Abstract
Parkinson’s disease (PD) is the second-most common neurodegenerative disease, affecting 10 million people worldwide. Neuroinflammation is one of the major pathologic processes in the development of PD. Neuroinflammation is promoted via the activation of TLRs present on immune cells in the brain. In addition, miRNA regulates TLR expression in neurodegenerative diseases. However, there is limited information on the miRNA that regulates TLR signaling genes in PD. In this study, we used GO, a bioinformatics tool that uses the representations for genes in an organism; PPI, which shows the physical interaction between proteins in an organism; and miRNet, a tool to navigate the complex relationships between miRNAs and their targets for deeper biologic understanding. To find out the potential TLR genes and regulatory miRNAs that play a role in neuroinflammation-induced PD. We acquired the gene expression profile, GSE26927, from the GEO Omnibus. DAVID bioinformatics and SHINY GO software were employed for GO analysis of DEGs, and the fold enrichment score for each pathway was verified. The TLR signaling pathways most deregulated genes (upregulated: log FC ≥ 2.0, downregulated: log FC ≤ – 2.0) were chosen for network analysis to identify crucial or hub genes. Subsequently, a miRNA-gene network was constructed using the miRNet tool. The foremost TLR signaling gene, distinguishing between PD and control samples, has been discerned. In the Protein–Protein Interaction (PPI) network, we identified genes with heightened connectivity, notably TLR4, exhibiting the highest degree of betweenness (degree = 22) in the TLR signaling pathway. Furthermore, in the miRNA-gene network, we unveiled the preeminent five miRNAs: hsa-miR-21-5p, hsa-miR-17-5p, hsa-miR-93-5p, hsa-miR-7-5p, and hsa-mir-92b-3p that interacted with the TLR signaling gene. The top ten TLR genes could be potential targets for new therapeutics. In addition, the identified potential miRNAs can strongly regulate the expression of TLR genes in PD and serve as therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Parkinson’s disease (PD) is the second-most common neurodegenerative disease, affecting 10 million people worldwide. Cardinal motor symptoms such as tremors, muscle rigidity, and bradykinesia characterize PD (Tysnes and Storstein 2017). The degeneration of dopaminergic neurons in the substantia nigra region of the PD brain is caused by various pathologic mechanisms, including neuroinflammation, oxidative stress, mitochondrial dysfunction, impaired proteostasis, abnormal inclusion of α-synuclein aggregation, and epigenetic changes (Simon et al. 2020). Our primary focus is on understanding neuroinflammation, which plays a significant role in the death of dopaminergic neurons in PD. Therefore, targeting neuroinflammation holds great promise for developing effective PD treatment (Pajares et al. 1687). Neuroinflammation is promoted by both the innate immune system and the adaptive immune system. Furthermore, the innate immune system plays a crucial role in regulating neuroinflammation in PD (Tan et al. 2020). Toll-like Receptors (TLRs) are pattern-recognition receptors expressed in immune cells. Recently, it has been reported that the activation of TLR present on neuroimmune cells promotes neuroinflammation in PD. The TLR present on the plasma membrane (TLR1, TLR2, TLR4, TLR5) and in endosomes (TLR7, TLR9, TLR8) of glial cells (microglia, astrocytes) significantly contributes to PD pathogenesis (Heidari et al. 2022). α-synuclein aggregates act as damage-associated molecular patterns (DAMPs) and activate TLRs, triggering the neuroinflammatory cascade observed in PD (Li et al. 2021). Reactive oxygen species (ROS), viruses, and bacteria in addition stimulate TLR. TLR activation encourages neuroinflammation via various downstream signaling, with TLR/MyD88/NF-kB being one of the primary pathways (Dutta et al. 2021). The adaptor proteins triggered by TLR signaling such as MyD88, TRIF and TIRAP activate the transcription factor NF-kB and increase the release of pro-inflammatory mediators (IL-16, IL-1, IL-18), that leads to neuroinflammation. Various researchers have attempted to inhibit TLR signaling in in-vitro and in-vivo PD models using natural inhibitors (e.g., curcumin, celastrol, etc.) (Yu et al. 2016), and ongoing research aims to identify standard TLR inhibitors for PD treatment (Nemutlu Samur et al. 2022).
