Abstract
Cholangiocarcinoma (CCA) is one of the most aggressive and lethal malignancies. Long noncoding RNAs (lncRNAs) are being found to play crucial roles in CCA progression. This work aims to investigate the roles of long intergenic non-protein coding RNA 667 (LINC00667) in progression of CCA. RT-qPCR and western blot were applied to detect gene expression. Clinical correlation and survival were analyzed by statistical methods. Overexpression and RNA interference approaches were used to investigate the effects of LINC00667 on CCA cells. Tumor xenograft assay was performed to detect the function of LINC00667 in vivo. Transcriptional regulation and competing endogenous RNA (ceRNA) mechanism were predicted via bioinformatics analysis. ChIP, luciferase reporter, and Ago2 RIP assays further confirmed the predicted results. Our data indicated that LINC00667 was highly expressed in CCA tissues and cells, and transcription factor Yin Yang 1 (YY1) induced LINC00667 expression in CCA cells. Up-regulated LINC00667 was significantly associated with lymph node metastasis, advanced TNM stage, and poor prognosis. Knockdown of LINC00667 suppressed the proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) of CCA cells, while overexpression of LINC00667 acquired opposite effects. Moreover, knockdown of LINC00667 inhibited tumor growth in vivo. In addition, LINC00667 was demonstrated to function as a ceRNA for miR-200c-3p, and then LINC00667 up-regulated pyruvate dehydrogenase kinase 1 (PDK1) to promote CCA development by inhibiting miR-200c-3p. These findings identified a pivotal role of LINC00667 in tumorigenesis and development of CCA. Targeting the YY1/LINC00667/miR-200c-3p/PDK1 axis may provide a new therapeutic strategy for CCA treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cholangiocarcinoma (CCA) is a group of digestive system malignant tumor that occurs at any location along the biliary tree [1]. The incidence of CCA has been rising over the past two decades in most countries, especially in Thailand with the highest incidence of 96 cases per 100,000 [2]. Early diagnosis of CCA is difficult due to lack specific symptoms. The resistance to conventional therapies further increases the difficulty of treatment. Surgical resection is the only curative approach, but most patients have lost the opportunity for radical resection when diagnosis. It is necessary to identify the key pathological mechanisms and effective therapeutic targets, thereby improving the prognosis of CCA.
Long noncoding RNAs (lncRNAs) belong to a class of non-protein coding transcripts that are longer than 200 nucleotides. The lncRNAs play crucial roles in regulating gene expression at different stages including chromatin remodeling, transcription, and post-transcription regulation [3]. MicroRNAs (miRNAs) are a class of small single-stranded noncoding RNAs with 20–24 nucleotides in length. The miRNAs regulate gene expression at the post-transcriptional level by inhibiting their target mRNAs [4]. Some lncRNAs were identified as competing endogenous RNAs (ceRNAs) to competitively bind miRNAs, thereby alleviating the miRNA-dependent inhibition of mRNAs [5]. Many lncRNAs and miRNAs have been shown to play vital roles in different physiological and pathological processes, especially in tumor development [6].
Long intergenic non-protein coding RNA 667 (LINC00667), located on chromosome 18p11.31, is a newly recognized lncRNA [7]. LINC00667 has been reported to be a cancer promoter in various types of tumors, such as glioma, lung cancer, and Wilms’ tumor [8,9,10]. Wang et al. [8] reported that LINC00667 was involved in glioma tumorigenesis by promoting cancer cell proliferation, migration and invasion via sponging miR-429. Yang et al. [9] demonstrated that LINC00667 acted as an oncogene in non-small cell lung cancer partly through serving as a ceRNA to suppress miR-143-3p expression. However, studies regarding the clinical value and biological role of LINC00667 in CCA have not been elucidated.
