Abstract
MicroRNAs (miRNAs) and autophagy exert an important role in hypoxia/reoxygenation (H/R)-induced cardiomyocyte injury. The current study aimed to explore the role of miRNA and autophagy in H/R-induced cardiomyocyte injury. Cardiomyocyte H9c2 was exposed to H/R to simulate H/R injury in vitro. The differentially expressed miRNAs were identified using quantitative RT-PCR (qPCR). Lactate dehydrogenase (LDH) activity was assayed to assess H/R injury. The role of miRNA and autophagy in regulating the viability and cell apoptosis was evaluated using cell counting kit-8 (CCK-8) assay, flow cytometry (FCM), and western blot. The autophagy activation was assessed through testing the number of light chain 3 (LC3) puncta and LC3-II expression using western blot and immunofluorescence analysis. In the present study, we found that the miR-542-5p expression and the autophagy activation were significantly increased in H9c2 cells after H/R injury. Functionally, forced expression of miR-542-5p further aggravated H/R injury in H9c2 cells, whereas miR-542-5p inhibition alleviated H/R injury as measured by the cell viability, LDH activity and cell apoptosis. miR-542-5p repressed autophagy activation, whereas miR-542-5p inhibition facilitated autophagy activation in H9c2 cells exposed to H/R as measured by the LC3 puncta number, LC3II, and p62 protein level. Especially, autophagy inhibition by specific inhibitor partially lessened the role of miR-542-5p inhibitor in alleviating H/R injury. Finally, the autophagy-related 7 (ATG7) was identified as a novel target gene of miR-542-5p in H9c2 cells. The current data suggest that miR-542-5p/autophagy pathway might be a potential target for the treatment of H/R-related heart diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myocardial infarction (AMI) is considered to be one of the leading causes of human morbidity and mortality [1, 2]. It is induced by myocardial ischemia and coronary artery occlusion, then followed by the decrease of oxygen supply and nutrients to cardiomyocytes, finally, leads to cardiomyocytes dysfunction and heart failure [3, 4]. Although many efficient treatments have been reported on animal modes, the translation of these treatments for the clinical setting of AMI has been discouraging [5]. One of the most common cardioprotective strategies is to restore the blood flow early to prevent lethal damage [6]. However, the sudden restoration of blood flow could lead to the development of several severe conditions include irreversible shock, myocardial infarction, and heart failure [7]. Besides, the efficiency of this strategy is highly dependent on the present rates of these hypoxia/reoxygenation (H/R)-induced cardiomyocyte injuries [8]. Since the mechanism of H/R-induced cardiomyocyte injury is not well understood, the understanding of H/R-induced cardiomyocyte injury is vital for research of AMI treatments.
The molecular mechanism related to H/R-induced cardiomyocyte injury has recently come to the spotlight in the research. MicroRNAs (miRNAs) are a class of non-coding, single-stranded RNA that consist of approximately 22 nucleotides, and it is associated with several biological processes, such as cell differential, proliferation, and apoptosis [9,10,11,12]. In recent years, data in maintaining numbers indicated that several miRNA is considered as crucial modulators of in H/R injuries, they could protect cardiomyocytes from H/R injury by targeting the FAS ligand, modulating SIRT7/p53 signaling, targeting SIRT1 and suppressing oxidative stress and NLRP3-mediated pyroptosis pathway, etc. [13,14,15]. Besides, these miRNAs that could also be potential treatment targets or diagnose biomarkers for relevant diseases [16, 17]. According to recent research, miR-542-5p could be a potential inhibitor for cellular autophagy, thus affect the normal function of a cell [18].
Autophagy, a form of programmed cell death, is an intracellular catabolic process that is vital for cell survival [19]. It employs lysosomes to degrade the dysfunction proteins and organelles, serving as a cell control mechanism [20]. It is reported that autophagy dysfunction is commonly seen in H/R injuries [21, 22], and many miRNAs can regulate autophagy by regulating TNFα expression, SIRT1-mediated autophagy, regulating the Akt/mTOR pathway and etc. [20, 23,24,25,26,27]. However, the mechanism for the role of miRNA-542-5p in H/R injury meditate H/R is still not clear.
In this study, we sought to explore the possible role of miR-542-5p in H/R injury. To address this issue, we establish a myocardial H/R H9C2 cell model to investigate how H/R injury effects cell autophagy. The result shows that the miR-542-5p expression and the autophagy activation were remarkably elevated in H9c2 cells after H/R injury. The autophagy-related 7 (ATG7) was identified as a novel target gene of miR-542-5p in H9c2 cells. Experiment data suggest miR-542-5p/autophagy pathway might be a potential target for the treatment of H/R-related heart diseases.