In addition, miRNAs are non-coding, single-stranded RNAs consisting of 21–24 nucleotides. They account for 1–5% of the human genome and regulate 30% of protein-coding genes (Ludwig et al. 2016). Accumulated evidence indicates that miRNA expression regulates the pathologic mechanisms of neurodegenerative diseases (Sharma and Lu 2018). In PD, miRNAs potentially regulate pathologic mechanisms such as α-synuclein aggregation, mitochondrial dysfunction, neuroinflammation, and oxidative stress. Dysregulated miRNAs promote PD pathogenesis, making targeting miRNAs a therapeutic approach in PD (Tryphena et al. 2023). Furthermore, miRNAs also serve as biomarkers for diagnosing PD (He et al. 2018a). Different dysregulated miRNAs contribute to PD development. miRNAs regulating microglia function in PD include miR-124, miR-195, miR-150, miR-155-5p, miR-7, etc. These miRNAs also regulate neuroinflammation in PD (Li et al. 2022). In addition, TLR signaling is regulated by different miRNAs. Research reveals that the adaptor protein MyD88 is regulated through overexpression of miR-203-5p or miR-149-5p in mouse macrophages stimulated with LPS, resulting in decreased expression of MyD88 protein (Arenas-Padilla and Mata-Haro 2018). Moreover, miRNAs interact with TLR and regulate neurodegeneration; examples include let-7, which can activate TLR-7 and cause neurodegeneration (Lehmann et al. 2012), and the downregulation of miR-93, which activates TLR4/MyD88/NF-kB signaling in vascular dementia (VD) (Wang et al. 2020). These studies collectively indicate that miRNAs regulate TLR in the Central Nervous System (CNS). More research is needed to uncover potential miRNAs regulating TLR genes in PD pathogenesis. Therefore, in this study, we conducted GEO2R analysis of microarray data of healthy and PD patients in the Gene Expression Omnibus (GEO) database. We constructed a Protein–Protein Interaction (PPI) network of Differentially Expressed Genes (DEGs) belonging to TLR signaling and miRNA-gene network of TLR signaling genes to identify potential miRNAs regulating TLR signaling in PD.
2 Materials and methods
2.1 Microarray data
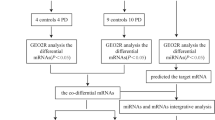
The microarray data were obtained from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) an international public repository of microarrays, second-generation sequencing, and other forms of high-throughput functional genomic datasets submitted by the research community. Our research utilized the GSE26927 dataset, encompassing microarray data derived from individuals with PD and control subjects (Durrenberger et al. 2012). Durrenberger et al. identified novel reference genes in human post-mortem tissue in this data series, while a separate study by the same authors demonstrated alterations in inflammatory genes across various neurologic diseases. The authors explicitly acknowledged the supportive nature of their findings regarding the involvement of the innate immune system in the pathogenesis of neurodegenerative diseases such as AD, PD, and ALS (Durrenberger et al. 2015). Consequently, we selected this study to investigate the role of innate immunity genes in neuroinflammation, focusing specifically on TLR and miRNAs implicated in the regulation of TLR genes in PD. The complete workflow of data processing is given in Fig. 1. The DEGs were analyzed for PPI and miRNA-gene network. All data used in this study are freely available, and this study did not involve animal experimentation.
2.2 GEO2R analysis
The GEO2R online analysis tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/) facilitated the examination of DEGs between PD and healthy patients. Employing the GEO query and limma R packages for genomic data analysis, we identified DEGs in PD (n = 12) versus Control (n = 8) samples, calculating p values and logFC values. DEGs meeting the criteria of p ≤ 0.05 and logFC ≥ 2.0 were categorized as upregulated, while those with p ≤ 0.05 and logFC ≤ – 2.0 were deemed downregulated (Hunt et al. 2022). Subsequently, these DEGs were subjected to gene Ontology analysis for functional annotation. Notably, Log FC (log fold changes) was utilized to quantify the extent of gene expression variations between disease and control samples (Jung et al. 2011).
2.3 Gene ontology of selected genes
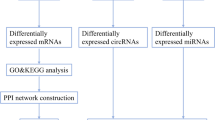
Gene ontology (GO) analysis was conducted to elucidate the functional roles of DEGs using SHINY GO (http://bioinformatics.sdstate.edu/go/) and the DAVID bioinformatics tool (https://david.ncifcrf.gov). The analysis aimed to identify specific biologic properties associated with the DEGs, including immune system response, locomotion, stimulus response, stress response, and behavior (Functional annotation) as shown in Fig. 2. Subsequently, DEGs related to the immune system were refined using the DAVID tool, with a specific focus on innate immunity. A heatmap of the shortlisted genes (innate immunity genes) was generated using SRplot (https://www.bioinformatics.com.cn/en). These shortlisted DEGs served as input genes for generating enrichment charts and visualizing KEGG pathways in the SHINY GO database (Botstein et al. 2000).