MiR-200c-3p is a cancer-associated miRNA that belongs to the miR-200 family. Studies have shown that miR-200c-3p participates in regulating diverse kinds of cancers [11, 12]. For instance, miR-200c-3p inhibited cell migration and invasion of renal carcinoma by sponging SLC6A1 [12]. Notably, urine miR-200c-3p was confirmed as potential non-invasive biomarker in patients with prostate cancer [13]. Pyruvate dehydrogenase kinase 1 (PDK1), an important glycolytic enzyme, can phosphorylate and inactivate pyruvate dehydrogenase, thereby suppressing pyruvate oxidation [14]. As a glycolytic enzyme, PDK1 has been identified to be closely associated with cancer cell proliferation, metastasis, and chemotherapy resistance [15, 16]. For instance, PDK1 promoted ovarian cancer metastasis by modulating tumor-mesothelial adhesion, invasion, and angiogenesis [15].
In this study, LINC00667 was up-regulated in CCA tissues and cells, and its up-regulation was correlated with shorter survival of CCA patients. Yin Yang 1 (YY1) directly bound to the LINC00667 promoter and induced its transcription. Furthermore, LINC00667 facilitated tumor proliferation, migration, invasion and epithelial-mesenchymal transition (EMT). For the mechanism, LINC00667 functioned as a ceRNA to up-regulate PDK1 by sponging miR-200c-3p. Our data disclose the biological function of LINC00667 in CCA progression, and suggest LINC00667 as a potential target for anti-cancer therapy.
Materials and methods
Patient samples
A total of 62 tumor tissues and paired adjacent tumor-free tissues surgically removed from CCA patients were collected in The 2nd Affiliated Hospital of Harbin Medical University. Cancer tissues and paired adjacent tumor-free tissues from CCA patients were immediately frozen in liquid nitrogen after the resection. Patients who have received neoadjuvant chemotherapy and radiotherapy were excluded. Written informed consents were acquired from all patients prior to the study. The protocol of this study was approved by the Ethics Committee of The 2nd Affiliated Hospital of Harbin Medical University (KY2018-301). To investigate the clinical significance of abnormal LINC00667 expression in CCA patients, the 62 CCA patients were classified into low LINC00667 expression group (< the median value) and high LINC00667 expression group (> the median value) according to the median value of LINC00667 expression in CCA tissues. Thus, through relatively high and low expression grouping, we can analyze the correlation between LINC00667 and clinicopathological characteristics as well as survival in CCA patients by appropriate statistical methods.
Cell culture and transfection
Four CCA cell lines containing CCLP-1, QBC939, RBE, and HCCC-9810 as well as a normal human biliary epithelial cell line HIBEC were involved in this study. RBE and HCCC-9810 were purchased from the Institute of Cell Biology of Chinese Academy of Sciences (Shanghai, China). CCLP-1, QBC939 and HIBEC were stored in our laboratory. DMEM or RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) was applied for cell cultivation in a humidified atmosphere of 5% CO2 and 37 °C. The small interfering RNA (siRNA) for LINC00667 (si-LINC00667-1, si-LINC00667-2), YY1 (si-YY1) and PDK1 (si-PDK1) with the corresponding negative control (si-NC), the short hairpin RNA (shRNA) for LINC00667 (sh-LINC00667) with the negative control (sh-NC), the recombinant pcDNA3.1 plasmid for overexpressing LINC00667 (oe-LINC00667) and YY1 (oe-YY1) with the corresponding empty vector (oe-NC), the miR-200c-3p mimics with negative control (NC mimics) and miR-200c-3p inhibitor with negative control (NC inhibitor) were all obtained from GenePharma (Shanghai, China) and transfected into cells using the Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s protocol. The siRNA sequences used in the present study are summarized in Table S1.