Materials and methods
Cell culture
H9c2 cardiomyocytes were acquired from the American Type Culture Collection (ATCC, USA). Cells were cultured in DMEM/F12 medium (Hyclone; USA), which were added with 10%(v/v) fetal bovine serum (Wisent; Australia) and 1% penicillin–streptomycin solution, at 37 ℃ in a controlled atmosphere of 95% air and 5% CO2. Experimental procedures were performed when the cells reach 80% confluency.
RNA extraction and quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA of the H9c2 cells was extracted using Trizol reagent (Invitrogen Life Technologies) following the manufacturer’s protocol. The quality of total RNA was determined by NanoDrop 200 (Manufacturer) and gel electrophoresis. The reverse transcription of miRNA from the qualified total RNA was carried out using Mir-X™ miRNA First-Strand Synthesis Kit (Clontech, Mountain View, USA) in accordance with the manufacturers’ protocols. An SYBR Premix Ex Taq kit (Takara, Beijing, China) was employed to prepare the qPCR reactions, and the reaction was performed in a Bio-Rad CFX96 PCR System (Bio-Rad, Hercules, USA), U6 was used as an internal control for miRNA. miR-542-5p, Forward: 5′-GCGGTCGGGGATCATCATGTC-3′; Reverse: 5′-ATCCAGTGCAGGGTCCGAGG-3′. U6, Forward: 5′-CGCTTCGGCAGCACATATAC-3′; Reverse: 5′-TTCACGAATTTGCGTGTCATC-3′.
H/R treatment
H9c2 cardiomyoblasts were seeded at the density of 1 × 104 cells/cm2 with fresh media for each experiment. Cells were subjected to H/R, as reported previously [28]. Briefly, cells were cultured in Tyrode’s solution (130 mM NaCl, 5 mM KCl, 10 mM HEPES, 1 mM MgCl2, and 1.8 mM CaCl2) at 37 ℃. To simulate ischemia, the cells were then placed in a hypoxia chamber (Adelbio, Clermont-Ferrand, France) containing 95% N2 and 5% CO2 for 5 h. The media were immediately replaced with standard DMEM/F12 media after hypoxia. Then, reoxygenation (0 h, 1 h, 3 h, 12 h, or 24 h) were executed in a normoxic incubator.
Flow cytometric analysis
Cells were added with 0.25% trypsin (Boster, Wuhan, China) and transferred to tubes. The cells were centrifuged and then washed with PBS three times. The experiment was performed according to the manual in the Annexin-V-FITC cell apoptosis detection kit (BioVision, Milpitas, CA).
Cell viability assay
Cell viability was measured by employing the cell count kit-8 (CCK-8; Beyotime Biotech, Jiangsu, China). The procedure was performed according to the manufacturer’s instructions. Cells were seeded in 96-well plates at the density of 29–10 cells/plate after H/R treatment. CCK- 8 reagents were added after the treatments were finalized. The number of viable cells was determined by measuring the absorbance at the wavelength of 450 nm for each well.
Immunofluorescence analysis
After H/R treatment, H9c2 cells were treated with 4% paraformaldehyde, 0.1% Triton X-100 and 2% bovine serum albumin solution subsequently. They were cultured overnight at 4 ℃ with primary antibodies to light chain 3 (LC3; 1:100; Novus Biologicals). FITC-conjugated secondary antibodies were then employed for labeling the cells for 1 h at 37 ℃. Cells were imaged by an LSM 710 confocal microscope (Carl Zeiss AG; Oberkochen, Germany).
Western bolt analysis
The cells were lysed after H/R treatment by adding sample buffer and boiling for 15 min. A BCA protein assay kit (Beyotime Institute of Biotechnology; China) was applied to measure the total protein concentration according to the manufacturer’s protocol. Proteins were isolated using SDS-PAGE gels and transferred to PVDF membranes (Millipore; Germany). Then, 5% nonfat milk was used to block the membranes for 1 h. Next, it was cultured with primary antibodies raised against LC3(1:1000, Novus Biologicals, U.S.), p62 (1:1000, Abcam, UK), cleaved caspase-3 (1:1000, Cell Signaling, U.S.), and bactin (1:1000, Transgene, U.S.) at 4 ℃ overnight. It is then treated with a horseradish peroxidase-conjugated second antibody (1:10,000, Promega, U.S.) for 1 h at 37 ℃. Membranes were cultured using an ECL kit and visualized using a chemiluminescence instrument (ImageQuant LAS 4000; GE Healthcare; Little Chalfont, UK).