DEGs of innate immunity in PD patient samples. A Volcano plot of DEGs in GSE26927: Common neuroinflammatory pathway in neurodegenerative, red color represents upregulated genes (log FC ≥ 2.0), blue color represents down regulated (log FC ≤ -2.0) gene and others are non-significant genes. B Top biologic process and percentages of DEGs in GSE26927 constructed by using SHINY GO and Panther software. C Top five category of DEGs involved in immune system functions, that include adaptive immunity, innate immunity, viral infection and apoptosis genes
2.4 PPI network construction
The STRING database (http://string-db.org/), complemented with heuristic methods of association and analysis, was utilized to explore the predicted PPI associations of both innate immune genes and TLR signaling genes expressed in PD. Interactions generated in STRING are sourced from various channels: High-throughput Lab Experiments, Co-expression, Automated Text Mining, and Previous knowledge in databases (Szklarczyk et al. 2011). Key parameters assessed in STRING include confidence score, interaction source, known and predicted interactions, network topology, functional enrichment, etc. PPI pairs were extracted with a confidence score of 0.4 (medium confidence). Subsequently, the PPI network was visualized using Cytoscape software (https://www.cytoscape.org/). Cytoscape, serves as a user-friendly interface for the construction and analysis of interaction networks obtained from the STRING database. Nodes, representing genes, with a high degree of connectivity were evaluated through the CytoHubba plugin within Cytoscape. In Cytoscape, the degree of a node refers to the number of edges connected to that node. The degree of a node is one of the important topologic parameters in the analysis of PPI networks, aiding in understanding centrality of genes, connectivity, identification of hubs, network visualization, and network dynamics. When compared to other topologic parameters, the degree of a node is more straightforward and computationally efficient. Therefore, the degree of each node, indicative of its level of interaction within the network, was meticulously calculated. Our study places particular emphasis on the top ten genes within the TLR pathway, recognizing them as central components crucial to our investigation.
2.5 miRNA-gene network analysis
To identify potential miRNAs regulating TLR signaling genes, we employed the miRNet (Chang et al. 2020) online database (https://www.mirnet.ca/), a miRNA-centric network visual analytics platform. miRNet serves as a freely available tool for exploring miRNA-gene networks, hosted on the Google Cloud Computing Engine with 64 GB RAM and 8 CPU cores (n2-highmem-8). We utilized four well-annotated databases (miRTarBase v8.0, TarBase v8.0, miRanda, and miRecords). In this study, we constructed a miRNA-gene interaction network through the miRNet platform. Specific parameters, including species, miRNA identifiers, and target gene identifiers, were employed to identify the top miRNA and gene nodes. The top ten genes related to TLRs were imported into the miRNet tool, resulting in the creation of the desired miRNA-gene network.
3 Results
3.1 Identification of DEGs related to innate immunity in PD
The DEGs identified through GEO2R analysis were subjected to gene ontology study, as illustrated in volcano plot of Fig. 2A. A significant proportion of these genes were associated with functional categories including response to stress, catabolic processes, locomotion, behavior, and immune system regulation, as depicted in Fig. 2B and outlined in detail in Supplementary Table 1. Specifically focusing on immune system-related DEGs, we narrowed down our investigation to innate immunity genes. After further filtration, we observed that 15% of these genes were dedicated to innate immunity, while 5% were associated with adaptive immunity. The remaining genes were classified into other categories, as outlined in Fig. 2C and detailed in Supplementary Table 2. The 321 genes identified under the category of innate immunity were subsequently chosen for gene ontology analysis to facilitate a more targeted exploration of innate immune responses aligned with our research objectives (Fig. 3). For enrichment analysis of innate immunity genes, we utilized SHINY GO and the results are presented in Supplementary Table 3.
3.2 Enrichment pathways and differential expression analysis
We conducted a comprehensive gene functional enrichment analysis using SHINY GO software to elucidate potential genes and pathways of significance. Our focus was primarily on identifying DEGs associated specifically with the Innate Immune System. This emphasis arises from the recognition that neuroinflammation in PD is intricately linked to the activation of brain immune cells, and the innate immune response serves as the initial and expeditious phase in initiating the neuroinflammatory cascade in PD. DEGs of the innate immune response were meticulously input into the online database, providing profound insight into their functional roles. Our analysis revealed the top 20 enrichment pathways, with notable distinctions in enrichment levels highlighted by Fold Enrichment (FE) values. Particularly, pathways such as complement and coagulation cascade (FE = 16) and TLR Signaling (FE = 16) emerged as highly enriched, underscoring their potential significance (as visualized in Fig. 4A). Conversely, less-enriched pathways, including those related to cancer, Salmonella infection, and HSV-1 infection, were also identified, serving as informative contrasts. Furthermore, our investigation delved deeper into the TLR signaling genes expressed within the context of PD and control samples (as depicted in Fig. 4B, supplementary Table 4). In addition, our exploration encompassed KEGG pathway analysis, revealing interconnections between TLR pathway genes and significant signaling pathways such as PI3-AKT signaling and JAK-STAT signaling. These interactions can potentially contribute significantly to the complex landscape of neuroinflammation, as visually represented in Fig. 5.