RNA extraction and real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from CCA tissues or cells using the TRIzol reagent (Invitrogen) according to the instructions of manufacturer. First-strand cDNA was synthesized from 1 μg of total RNA by PrimeScript RT Master Mix kit (Takara, Shiga, Japan). Then real-time PCR was conducted by SYBR PrimeScript PLUS RT-PCR kit (Takara) on the ABI7500 PCR instrument (Applied Biosystems, Foster City, CA, USA). The relative fold change for gene expression was calculated using \(2^{{ - \Delta \Delta C_{{\text{t}}} }}\) method. GAPDH and U6 were used as endogenous controls to normalize mRNA/lncRNA and miRNA expressions, respectively. Primers used in the present study are summarized in Table S1.
Western blot analysis
CCA cells were lysed in cold RIPA buffer (Beyotime, Beijing, China), and the protein concentration was quantified by BCA kit (Beyotime). The protein lysates were separated in 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and then transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% non-fat milk and then treated with primary antibody overnight at 4 °C. Following extensive washing with Tris-buffered saline with 0.1% Tween-20 (TBST) three times, the membranes were further incubated with secondary antibody (Abcam, Cambridge, MA) at room temperature. After further washing with TBST three times, the protein bands were detected using BeyoECL kit (Beyotime) according to the manufacturer’s guidelines. GAPDH was employed as an internal control. The antibodies against YY1, PDK1, E-cadherin, N-cadherin, vimentin and GAPDH were all obtained from Abcam.
CCK-8 cell proliferation assay
Cell counting kit-8 (CCK-8) was utilized to detect the cell proliferative ability. In brief, transfected cells were seeded into 96-well plates. At five time points (0, 24, 48, 72, and 96 h), 10 μl CCK-8 solution (Dojindo, Kumamoto, Japan) was added into each well at the same time of the day. Then, the absorbance of each well was measured at 450 nm by a microplate reader.
5-ethynyl-2′-deoxyuridine (EdU) assay
Transfected cells were seeded into 96-well plates and incubated for 24 h. Then, each well was incubated with 100 μl diluted EdU solution (Ribobio, Guangzhou, China) for 2 h. The cells were fixed with paraformaldehyde and stained with Apollo 567 and Hoechst 33,342 according to the instructions of manufacturer. The numbers of EdU-positive cells were detected under a fluorescence microscope.
Colony formation assay
Cell proliferation was also analyzed using the colony formation assay. In brief, equal numbers of CCA cells from the different treatment groups were seeded into 6-well plates and cultured with complete medium for 14 days. Then, the cell colonies were fixed with paraformaldehyde and stained with crystal violet (Beyotime). The numbers of stained colonies were counted.
Wound healing migration assay
After transfection, CCA cells were seeded into 6-well plates and cultured until more than 90% confluence. A linear wound was created by scratching the cell monolayer with a sterile pipette tip. The floating cells were washed off with PBS and the adherent cells were cultured with serum-free medium. An inverted optical microscope was used to monitor the closure of the wound at 0 and 48 h.
Transwell assay
The transfected cells were collected and re-suspended in serum-free medium. Then, the re-suspended cells were seeded into transwell chambers (Corning Incorporated, Corning, NY, USA) with or without pre-packed Matrigel (BD Biosciences, San Jose, CA, USA), and the chambers were inserted into 24-well plates containing complete medium. After incubation for 24 h, the cells remaining on the upper membrane surface were removed using a cotton swab, and the cells invading into the lower membrane surface were fixed with paraformaldehyde and stained with crystal violet. Finally, the cells were observed and counted using an inverted microscope.
Subcellular fractionation assay
Nuclear and cytosolic fractions were separated using a PARIS kit (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The expression levels of GAPDH, U6 and LINC00667 in the nucleus and cytoplasm of CCA cells were detected by RT-qPCR assays.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay certified the interplay of YY1 on LINC00667 promoter using the EZ-ChIP kit (Millipore). The cross-linked chromatin was obtained using formaldehyde. The separation of chromatin by sonication was conducted for the generation of chromatin fragments. Afterwards, the lysates were further incubated with IgG and anti-YY1 (Abcam) to generate chromatin precipitations. Finally, RT-qPCR was used to examine the relative enrichment of DNA fragments containing the putative YY1 binding sites in the LINC00667 promoter region. Primers are summarized in Table S1.
