diffraction and SHELXL crystallographic software of molecular structure. The synthesis procedure of Eopz, complexes 1 and 2 seen in Scheme 1.
Synthesis of Eopzc
To solution of ethanol (50 mL) and 3-pyrazolecarboxylic acid (1.112 g 10 mmol) at 80 °C was added concentrated sulfuric acid (2 mL), and after 2 h of stirring the ethyl 1H-pyrazole-3-carboxylate was obtained (1.350 g, 9.6 mmol, 96 %). To 20 mL three-necked round bottomed flask equipped with a refluxing condenser was added 1H-pyrazole-3-carboxylate (0.7 g, 5 mmol), ethyl bromoacetate (0.835 g, 0.6 mmol) in 30 mL anhydrous acetone. This suspension was stirred at 60 °C and the reaction was monitored by TLC. When the 1H-pyrazole-3-carboxylate was consumed, the yellow brown reaction mixture was concentrated to dryness. The crude product was purified by column chromatography on silica gel with petroleum ether/ethyl acetate (1/1) as eluent to give the liquid state product Eopzc in yield of 55 % (0.622 g, 2.7 mmol). 1H NMR (DMSO-d6, 400 MHz): δ 7.60 (d, J = 2 Hz, 1H), 6.92 (d, J = 2 Hz, 1H), 5.29 (s, 2H), 4.28–4.22 (m, 2H), 4.15–4.10 (m, 2H), 1.27–1.24 (m, 3H), 1.20–1.16 (m, 3H); 13C NMR (DMSO-d6, 100 MHz): 168.29, 159.50, 139.01, 132.95, 111.87, 61.55, 61.48, 53.77, 14.44, 14.40.
Synthesis of [Pd(HL)2] (1)
A mixture of PdCl2 (0.0354 g, 0.2 mmol), Eopzc (0.0226 g, 0.1 mmol), KOH (0.0112 g 0.2 mmol), water (5 mL) and ethanol (10 mL) was stirred for 10 min at room temperature with the pH value of 8.0. The resulting solution was put into a Teflon-lined autoclave. The reaction mixture was heated at 110 °C for 4 days, followed by slow cooling to room temperature and phase pure crystals of 1 were obtained by manual separation (yield: 0.0135 g, ca. 30.36 % based on Eopzc ligand). Anal. Calcd for C12H10N4O8Pd (M r = 444.64): C, 32.39; H, 2.25; N, 12.60 %, Found: C, 32.28; H, 2.32; N, 12.53 %. IR (cm−1): 3238 (w), 1722 (s), 1640 (s), 1340 (s), 1266 (w), 1240 (s), 1211 (s), 830 (m), 782 (w).
Synthesis of [PtL2] (2)
The synthetic method of 2 is similar to 1, only the salt PdCl2 replaced by K2PtCl6. Finally, phase pure crystals of 2 were obtained by manual separation (yield: 0.0112 g, ca. 21.08 % based on Eopzc ligand). Anal. Calcd for C12H8N4O8Pt (M r = 531.31): C, 27.10; H, 1.51; N, 12.05 %, Found: C, 26.05; H, 1.62; N, 11.08 %. IR (cm−1): 3124 (w), 1734 (m), 1654 (s), 1340 (s), 1266 (w), 1238 (s), 1212 (m), 834 (m), 780 (w).
Crystal structure determination
The diffraction data of two complexes were collected on Agilent G8910A CCD diffractometer with graphite monochromated Mo–Kα radiation (λ = 0.71073 Ǻ) and using the ω–θ scan mode in the ranges 3.3º ≤ θ ≤ 29.0º (1), 4.1º ≤ θ ≤ 29.2º (2), respectively. Raw frame data were integrated with the SAINT program. The structures were solved by direct methods using SHELXS-97 and refined by full-matrix least-squares on F 2 using SHELXS-97 [12]. An empirical absorption correction was applied with the program SADABS [12]. All non-hydrogen atoms were refined anisotropically. All hydrogen atoms were positioned geometrically and refined as riding. Calculations and graphics were performed with SHELXTL [12]. The crystallographic details are provided in Table 1. Selected bond distances and angles of the complexes 1 and 2 are listed in Table 2. The hydrogen bonds and angles of complexes 1 and 2 have been listed in Table 3. Crystallographic data for the structural analysis have been deposited with the Cambridge Crystallographic Data Centre (CCDC numbers: 1061337 and 1061338).