Enrichment analysis of Innate immunity genes and DEGs belong to TLR signaling contributing to neuroinflammation. A Enrichment chart of biologic process of innate immunity genes. Enrichment chart based on the FE (Fold enrichment) value, the FE value TLR signaling pathway is 16 and – log10(FDR) = 10 therefore TLR pathway belong to highly enriched pathways. B Heat map of TLR signaling genes expressed between PD and control sample; row side represents name of genes and columns represent sample ID of patients
DEGs of TLR signaling and its association with neuroinflammatory pathway. KEGG pathway of TLR signaling representing association with NF-KB, AKT and JAK-STAT pathways (https://david.ncifcrf.gov)
3.3 PPI network analysis of DEGs
The PPI analysis of DEGs within the Innate Immune System Genes Pathway was conducted using the STRING tool (Fig. 6). Notably, the network comprising 314 nodes representing genes related to innate immunity exhibited a dense connectivity, with 18 nodes associated with the TLR pathway within the PPI network. Our focus was on identifying the top hub genes in this network, leading to the recognition of the top ten hub genes based on their degree of connectivity (Fig. 7A). To assess the significance of these hub genes, key parameters related to PPI, such as Rank, Score, and Degree of Betweenness, were computed using Cytoscape. The expression level of the identified gene in PD or neuroinflammation was also found in the published literature to support the present study’s research findings (Table 1). The findings highlighted TLR4 as the most interactive gene, with a connectivity degree of 22, followed by MyD88 (degree = 21), TLR9 (degree = 17), TLR2 (degree = 17), IRAK1 (degree = 16), IRAK4 (degree = 13), TRAF6 (degree = 13), IRAK3 (degree = 9), IRF7 (degree = 8), and MAP37K (degree = 8). A higher degree implies more extensive interactions within the network, while the score reflects gene expression, and rank denotes the gene’s position in the network (Fig. 7B).
PPI network analysis of TLR signaling genes. A) TLR signaling gene cluster, confidence score = 0.4, number node 18 and expected number of edges = 36, p value: < 1.0e-16 (In Predicted Interactions, green is from gene neighborhood analyses, red are gene fusions events, and blue are from gene co-occurrence. The other remaining interactions are; Olive = text-mining, black = co-expression, Navy Blue = protein homology. B). Top ten hub genes in PPI network of Toll-like Receptors signaling analyzed in cytoscape software
3.4 Potential miRNA regulating TLR signaling in PD
The potential miRNA regulating TLR pathway was predicted using the mirNet online database, which is connected with the miRDB, Target Scan, and miRTar base databases. In the results of the miRNA-gene network, we identified 53 miRNAs, 18 genes, and 158 edges in the network (Fig. 8). Further, we identified the top miRNAs targeting the network’s highest number of genes. Among all miRNAs, hsa-miR-21-5p interacted with six proteins or genes (IRAK1, TLR3, TLR4, TBK1, IRAK4, MyD88), hsa-miR-17-5p interacted with four genes (TLR4, IRAK1, IRAK4, TLR7), hsa-miR-93-5p interacted with two genes (IRAK4, TLR7), hsa-miR-7-5p interacted with two genes (TLR4, FADD), and hsa-mir-92b-3p with TLR3, TIRAP (Table 2).
4 Discussion
Neuroinflammation is a common process in neurodegenerative diseases such as AD, PD, HD, and ALS. Neuroinflammation is regulated by both peripheral and central immune systems in all neurodegenerative diseases. Furthermore, innate immunity has a great role in neuroinflammation-induced neurodegeneration. In the present study, we focused on PD, which is the second-most prevalent neurodegenerative disease globally, impacting millions of individuals. Various preclinical research studies provide evidence of the link between innate immunity and PD because α-synuclein aggregates act as DAMP for PRRs (Kouli et al. 2019). TLR are the type of PRRs present in peripheral and central immune cells (Pei et al. 2007). TLRs activated by α-synuclein aggregates lead to increases in M1 microglia phenotypes and lower the M2 microglia phenotype and the production of pro-inflammatory (IL-18, IL-1β) and anti-inflammatory mediators (IL-4, IL-6, IL-10), that will ultimately cause neuroinflammation and after that neurodegeneration in PD (Lazdon et al. 2020).There are different types of TLR expressed on the microglia, but potential TLR gene need to be explored that contributes to PD pathogenesis. Similarly, researchers also target the TLR3 and TLR4 receptors to elevate neuroinflammation in PD.