Luciferase reporter assay
The sequence of miR-542-5p binding sites in ATG7 was synthesized by Genechem (Shanghai, China). The wild type (WT) and mutant type (MUT) binding site sequences were digested by restriction endonuclease, and the target fragment was cloned into dual luciferase reporter vector (Promega, Madison, WI, USA). The 293 T cells were plated into 96-well plates and kept in DMEM medium supplemented with 10% FBS. When reaching 80% confluence, the correctly sequenced luciferase reporter plasmids (WT and MUT) were co-transfected with mimics NC and miR-542-5p mimics into 293 T cells using Lipofectamine 2000 reagent (Invitrogen). After 48‐h transfection, the cells were lysed. Luciferase activity was determined with Dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Measurement of CK level and LDH activity
Serum and culture supernatant were collected subsequent to H/R treatment, The CK level and LDH activity were measured by CK activity assay kit and LDH activity assay kit (Sigma, USA), respectively, after completing the above protocol. The results were interpreted strictly according to the manufacturer’s instructions.
RNA immunoprecipitation (RIP) assay
RIP assay was carried out using the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Merck Millipore, Billerica, MA, USA) following the instructions of the manufacturer. In brief, H9c2 cells were gathered and lysed using NP-40 lysis buffer (Solarbio) supplemented with PMSF (1 mmol/L), DTT (1 mmol/L), Protease Inhibitor Cocktail (1%) and RNase inhibitor. After that, the cells were incubated with RIP buffer supplemented with magnetic beads bound with human anti- Ago2 antibody or normal IgG for 4 h at 4 °C. Precipitates were digested by Proteinase K buffer, and then co-immunoprecipitated RNA was isolated for qRT-PCR.
Statistical analysis
All experiments were repeated three times at a minimum. The data were presented in the form of the means ± S.D. One-way ANOVA, all pair-wise multiple comparison procedures using Bonferroni’s test were carried out. A P value of < 0.05 was considered statistically significant. The statistical analysis was performed in SPSS version 20.0 (SPSS, Chicago, IL, USA).
Results
Upregulated miR-542-5p aggravated H/R injury in H9c2 cells
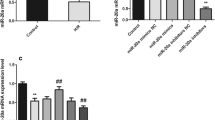
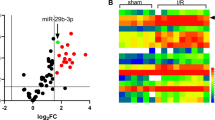
A recent study showed that a large number of miRNAs were differentially expressed in H9c2 cells after hypoxia treatment (Supporting Table S1) [29]. To identify the functional miRNAs in H/R-induced cardiomyocyte injury, the top 10 dysregulated miRNAs (Supporting Table S2) were selected to verify in H9c2 cells exposed to H/R. As shown in Fig. 1a, the expression of miR-542-5p, miR217-5p and miR-411-5p was significantly upregulated in H9c2 cells exposed to H/R compared to control group. Here the most significantly upregulated miRNA, miR-542-5p, was selected to further functional study. Figure 1b also showed that the expression of miR-542-5p was increased in H/R-treated H9c2 cells at different time point.
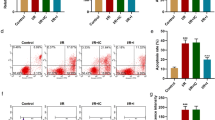
Upregulated miR-542-5p aggravated H/R injury in H9c2 cells. a The expression of the top 10 dysregulated miRNAs were detected in H9c2 cells after hypoxia treatment by qRT-PCR. b The expression of miR-542-5p was detected in H/R-treated H9c2 cells at different time point by qRT-PCR. *P < 0.05. (N = 3). c The cell viability of Hc92 cells after miR-542-5p overexpression or inhibition with H/R treatment was detected by CCK-8 assay. *P < 0.05. (N = 3). d Cell membrane integrity of H9c2 cells after miR-542-5p overexpression or inhibition with H/R treatment was measured by assessing LDH activity. *P < 0.05. **P < 0.01. (N = 3). e Cell apoptosis of H9c2 cells after miR-542-5p overexpression or inhibition with H/R treatment was measured by flow cytometric analysis. *P < 0.05. **P < 0.01. (N = 3). f The expression of cleaved caspase-3 protein in H9c2 cells after miR-542-5p overexpression or inhibition with H/R treatment was assessed by western bolt analysis. (N = 3)
The function of miR-542-5p on regulating H/R injury in H9c2 cells was then assessed. As shown in Fig. 1c, cell viability was reduced after H/R treatment and miR-542-5p overexpression resulted in an aggravation of cell viability loss, whereas miR-542-5p inhibition exerted an amelioration of H/R-induced cell viability loss (Supporting Figure S1). Cell membrane integrity was measured by assessing LDH activity. Figure 1d showed that the LDH release was induced following H/R and miR-542-5p overexpression further facilitated LDH release, whereas the increase was markedly repressed by miR-542-5p inhibition. Meanwhile, miR-542-5p overexpression aggravated H/R-induced cell apoptosis, while miR-542-5p inhibition attenuated H/R-induced cell apoptosis (Fig. 1e, f). These results demonstrate that upregulated miR-542-5p aggravated H/R injury in H9c2 cells.