Results and discussion
Crystal structure of [Pd(HL)2] (1)
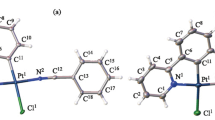
Complex 1 crystallizes in the monoclinic system with the P2/c space group, and each asymmetric unit consists of one-half divalent Pd(II) ion and one deprotonated ligand (HL), shown in Figure S1. The molecular structure is illustrated in Fig. 1. The structure consists of discrete centrosymmetric [Pd(HL)2] molecules linked by intermolecular forces. 1 is a monomeric molecule containing Pd(II) coordinated in a square planar environment, by two oxygen atoms from carboxyl groups and two nitrogen atoms in the pyrazole rings. The angles N1-Pd1-O1 and N1i-Pd1-O1i are 80.60 (7)º, which means that the metallic atoms lie in the centre of the plane determined by the two nitrogen atoms and the two oxygen atoms. HL ligand behaves as a bidentate ligand and uses only one of its two carboxyl groups, forming a five membered metallocycle, which has a planar configuration. The bond distances Pd–N (1.9894 (19) Å) are similar to those of Pd–Npz (1.979 (3) Å) [13]. And these distances are consistent with previously described values (1.960 (10)–2.125 (10) Å) and (1.968 (2)–2.153 (2) Å) [14, 15], respectively. The Pd–O bond lengths 2.0042 (16) and 2.0043 (16) Å, can be regarded as normal compared with the distances found in the literature (1.9877 (19)–2.014 (2) Å) [16].
There exists intermolecular hydrogen bonds O4–H4···O2ii, C3–H3A···O2iii, C4–H4A···O3iv and C5–H5A···O3iv constructing the complex 1 into 3D structure as shown in Fig. 2 (Symmetry codes: (ii) x − 1, −y + 1, z − 1/2; (iii) –x + 1, −y + 2, −z + 1; (iv) x, y + 1, z).
Crystal structure of [PtL2] (2)
Single-crystal X-ray diffraction analysis of complex 2 reveals that the asymmetric unit consists of one-half Pt(IV) ion and L2− which deprotonated two H atoms from carboxyl groups, shown in Figure S2. The molecular structure is illustrated in Fig. 3. The coordination geometry around Pt(IV) is a distorted tetrahedron with bond angles N1–Pt1–O1 77.0 (10)º and N1i–Pt1–O1 101.4 (10)º, and the dihedral angles of [C1, O1, Pt1, N1, C2] and [C1A, O1A, Pt1, N1A, C2A] is 62.20 (3)º. The metal atom is coordinated to two H2L ligands via two pyrazole nitrogen and two carboxyl oxygen. H2L behaves as a bidentate ligand and uses only one of its two carboxyl groups, forming a five-membered metallocycle, which has a planar configuration. The bond distances Pt–Npz (1.982 (11) Å) are shorter than those of Pt–Npz (2.014(3) Å) reported [17–19]. The Pt–O bond lengths (2.019 (7) Å) can be regarded as normal compared with the distances found in the literature (2.010(6)–2.035 (5) Å) [20]. The N1–Pt–N1A bite angle, 93.2 (7)º, is comparable to that found in the structure with the ligands 3, 5- bis(2-pyridyl)pyrazole (pz1) [Cu4(pz1)4(ClO4)4]∙2H2O, 80.3(2)º [21].
In this complex, the centroid–centroid separations of the pyrazole rings from adjacent H2L are 4.596 (9) Å, showing the existence of intramolecular weak π···π stacking interactions. The adjacent [PtL2] units are linked together through intermolecular C–H···O H-bonding interactions between the uncoordinated O atoms of carboxyl groups and H-atoms of pyrazole rings (C4–H4A···O3i, ii, C5–H5B···O3i, symmetry codes: (i) x, −1 + y, z, (ii) 1 − x, −1 + y, 1 − z), thus 2D layers are formed in Fig. 4. The uncoordinated O2 and O4 further linked the layers into 3D structure by intermolecular contacts with distance O···O is 1.924 (12) Ǻ.