As we understood from above Innate immunity and TLR signaling have a great role in neuroinflammation and PD progression. Previously, Durrenberger et al. did work on neuroinflammatory genes involved in neurodegenerative diseases. The authors used the microarray technique to check dysregulated genes between normal patients and diseased patients. They reported significant changes in inflammatory genes and the results of study prove the role of innate immunity in the progression of all neurodegenerative diseases such as AD, PD, HD, and ALS. The data of this study was available in the GEO database with series number GSE26927 (Durrenberger et al. 2015). So, the present study is completely bioinformatics in which we did gene ontology of DEGs obtained through GEO2R analysis of the normal patient sample and PD patient sample of GSE26927 series. The DEGs shortlisted based on the functional annotation or biologic process, so we were selected DEGs that play a role in immune system function and further filtered to select only innate immunity-related DEGs (Fig. 3). The gene enrichment chart of the dysregulated gene of the innate immune system was created in SHINY GO which showed the top 20 signaling pathways (Fig. 4). The top 20 signaling pathways include highly enriched pathways complement and coagulation cascade, TLR signaling pathway, RIG-like receptor pathway, cytosolic DNA sensing pathway, NF-kB pathway, Nod-like receptor pathway, and remaining infectious diseases. These mentioned pathways have a role in the pathogenesis of PD; Ma, Shi-Xun, et al. give evidence on the involvement of complement and coagulation pathway in PD, authors prove that complement fragment C3 significantly increase in the mid-brain of PD mouse and mediate neurodegeneration-induced by α-synuclein (Ma et al. 2021). Similarly, RIG-like receptors (Kaur et al. 2019), Nod-like receptor regulate neuroinflammation (Liu et al. 2023) and cytosolic DNA sensing pathway mediated mitochondrial damage reported in the zebrafish model of PD (Matsui et al. 2021).
In this study, we focused on the TLR signaling pathway, we showed the contribution of the TLR signaling pathway in the pathogenesis of PD. To find out the potential TLR gene that contributes to neuroinflammation in PD. We construct a PPI network of genes participating in the KEGG pathway of TLR signaling and identified top ten hub genes are TLR3, IRAK1, IRF7, TLR4, TLR9, MYD88, IRAK4, MAP3K7, TLR9, TRAF6 (Table 1). These genes have great contributions to PD pathogenesis such as TLR4 and TLR9 activated in synucleinopathies and promote neuroinflammation in PD (Kouli et al. 2019), MYD88-dependent pathway activate M1 microglia and promote neuroinflammation in MPTP-treated mice model (Drouin-Ouellet et al. 2011). Furthermore, IRAK4 is major inflammatory protein; its upregulation activate IRF7, MAPK and NLRP3 proteins, which will lead to neuroinflammation in PD (Lee et al. 2021). Hence, the identified gene could act as target for new therapeutics to control neuroinflammation in PD, especially α-synuclein mediated neuroinflammation. Similarly, we also identified miRNAs that could be act as regulator for identified genes. The miRNA-gene network results suggested that the top five significant miRNAs such as hsa-miR-21-5p, hsa-miR-17-5p, hsa-miR-93-5p, hsa-miR-7-5p and hsa-mir -92b-3p (Table 2). The top five miRNAs has role in PD progression and diagnosis, like has-miR-17-5p downregulation contribute in PQ-induced neurodegeneration of dopaminergic neurons (Wang et al. 2018), hsa-miR-7-5p regulate CXCL12 expression in MPP+ induced neuroinflammation (Lian et al. 2021), hsa-miR-93-5p and hsa-mir -92b-3p level increases in PD therefore in act as biomarker for PD (Vallelunga et al. 2021). Moreover, identified miRNAs have a role in pathogenesis and diagnosis of different disease other than PD (Fig. 9). From all the identified miRNAs, hsa-miR-21-5p interacted with a greater number of node or gene in network. The research studies also found that the level of expression of hsa-miR-21-5p significantly increases in PD patients sample and SH-SY-5Y cells exposed with synuclein peptide (Alvarez-Erviti et al. 2013). Furthermore, an in-vitro study by Yelamanchili, Sowmya V. et al. provided the evidence on link between TLR gene and miR-21-5p, authors observed that miR-21-5p present in vesicles promote neurotoxicity (Yelamanchili et al. 2015). Furthermore, miR-21 is also known as inflammamiR because it targets NLRP3 and NF-kB pathway and it also regulate neuroinflammation in age related diseases (Olivieri et al. 2021) and miR-21 can be also act as biomarker in AD and PD (Burgos et al. 2014). Hence, it indicates that miR-21 expression linked with TLR signaling pathway, so therefore we can say that miR-21 could be potential miRNA that can regulate TLR signaling gene in PD.