miR-542-5p inhibited autophagy induced by H/R injury
As a survival mechanism during cellular stress, autophagy exerts a crucial role in regulating H/R-induced cardiomyocyte injury [30, 31]. We thus investigated whether miR-542-5p regulated autophagy activation in H9c2 cells exposed to H/R. H9c2 cells were treated with H/R in the presence or absence of miR-542-5p mimics or inhibitor, and then the autophagy activation was assessed. Figure 2a showed that following miR-542-5p treatment, there was a significant upregulation of green puncta representing autophagic vacuoles in H9c2 cells exposed to H/R and miR-542-5p inhibition resulted in an aggravation of autophagy activation, whereas miR-542-5p overexpression exerted an amelioration of H/R-induced autophagy activation in H9c2 cells. Figure 2b showed that miR-542-5p reduced the ratio of LC3-II/β-actin, whereas miR-542-5p inhibition resulted in an increase of ratio of LC3-II/β-actin, indicating miR-542-5p negatively regulated autophagy activation in H9c2 cells exposed to H/R. p62/SQSTM1 protein is an autophagy substrate [32] and then the autophagy flux was validated by measuring the change of p62 protein expression. Figure 2c showed that miR-542-5p repressed autophagy flux, whereas miR-542-5p inhibition resulted in an increase of autophagy flux.
miR-542-5p inhibited autophagy induced by H/R injury. a The autophagy activation in H9c2 cells treated with H/R in the presence or absence of miR-542-5p mimics or inhibitor was assessed by immunofluorescence analysis, LC3B staining (green) and nuclei staining (blue). (N = 3). b The expression of cleaved LC3-I and LC3-II protein in H9c2 cells treated with H/R in the presence or absence of miR-542-5p mimics or inhibitor were assessed by western bolt analysis. *P < 0.05. (N = 3). c The expression of cleaved p62 protein in H9c2 cells treated with H/R in the presence or absence of miR-542-5p mimics or inhibitor were assessed by western bolt analysis. *P < 0.05. **P < 0.01. (N = 3)
miR-542-5p inhibition alleviated H/R injury by the mediation of autophagy
To explore the effect of miR-542-5p/autophagy pathway on regulating H/R injury, miR-542-5p was inhibited in the presence or absence of autophagy inhibitor (3-MA) and H/R injury in H9c2 cells was assessed. Figure 3a showed that cell viability was increased in H9c2 exposed to H/R after miR-542-5p inhibition, whereas autophagy inhibition partially alleviated the protective role of miR-542-5p inhibition in H/R injury. Figure 3b showed that the LDH release was repressed following miR-542-5p inhibition, whereas the effect was partially destroyed because of autophagy inhibition. Furthermore, miR-542-5p inhibition alleviated H/R-induced cell apoptosis, while the effect was also partially repressed because of autophagy inhibition (Fig. 3c). These data suggest that autophagy exerted as a protective role in H/R injury and miR-542-5p inhibition alleviated H/R injury by the mediation of autophagy.