Fluorescent spectra
Both the coordinated metals in complexes 1 and 2 have d10 electron configuration. The luminescence spectra of 1 and 2 and the free ligand Eopzc were investigated in the liquid state at room temperature (Fig. 5). The luminescence spectrum of Eopzc shows emission at λ max = 327 nm (λ ex = 298 nm). The emissions of 1 and 2 are also at 327 nm (λ ex = 298 nm) assigned to intraligand (π–π*) fluorescence, which is modified by the Pd(II) and Pt(IV) ions. On comparison with the free ligand, the emissions of the complexes are much weaker than that of the ligand. This could be influenced by the change of the electron conformation when the ligands coordinate to the metals, and the hydrogen bonding interactions among the complexes.
Hirshfeld surface
Hirshfeld surface and fingerprint plot analysis could offer rapid reckonable understanding into the intermolecular interactions in complex molecular solids as well as crystal structures by color-coding short or long contacts, which acts as an easier and considerably faster graphical tool which is based on 3D Hirshfeld surface and 2D fingerprint plots, and gives a quantitative summary of the nature and type of intermolecular contacts experienced by the molecule in complexes 1 and 2. The Hirshfeld surface is mapped with d norm, and 2D fingerprint plots presentedin this paper were generated using CrystalExplorer 3.1 [22]. In this section, we study the molecular Hirshfeld surface of H2L in the complexes, to elucidate the features of different supramolecular synthons. The 3D Hirshfeld surface and 2D fingerprint plots of 1 and 2 are shown in Fig. 6. They clearly show the influences of different relationship on the intermolecular interactions of the complexes. The large and deep red spots on the 3D Hirshfeld surfaces indicate the close-contact interactions, which are mainly responsible for the significant hydrogen bonding contacts, while the blue color points in the 2D fingerprint plots are indicative of short contacts of H···H, O···O, and H···O interactions [23–27].
For complex 1, H···H interactions, which are reflected in the middle of scattered points in the 2D fingerprint plots, have the most significant contribution (11.6 %) to the total Hirshfeld surface. The O···H hydrogen bonding intermolecular interaction appear as a fork in the 2D fingerprint plots, which has 49.4 % contribution to the total Hirshfeld surface and match with O–H···O hydrogen bonds in 1. For complex 2, O···O interaction appear as a spike in the 2D fingerprint plots, which has 15.2 % contribution to the total Hirshfeld surface. The main interaction of 2 is also O···H contact and match the C–H···O hydrogen bonds, as shown in Fig. 7.
Conclusions
In summary, we have presented two precious metal complexes [Pd(HL)2] (1) and [PtL2] (2) by one-pot in situ hydrolysis of Eopzc as ligand. Their molecular structures have been characterized by single-crystal X-ray diffraction, elemental analyses, and IR spectra. The formation of the two complexes demonstrates that the 1-(carboxymethyl)-1H-pyrazole-3-carboxylic acid ligand is potential multidentate chelating ligands, due to the carboxyl group that displays multi coordination modes. As shown in the liquid-state fluorescent emission spectra, two similar emission bands can be observed in the two complexes which are much weaker than the ligand Eopza, and this phenomenon may be caused by the change of the electron conformation when the ligands coordinate to the metals. Hirshfeld surface analysis was studied on complexes 1 and 2, and reveals that the most interactions in both complexes are O···H contacts. On the basis of the present work, our group will study antibacterial based on the palladium and platinum complexes.