Role of identified miRNAs in different pathologic condition. miRNA-gene network constructed in cytoscape and analyzed by cytohubba, and top miRNA mentioned are miR-21-5p (immunoregulatory role), miR-92b-3p (act as biomarker in breast cancer), miR-17-5p (promote Aβ deposition in AD), miR-93-5p (regulate tumorigenesis) and miR-7-5p (enhances cerebral reperfusion injury)
5 Conclusion
In conclusion, our data demonstrated that TLR signaling plays a role in PD pathogenesis. Top ten hub genes (TLR4, TLR9, MYD88, IRAK4, MAP3K7, TLR9, TRAF6, TLR3, IRAK1, IRF7) could be potential target for new therapeutic. In addition, identified potential miRNAs (hsa-miR-21-5p, hsa-miR-17-5p, hsa-miR-93-5p, hsa-miR-7-5p, and hsa-mir-92b-3p) can strongly regulate the expression of TLR signaling genes in PD and act as a therapeutic target however, further in-vitro and in-vivo study required in future.
Data availability
The GEO database from NCBI (Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo/) was used to access the GSE26927 dataset.
Abbreviations
- PD:
-
Parkinson’s disease
- TLRs :
-
Toll-like Receptors
- VD:
-
Vascular dementia
- DEGs:
-
Differentially Expressed Genes
- GEO:
-
Gene Expression Omnibus
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DAVID:
-
Database for Annotation, Visualization, and Integrated Discovery
- STRING:
-
Search Tool for Recurring Instances of Neighbouring Genes
- PPI:
-
Protein–Protein interaction
- PRRs:
-
Pattern recognition receptor
- TLR4 :
-
Toll-like receptor 4
- MyD88 :
-
Myeloid differentiation primary response 88
- IRAK1 :
-
Interleukin-1 receptor-associated kinase 1
- TRAF6 :
-
Tumor necrosis factor receptor-associated factor 6
- IRF7 :
-
Interleukin regulatory factor 7
- FE:
-
Fold enrichment
References
Alvarez-Erviti L, Seow Y, Schapira AHV, Rodriguez-Oroz MC, Obeso JA, Cooper JM (2013) Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson’s disease. Cell Death Dis 4(3):e545–e545
Arenas-Padilla M, Mata-Haro V (2018) Regulation of TLR signaling pathways by microRNAs: implications in inflammatory diseases. Cent Eur J Immunol 43(4):482–489
Botstein D, Cherry JM, Ashburner M, Ball CA, Blake JA, Butler H et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29
Burgos K, Malenica I, Metpally R, Courtright A, Rakela B, Beach T et al (2014) Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 9(5):e94839
Cao C, Ding J, Cao D, Li B, Wu J, Li X et al (2022) TREM2 modulates neuroinflammation with elevated IRAK3 expression and plays a neuroprotective role after experimental SAH in rats. Neurobiol Dis 171:105809
Chang L, Zhou G, Soufan O, Xia J (2020) miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res 48(W1):W244–W251
Chung J-Y, Park HR, Lee S-J, Lee S-H, Kim JS, Jung Y-S et al (2013) Elevated TRAF2/6 expression in Parkinson’s disease is caused by the loss of Parkin E3 ligase activity. Lab Investig 93(6):663–676
Conte C, Ingrassia A, Breve J, Bol JJ, Timmermans-Huisman E, van Dam A-M et al (2023) Toll-like receptor 4 is upregulated in Parkinson’s disease patients and co-localizes with pSer129αSyn: a possible link with the pathology. Cells 12(10):1368
Drouin-Ouellet J, Gibrat C, Bousquet M, Calon F, Kriz J, Cicchetti F (2011) The role of the MYD88-dependent pathway in MPTP-induced brain dopaminergic degeneration. J Neuroinflamm 8(1):1–12
Durrenberger PF, Fernando FS, Magliozzi R, Kashefi SN, Bonnert TP, Ferrer I et al (2012) Selection of novel reference genes for use in the human central nervous system: a BrainNet Europe study. Acta Neuropathol 124:893–903
Durrenberger PF, Fernando FS, Kashefi SN, Bonnert TP, Seilhean D, Nait-Oumesmar B et al (2015) Common mechanisms in neurodegeneration and neuroinflammation: a BrainNet Europe gene expression microarray study. J Neural Transm 122:1055–1068
Dutta D, Jana M, Majumder M, Mondal S, Roy A, Pahan K (2021) Selective targeting of the TLR2/MyD88/NF-κB pathway reduces α-synuclein spreading in vitro and in vivo. Nat Commun 12(1):5382