miR-542-5p inhibition alleviated H/R injury by the mediation of autophagy. a Cell viability was assessed using CCK-8 assay in H9c2 exposed to H/R and miR-542-5p inhibition in the presence or absence of autophagy inhibitor (3-MA). *P < 0.05. (N = 3). b Cell membrane integrity was measured in H9c2 exposed to H/R and miR-542-5p inhibition in the presence or absence of autophagy inhibitor (3-MA) by assessing LDH activity. *P < 0.05. (N = 3). c Cell apoptosis was measured in H9c2 exposed to H/R and miR-542-5p inhibition in the presence or absence of autophagy inhibitor (3-MA) by flow cytometric analysis. *P < 0.05. (N = 3)
miR-542-5p inhibited ATG7 expression in H9c2 cells
miRNAs play importantly biological roles by binding to 3′-untranslated region (UTR) of target gene to repress gene expression. In the study the target genes of miR-542-5p were screened using Targetscan software (http://www.targetscan.org/vert_71/). Four autophagy-related genes, ATG9B, ATG7, ATG4B, and ATG14, were the potential target genes of miR-542-5p. To assess whether miR-542-5p repressed the expression of these autophagy-related genes, the luciferase reporter plasmids containing 3′-UTR of ATG9B, ATG7, ATG4B, and ATG14 (ATG9B-3′UTR-LUC, ATG7-3′UTR-LUC, ATG4B-3′UTR-LUC, and ATG14-3′UTR-LUC) were constructed, respectively. The results from luciferase reporter assay showed that miR-542-5p specifically repressed the luciferase expression of ATG7-3′UTR-LUC (Fig. 4a), whereas mutation of 4 nucleotides in ATG7-3′-UTR resulted in complete abolition of the repressive role (Fig. 4b, c). Furthermore, miR-542-5p overexpression markedly inhibited ATG7 protein expression in H9c2 cells (Fig. 4d, e). The RIP assay was further performed to explore the binding of miR-542-5p and ATG7 in H9c2 cells. As shown in Fig. 4f, miR-542-5p and ATG7 were both enriched in Ago2-coating beads compared to IgG control group. In addition, we identified ATG7 is enriched in cell transfected with miR-542-5p mimics compared with miRcont treated group through RIP assay. The present data demonstrate that H/R-induced increase of miRNA-542-5p aggravated cardiomyocytes injury by inhibiting autophagy.
miR-542-5p inhibited ATG7 expression in H9c2 cells. a Luciferase reporter assay was performed to assess whether miR-542-5p repressed the expression of four autophagy-related genes (ATG9B, ATG7, ATG4B, and ATG14). b Schematic representation of the miR-542-5p site in ATG7-3′UTR. c The 3′UTR reporter assay was performed in H9c2 cells overexpressed with miR-542-5p. pGL3- ATG7-3′-UTR-WT or pGL3-ATG7-3′-UTR-Mutation was co-transfected with pRL-TK. Luciferase assays were performed 48 h after transfection. Firefly luciferase activity was normalized to the Renilla luciferase control. **P < 0.01 (N = 3). d and e The expression of ATG7 protein in H9c2 cells overexpressed with miR-542-5p was assessed by western bolt analysis. **P < 0.01 (N = 3). f Anti-Argonaute 2 (AGO2) RNA immunoprecipitation (RIP) assays were used in H9c2 cells to determine ATG7 and miR-542-5p RNA enrichment in immunoprecipitated (IP) complex. Anti-immunoglobulin G (IgG) was used as the control. ATG7 and miR-542-5p were enriched preferentially in miRNA ribonucleoprotein complexes (miRNPs) containing AGO2 compared with anti-IgG immunoprecipitates. **P < 0.01, ***P < 0.001 (N = 3)
The function of miR-542-5p on H/R injury in H9c2 cells was blocked by ATG7
To explore the effect of ATG7 on regulating H/R injury, miR-542-5p was upregulated in the presence or absence of ATG7 and H/R injury in H9c2 cells was assessed. Figure 5a showed that cell viability was decreased in H9c2 exposed to H/R after miR-542-5p overexpression, whereas the effect was partially blocked by ATG7 overexpression. Figure 5b showed that the LDH release was increased following miR-542-5p treatment, whereas the effect was partially restored because of ATG7 overexpression. Furthermore, miR-542-5p mimic aggravated H/R-induced cell apoptosis, while the effect was also partially suppressed because of ATG7 overexpression. (Fig. 5c). These data suggest that miR-542-5p aggravated H/R injury by the mediation of autophagy through mediating ATG7.
The function of miR-542-5p on H/R injury in H9c2 cells was blocked by ATG7. a Cell viability was assessed using CCK-8 assay in H9c2 exposed to H/R in the presence or absence of miR-542-5p mimic and ATG7. *P < 0.05 (N = 3). b Cell membrane integrity was measured in H9c2 exposed to H/R in the presence or absence of miR-542-5p mimic and ATG7 by assessing LDH activity. *P < 0.05 (N = 3). c Cell apoptosis was measured in H9c2 exposed to H/R in the presence or absence of miR-542-5p mimic and ATG7 by flow cytometric analysis. *P < 0.05 (N = 3)
Discussion
In the present study, we explored the role of miR-542-5p in regulating autophagy and H/R injury in H9c2 cells. Our data verify that (i) Upregulated miR-542-5p aggravated H/R injury in H9c2 cells, (ii) miR-542-5p inhibited autophagy induced by H/R injury, (iii) miR-542-5p inhibition alleviated H/R injury by the mediation of autophagy, (iv) miR-542-5p inhibited ATG7 expression in H9c2 cells. (v) The function of miR-542-5p on H/R injury in H9c2 cells was blocked by ATG7. These results identified the important role of miR-542-5p/autophagy pathway in H/R-induced cardiomyocyte injury and indicated that pharmacological intervention for miR-542-5p/autophagy pathway might be an effective way to treat H/R-related heart diseases.