References
G. La Monica, G.A. Ardizzoia, Prog. Inorg. Chem. 46, 151 (1997)
J. Pérez, L. Riera, Eur. J. Inorg. Chem. 33, 4913 (2009)
M.A. Halcrow, Dalton Trans. 2059 (2009)
J. Klingele, S. Dechert, F. Meyer, Coord. Chem. Rev. 253, 2698 (2009)
R. Mukherjee, Coord. Chem. Rev. 203, 151 (2000)
R.A. de Souza, A. Stevanato, O. Treu-Filho, A.V.G. Netto, A.E. Mauro, E.E. Castellano, I.Z. Carlos, F.R. Pavan, C.Q.F. Leite, Eur. J. Med. Chem. 45, 4863 (2010)
R.Y. Mawo, D.M. Johnson, J.L. Wood, I.P. Smoliakova, J. Organomet. Chem. 693, 33 (2008)
R.W.Y. Sun, D.L. Ma, E.L.M. Wong, C.M. Che, Dalton Trans. 4883 (2007)
J.M. Lehn, Science 260, 1762 (1993)
M. Guerrero, J. Pons, J. Ros, J. Organomet. Chem. 695, 1957 (2010)
H. Zhao, Z.J. Chu, G.Y. Gao, H.H. Huang, Z.R. Qu, Polyhedron 83, 143 (2014)
G.M. Sheldrick, Acta Cryst. C71, 3–8 (2015)
J. Pons, G. Aragay, J. Garcí-Antón, T. Calvet, M. Font-Bardia, J. Ros, Inorg. Chim. Acta 363, 911 (2010)
J. Pons, A. Chadghan, J. Casabó, A. Alvarez-Larena, J.F. Piniella, J. Ros, Inorg. Chem. Commun. 3, 296 (2000)
M.D. Ward, J.S. Fleming, E. Psillakis, J.C. Jeffery, J.A. McCleverty, Acta Cryst. C54, 609 (1998)
N.T.S. Phan, D.H. Brown, H. Adams, S.E. Spey, P. Styring, Dalton Trans. 1348 (2004)
A.J. Canty, N.J. Minchin, J.M. Patrick, A.H. White, J. Chem. Soc., Dalton Trans. 1253 (1983)
A.J. Canty, T. Honeyman, B.W. Skelton, A.H. White, J. Organomet. Chem. 430, 245 (1992)
F.A. Allen, O. Kennard, Chem. Des. Autom. News 8, 31 (1993)
C. Tessier, F.D. Rochon, Inorg. Chim. Acta 373, 22 (2001)
M. Munakata, L.P. Wu, M. Yamamoto, T. Kurodasowa, M. Maekawa, S. Kawata, S. Kitagawa, J. Chem. Soc., Dalton Trans. 4099 (1995)
S.K. Wolff, D.J. Grimwood, J.J. McKinnon, M.J. Turner, D. Jayatilaka, M.A. Spackman, Crystal Explorer version 3.1, 2013 (University of Western Australia: Perth)
J.J. McKinnon, M.A. Spackman, A.S. Mitchell, Acta Cryst. Sect. B60, 627 (2004)
D. Spackman, Jayatilaka. Cryst. Eng. Comm. 11, 19 (2009)
J.J. McKinnon, D. Jayatilaka, M.A. Spackman, Chem. Commun. 3814 (2007)
M. Pourayoubi, S. Shoghpour Bayraq, A. Tarahhomi, M. Nečas, K. Fejfarova, M. Dušek, J. Organomet. Chem. 751, 508 (2014)
A. Tarahhomi, M. Pourayoubi, J.A. Golen, P. Zargaran, B. Elahi, A.L. Rheingold, M.A. Leyva Ramírez, T. Mancilla Percino, Acta Cryst. B69, 260 (2013)
Acknowledgments
We gratefully acknowledge the financial support of the National Natural Science Foundation of China (20801012) and the financial support from Jiangsu Ainaji Neoenergy Science and Technology Co., Ltd (8507040091).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
13738_2015_797_MOESM1_ESM.doc
CCDC 1061337 and 1061338 contain the supplementary crystallographic data for complexes 1 and 2. These data can be obtained free of charge from the Cambridge Crystallographic Data Centre via http://www.ccdc.cam.ac.uk/data_request/cif. (DOC 206 kb)
Rights and permissions
About this article
Cite this article
Feng, C., Li, XJ., Zhang, D. et al. Two new palladium and platinum complexes with a bidentate pyrazole-based ligand: crystal structure, fluorescence and Hirshfeld surface analysis. J IRAN CHEM SOC 13, 823–830 (2016). https://doi.org/10.1007/s13738-015-0797-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-015-0797-3