Dzamko N, Gysbers A, Perera G, Bahar A, Shankar A, Gao J et al (2017) Toll-like receptor 2 is increased in neurons in Parkinson’s disease brain and may contribute to alpha-synuclein pathology. Acta Neuropathol 133:303–319
He R, Yan X, Guo J, Xu Q, Tang B, Sun Q (2018a) Recent advances in biomarkers for Parkinson’s disease. Front Aging Neurosci 10:305
He J, Zhong W, Zhang M, Zhang R, Hu W (2018b) P38 mitogen-activated protein kinase and Parkinson’s disease. Transl Neurosci 9(1):147–153
Heidari A, Yazdanpanah N, Rezaei N (2022) The role of Toll-like receptors and neuroinflammation in Parkinson’s disease. J Neuroinflammation 19(1):1–21
Herrán E, Requejo C, Ruiz-Ortega JA, Aristieta A, Igartua M, Bengoetxea H et al (2014) Increased antiparkinson efficacy of the combined administration of VEGF- and GDNF-loaded nanospheres in a partial lesion model of Parkinson’s disease. Int J Nanomed 9(1):2677–2687
Hunt GP, Grassi L, Henkin R, Smeraldi F, Spargo TP, Kabiljo R et al (2022) GEOexplorer: a webserver for gene expression analysis and visualisation. Nucleic Acids Res 50(W1):W367–W374
Jung K, Friede T, Beißbarth T (2011) Reporting FDR analogous confidence intervals for the log fold change of differentially expressed genes. BMC Bioinform 12:1–9
Kaur R, Mehan S, Singh S (2019) Understanding multifactorial architecture of Parkinson’s disease: pathophysiology to management. Neurol Sci 40:13–23
Kouli A, Horne CB, Williams-Gray CH (2019) Toll-like receptors and their therapeutic potential in Parkinson’s disease and α-synucleinopathies. Brain Behav Immun 81:41–51
Lazdon E, Stolero N, Frenkel D (2020) Microglia and Parkinson’s disease: footprints to pathology. J Neural Transm 127:149–158
Lee JH, Kim HJ, Kim JU, Yook TH, Kim KH, Lee JY et al (2021) A novel treatment strategy by natural products in NLRP3 inflammasome-mediated neuroinflammation in Alzheimer’s and Parkinson’s disease. Int J Mol Sci 22(3):1324
Lehmann SM, Krüger C, Park B, Derkow K, Rosenberger K, Baumgart J et al (2012) An unconventional role for miRNA: let-7 activates toll-like receptor 7 and causes neurodegeneration. Nat Neurosci 15(6):827–835
Lei K, Zhang L, He Y, Sun H, Tong W, Xu Y et al (2020) Immune-associated biomarkers for early diagnosis of Parkinson’s disease based on hematological lncRNA–mRNA co-expression. Biosci Rep 40(12):BSR20202921
Li Y, Xia Y, Yin S, Wan F, Hu J, Kou L et al (2021) Targeting microglial α-synuclein/TLRs/NF-kappaB/NLRP3 inflammasome axis in Parkinson’s disease. Front Immunol 12:719807
Li S, Bi G, Han S, Huang R (2022) MicroRNAs play a role in Parkinson’s disease by regulating microglia function: from pathogenetic involvement to therapeutic potential. Front Mol Neurosci 14:744942
Lian H, Wang B, Lu Q, Chen B, Yang H (2021) LINC00943 knockdown exerts neuroprotective effects in Parkinson’s disease through regulates CXCL12 expression by sponging miR-7-5p. Genes Genom 43:797–805
Liu Z, Shen C, Li H, Tong J, Wu Y, Ma Y et al (2023) NOD-like receptor NLRC5 promotes neuroinflammation and inhibits neuronal survival in Parkinson’s disease models. J Neuroinflamm 20(1):1–21
Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C et al (2016) Distribution of miRNA expression across human tissues. Nucleic Acids Res 44(8):3865–3877
Ma S-X, Seo BA, Kim D, Xiong Y, Kwon S-H, Brahmachari S et al (2021) Complement and coagulation cascades are potentially involved in dopaminergic neurodegeneration in α-synuclein-based mouse models of Parkinson’s disease. J Proteome Res 20(7):3428–3443
Maatouk L, Compagnion A-C, Sauvage M-AC, Bemelmans A-P, Leclere-Turbant S, Cirotteau V et al (2018) TLR9 activation via microglial glucocorticoid receptors contributes to degeneration of midbrain dopamine neurons. Nat Commun 9(1):2450