Autophagy is a dynamic process by which cytoplasmic proteins and organelles are degraded through lysosomes and recycled for sustaining cellular metabolism [33, 34]. Therefore, autophagy is generally thought to be a survival mechanism in cellular stress, and autophagy plays crucial roles in multiple physiopathologic processes including development [35], tumor progression [36] and H/R-related heart diseases [37, 38]. Although emerging studies showed that the activation of autophagy is enhanced in cardiomyocytes during H/R [39], whether autophagy exerts as a protective or poisonous role in H/R-induced cardiomyocyte injury remains unclear at the present time. On one hand, Nakai et al. [40] demonstrated that cardiomyocyte-specific ATG5 deficiency totally prevents autophagy activation and results in a marked reduction of cardiac performance and subsequent cardiac hypertrophy and left ventricular dilation. Mild‑to‑moderate H/R or I/R-induced autophagy activation prevents cardiomyocyte injury via degrading damaged organelles and thus is protective against H/R or I/R injury [41, 42]. On the other hand, several studies showed that autophagy activation exacerbates myocardial injury after H/R or I/R, suggesting that over-activation of autophagy is harmful to cardiomyocytes [43]. Zhang et al. [44] reported that overexpression of miR-27a-5p alleviates hypoxia-induced cardiomyocyte injury by repressing autophagy.
Is autophagy protective or destructive in cardiomyocytes subjected to H/R? It is acceptable that cardiomyocyte survival and function largely depend on the existence of basal level of autophagy, whereas over-activation of autophagy will aggravate cardiomyocyte injury during H/R or I/R. However, the appropriate degree of autophagy activation is difficult to determine. Perhaps the degree of autophagy activation is not a critical factor to its protective or destructive role in cardiomyocytes. Autophagy is activated to protect against H/R injury when ‘normal’ cardiomyocytes encounter H/R stress, and the activation becomes higher and higher if the H/R stress is not eliminated. Finally, some cardiomyocytes are damaged and the autophagy activation is high in ‘damaged’ cardiomyocytes. Over-activated autophagy still exerts a pro-survival role in ‘damaged’ cardiomyocytes, and thus this phenomenon misleads us to believe that autophagy is destructive in cardiomyocytes subjected to H/R or I/R.
In the study, we demonstrated that following miR-542-5p treatment, there was a significant activation of autophagy in H9c2 cells exposed to H/R. Autophagy plays a protective role against H/R injury. Cell viability was increased in H9c2 exposed to H/R after miR-542-5p inhibition, whereas autophagy inhibition partially alleviated the protective role of miR-542-5p inhibition in H/R injury. Autophagy inhibition also destroyed miR-542-5p inhibitor-induced the decrease of LDH release.
At present, our research still has some limitations. In the current study, all of results are derived from cell experiments, we need to further confirmed our results in animal model.
Taken together, the current data demonstrate that autophagy exerted as a protective role in H/R injury and miR-542-5p inhibition alleviated H/R injury by the mediation of autophagy.
References
Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS. Heart disease and stroke statistics–2012 update: a report from the American Heart Association. Circulation. 2012;125:e2–220.
Santos-Gallego CG, Picatoste B, Badimón JJ. Pathophysiology of acute coronary syndrome. Curr Atheroscler Rep. 2014;16:401.
Chung S-C, Gedeborg R, Nicholas O, James S, Jeppsson A, Wolfe C, Heuschmann P, Wallentin L, Deanfield J, Timmis A. Acute myocardial infarction: a comparison of short-term survival in national outcome registries in Sweden and the UK. Lancet. 2014;383:1305–12.
Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med. 2017;376:2053–64.
Davidson SM, Ferdinandy P, Andreadou I, Botker HE, Heusch G, Ibanez B, Ovize M, Schulz R, Yellon DM, Hausenloy DJ, Garcia-Dorado D, Action CC. Multitarget strategies to reduce myocardial ischemia/reperfusion injury: JACC review topic of the week. J Am Coll Cardiol. 2019;73:89–99.
Ovize M, Kloner RA, Hale SL, Przyklenk K. Coronary cyclic flow variations “precondition” ischemic myocardium. Circulation. 1992;85:779–89.
Zuo Z, Zuo PF, Sheng ZL, Wang X, Ding JD, Ma GS. Tetramethylprazine attenuates myocardial ischemia/reperfusion injury through modulation of autophagy. Life Sci. 2019;239:117016.
Huang ZQ, Xu W, Wu JL, Lu X, Chen XM. MicroRNA-374a protects against myocardial ischemia-reperfusion injury in mice by targeting the MAPK6 pathway. Life Sci. 2019;232:116619.
Wang N, Zhou Z, Liao X, Zhang T. Role of microRNAs in cardiac hypertrophy and heart failure. IUBMB Life. 2009;61:566–71.
Boon RA, Dimmeler S. MicroRNAs in myocardial infarction. Nat Rev Cardiol. 2015;12:135.
Hwang H, Mendell J. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Lu Y, Xi J, Zhang Y, Li C, Chen W, Hu X, Zhang M, Zhang F, Wei H, Li Z, Wang Z. MicroRNA-214-5p protects against myocardial ischemia reperfusion injury through targeting the FAS ligand. Arch Med Sci. 2019;16(5):1119–29.
Sun M, Zhai M, Zhang N, Wang R, Liang H, Han Q, Jia Y, Jiao L. MicroRNA-148b-3p is involved in regulating hypoxia/reoxygenation-induced injury of cardiomyocytes in vitro through modulating SIRT7/p53 signaling. Chem Biol Interact. 2018;296:211–9.
Ding S, Liu D, Wang L, Wang G, Zhu Y. Inhibiting microRNA-29a protects myocardial ischemia-reperfusion injury by targeting SIRT1 and suppressing oxidative stress and NLRP3-mediated pyroptosis pathway. J Pharmacol Exp Ther. 2020;372:128–35.
Lin H, Ewing LE, Koturbash I, Gurley BJ, Miousse IR. MicroRNAs as biomarkers for liver injury: current knowledge, challenges and future prospects. Food Chem Toxicol. 2017;110:229–39.
Mirzaei H, Momeni F, Saadatpour L, Sahebkar A, Goodarzi M, Masoudifar A, Kouhpayeh S, Salehi H, Mirzaei HR, Jaafari MR. MicroRNA: relevance to stroke diagnosis, prognosis, and therapy. J Cell Physiol. 2018;233:856–65.
Cheng D-D, Yu T, Hu T, Yao M, Fan C-Y, Yang Q-C. MiR-542-5p is a negative prognostic factor and promotes osteosarcoma tumorigenesis by targeting HUWE1. Oncotarget. 2015;6:42761.
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069.
Zheng Y, Gu S, Li X, Tan J, Liu S, Jiang Y, Zhang C, Gao L, Yang HT. Berbamine postconditioning protects the heart from ischemia/reperfusion injury through modulation of autophagy. Cell Death Dis. 2017;8:e2577.
Schiattarella GG, Hill JA. Therapeutic targeting of autophagy in cardiovascular disease. J Mol Cell Cardiol. 2016;95:86–93.
Nishida K, Otsu K. Autophagy during cardiac remodeling. J Mol Cell Cardiol. 2016;95:11–8.
Hu Z, Cai HY, Luo YY, Xiao JM, Li L, Guo T. Effect of varying hypoxia reoxygenation times on autophagy of cardiomyocytes. Acta Cir Bras. 2018;33:223–30.
Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, Wang ZV, Morales C, Luo X, Cho G, Jiang N, Jessen ME, Warner JJ, Lavandero S, Gillette TG, Turer AT, Hill JA. Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation. 2014;129:1139–51.
Huang Z, Wu S, Kong F, Cai X, Ye B, Shan P, Huang W. MicroRNA-21 protects against cardiac hypoxia/reoxygenation injury by inhibiting excessive autophagy in H9c2 cells via the Akt/mTOR pathway. J Cell Mol Med. 2017;21:467–74.
Zhao P, Zhang BL, Liu K, Qin B, Li ZH. Overexpression of miR-638 attenuated the effects of hypoxia/reoxygenation treatment on cell viability, cell apoptosis and autophagy by targeting ATG5 in the human cardiomyocytes. Eur Rev Med Pharmacol Sci. 2018;22:8462–71.
Huang Z, Wu S, Kong F, Cai X, Ye B, Shan P, Huang W. Micro RNA-21 protects against cardiac hypoxia/reoxygenation injury by inhibiting excessive autophagy in H9c2 cells via the Akt/mTOR pathway. J Cell Mol Med. 2017;21:467–74.
Paillard M, Tubbs E, Thiebaut P-A, Gomez L, Fauconnier J, Crola Da Silva C, Teixeira G, Mewton N, Belaidi E, Durand A. Depressing mitochondria-reticulum interactions protects cardiomyocytes from lethal hypoxia-reoxygenation injury. Circulation. 2013;128:1555–65.