Matsui H, Ito J, Matsui N, Uechi T, Onodera O, Kakita A (2021) Cytosolic dsDNA of mitochondrial origin induces cytotoxicity and neurodegeneration in cellular and zebrafish models of Parkinson’s disease. Nat Commun 12(1):3101
Nemutlu Samur D, Akçay G, Yıldırım S, Özkan A, Çeker T, Derin N et al (2022) Vortioxetine ameliorates motor and cognitive impairments in the rotenone-induced Parkinson’s disease via targeting TLR-2 mediated neuroinflammation. Neuropharmacology 208:108977
Olivieri F, Prattichizzo F, Giuliani A, Matacchione G, Rippo MR, Sabbatinelli J et al (2021) miR-21 and miR-146a: the microRNAs of inflammaging and age-related diseases. Ageing Res Rev 70:101374
Pajares M, Rojo AI, Manda G, Boscá L, Cuadrado A (2020) Inflammation in Parkinson’s disease: mechanisms and therapeutic implications. Cells 9(7):1687
Pei Z, Pang H, Qian LI, Yang S, Wang T, Zhang W et al (2007) MAC1 mediates LPS-induced production of superoxide by microglia: the role of pattern recognition receptors in dopaminergic neurotoxicity. Glia 55(13):1362–1373
Sharma S, Lu H-C (2018) microRNAs in neurodegeneration: current findings and potential impacts. J Alzheimer’s Dis Park. https://doi.org/10.4172/2161-0460.1000420
Simon DK, Tanner CM, Brundin P (2020) Parkinson disease epidemiology, pathology, genetics, and pathophysiology. Clin Geriatr Med 36(1):1–12
Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P et al (2010) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 39(suppl_1):D561–D568
Tan E-K, Chao Y-X, West A, Chan L-L, Poewe W, Jankovic J (2020) Parkinson disease and the immune system—associations, mechanisms and therapeutics. Nat Rev Neurol 16(6):303–318
Tryphena KP, Singh G, Jain N, Famta P, Srivastava S, Singh SB et al (2023) Integration of miRNA’s theranostic potential with nanotechnology: promises and challenges for Parkinson’s disease therapeutics. Mech Ageing Dev 211:111800
Tysnes O-B, Storstein A (2017) Epidemiology of Parkinson’s disease. J Neural Transm 124(8):901–905
Vallelunga A, Iannitti T, Capece S, Somma G, Russillo MC, Foubert-Samier A et al (2021) Serum miR-96-5P and miR-339-5P are potential biomarkers for multiple system atrophy and Parkinson’s disease. Front Aging Neurosci 13:632891
Wang Q, Liu Y, Zhou J (2015) Neuroinflammation in Parkinson’s disease and its potential as therapeutic target. Transl Neurodegener 4(1):19
Wang Q, Zhan Y, Ren N, Wang Z, Zhang Q, Wu S et al (2018) Paraquat and MPTP alter microRNA expression profiles, and downregulated expression of miR-17-5p contributes to PQ-induced dopaminergic neurodegeneration. J Appl Toxicol 38(5):665–677
Wang L, Yang J-W, Lin L-T, Huang J, Wang X-R, Su X-T et al (2020) Acupuncture attenuates inflammation in microglia of vascular dementia rats by inhibiting miR-93-mediated TLR4/MyD88/NF-κB signaling pathway. Oxid Med Cell Longev 2020:1–15
Yelamanchili SV, Lamberty BG, Rennard DA, Morsey BM, Hochfelder CG, Meays BM et al (2015) MiR-21 in extracellular vesicles leads to neurotoxicity via TLR7 signaling in SIV neurological disease. PLoS Pathog 11(7):e1005032
Yu S, Wang X, He X, Wang Y, Gao S, Ren L et al (2016) Curcumin exerts anti-inflammatory and antioxidative properties in 1-methyl-4-phenylpyridinium ion (MPP+)-stimulated mesencephalic astrocytes by interference with TLR4 and downstream signaling pathway. Cell Stress Chaperones 21:697–705
Acknowledgements
The authors acknowledge National Institute of Pharmaceutical Education and Research (NIPER)-Hyderabad for providing the necessary facilities and resources to prepare this manuscript.
Funding
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Singh, G., Khatri, D.K. MicroRNA-gene regulatory network of TLR signaling in neuroinflammation-induced Parkinson’s disease: a bioinformatics approach. Netw Model Anal Health Inform Bioinforma 13, 7 (2024). https://doi.org/10.1007/s13721-024-00445-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13721-024-00445-6