Zhang J, Ma J, Long K, Qiu W, Wang Y, Hu Z, Liu C, Luo Y, Jiang A, Jin L, Tang Q, Wang X, Li X, Li M. Overexpression of exosomal cardioprotective miRNAs mitigates hypoxia-induced H9c2 cells apoptosis. Int J Mol Sci. 2017;18(4):711.
Zuo Y, Zhang J, Cheng X, Li J, Yang Z, Liu X, Gu E, Zhang Y. Enhanced autophagic flux contributes to cardioprotection of remifentanil postconditioning after hypoxia/reoxygenation injury in H9c2 cardiomyocytes. Biochem Biophys Res Commun. 2019;514:953–9.
Li X, Xie X, Yu Z, Chen Y, Qu G, Yu H, Luo B, Lei Y, Li Y. Bone marrow mesenchymal stem cells-derived conditioned medium protects cardiomyocytes from hypoxia/reoxygenation-induced injury through Notch2/mTOR/autophagy signaling. J Cell Physiol. 2019;234:18906–16.
Berliocchi L, Russo R, Maiaru M, Levato A, Bagetta G, Corasaniti MT. Autophagy impairment in a mouse model of neuropathic pain. Mol Pain. 2011;7:83.
Ni HM, Bockus A, Wozniak AL, Jones K, Weinman S, Yin XM, Ding WX. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204.
Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–67.
Chai P, Ni H, Zhang H, Fan X. The evolving functions of autophagy in ocular health: a double-edged sword. Int J Biol Sci. 2016;12:1332–40.
Bryant KL, Stalnecker CA, Zeitouni D, Klomp JE, Peng S, Tikunov AP, Gunda V, Pierobon M, Waters AM, George SD, Tomar G, Papke B, Hobbs GA, Yan L, Hayes TK, Diehl JN, Goode GD, Chaika NV, Wang Y, Zhang GF, Witkiewicz AK, Knudsen ES, Petricoin EF 3rd, Singh PK, Macdonald JM, Tran NL, Lyssiotis CA, Ying H, Kimmelman AC, Cox AD, Der CJ. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019;25:628–40.
Gao G, Chen W, Yan M, Liu J, Luo H, Wang C, Yang P. Rapamycin regulates the balance between cardiomyocyte apoptosis and autophagy in chronic heart failure by inhibiting mTOR signaling. Int J Mol Med. 2020;45(1):195–209.
Chen Q, Zhou Y, Richards AM, Wang P. Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways. Biochem Biophys Res Commun. 2016;474:168–74.
Matsui Y, Takagi H, Qu X, Abdellatif M, Sakoda H, Asano T, Levine B, Sadoshima J. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007;100:914–22.
Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, Nishida K, Hori M, Mizushima N, Otsu K. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–24.
Hamacher-Brady A, Brady NR, Gottlieb RA. The interplay between pro-death and pro-survival signaling pathways in myocardial ischemia/reperfusion injury: apoptosis meets autophagy. Cardiovasc Drugs Ther. 2006;20:445–62.
Ren Z, Xiao W, Zeng Y, Liu MH, Li GH, Tang ZH, Qu SL, Hao YM, Yuan HQ, Jiang ZS. Fibroblast growth factor-21 alleviates hypoxia/reoxygenation injury in H9c2 cardiomyocytes by promoting autophagic flux. Int J Mol Med. 2019;43:1321–30.
Lavandero S, Troncoso R, Rothermel BA, Martinet W, Sadoshima J, Hill JA. Cardiovascular autophagy: concepts, controversies, and perspectives. Autophagy. 2013;9:1455–66.
Zhang J, Qiu W, Ma J, Wang Y, Hu Z, Long K, Wang X, Jin L, Tang Q, Tang G, Zhu L, Li X, Shuai S, Li M. miR-27a-5p attenuates hypoxia-induced rat cardiomyocyte injury by inhibiting Atg7. Int J Mol Sci. 2019;20(10):2418.
Acknowledgements
This work was supported by Shanghai Municipal Jiading District New Key Subject Program (2017-ZD-03), Shanghai Municipal Jiading District Health Commission Foundation (2019-KY-09) and Natural Science Foundation of Shanghai (19ZR1444900).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare no potential conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, F., Min, X., Hu, Sy. et al. Hypoxia/reoxygenation-induced upregulation of miRNA-542-5p aggravated cardiomyocyte injury by repressing autophagy. Human Cell 34, 349–359 (2021). https://doi.org/10.1007/s13577-020-00466-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00466-z