Luciferase reporter assay
For dual-luciferase assay, the wild type and mutation type of LINC00667 promoter fragment, and LINC00667 fragment or PDK1 fragment were synthesized and inserted into a pmirGLO reporter vector (Promega, Madison, WI, USA), respectively. CCA cells were seeded into 96-well plates and co-transfected with corresponding plasmids and miRNA mimics using the Lipofectamine 3000 reagent. Luciferase activity was measured using the dual-luciferase reporter assay kit (Promega) after 48 h of incubation according to the manufacturer’s instructions.
RNA immunoprecipitation (RIP) assay
RIP assay was performed using an EZ-Magna RIP kit (Millipore) according to the manufacturer’s instructions. Transfected cells were lysed in RIP lysis buffer and incubated with magnetic beads conjugated with normal IgG control or anti-Ago2 antibody (Millipore). The RNAs in the immunoprecipitations were isolated by Proteinase K and analyzed by RT-qPCR.
Tumor xenograft assay
For the xenograft tumor, 5 × 106 CCLP-1 cells transfected with sh-LINC00667 or sh-NC were subcutaneously injected into the posterior flank of 5-week-old female BALB/c nude mice (n = 6 per group). The tumor volumes were calculated every 3 days by the formula 0.5 × length × width2. After 21 days, all mice were euthanized by dislocation of cervical vertebra, and then the tumor weights were measured. All nude mice were purchased from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) and fed in specific-pathogen-free environment. All animal experiments were approved by the Animal Care and Use Committee of The 2nd Affiliated Hospital of Harbin Medical University (SYDW2018-114).
Statistical analysis
Experimental data were expressed as mean ± standard deviation and evaluated using SPSS 21.0 (IBM, Chicago, IL, USA) and GraphPad Prism 7.0 software (GraphPad Software, San Diego, CA, USA). One-way analysis of variance, two‐tailed Student’s t-test, or Pearson’s correlation analysis was used to estimate the obvious differences between groups. The relationship between LINC00667 expression and clinical characteristics was evaluated using the chi-squared test. Receiver operating characteristic curve (ROC) analysis was used to evaluate the performance of prognostic prediction of LINC00667 expression in CCA tissues. LINC00667 expression values in CCA tissues were selected as independent variable in ROC analysis. The dependent variable was a binary variable, so survival > 3 years was defined as good prognosis, and survival ≤ 3 years was defined as poor prognosis. Hence, the value of area under curve (AUC) and the sensitivity and specificity of optimal LINC00667 cut-off value could evaluate the performance of LINC00667 prognostic prediction in CCA. Each experiment was repeated at least three times. P value < 0.05 was indicative of statistically significant difference.
Results
LINC00667 was up-regulated in CCA and predicted a poor outcome for CCA patients
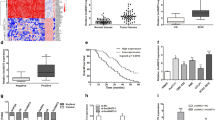
To elucidate the functions of LINC00667 on CCA progression, experiments were carried out to observe the expression of LINC00667 in CCA tissues and cells. Data from The Cancer Genome Atlas (TCGA) database exhibited that LINC00667 expression was significantly enhanced in CCA (P < 0.001, Fig. 1a). In addition, results from RT-qPCR showed that the expression of LINC00667 was also remarkably up-regulated in CCA tissues compared with paired adjacent tumor-free tissues (Fig. 1b). For LINC00667 expression at cell level, we confirmed that LINC00667 expression was up-regulated in CCLP-1, QBC939, RBE, and HCCC-9810 cells compared with that in normal HIBEC (Fig. 1c). Afterwards, we further explored the correlation between LINC00667 expression and clinicopathological features by χ2 test. Using the median value of LINC00667 expression in CCA tissues as the cut-off value, we divided the 62 CCA patients into high LINC00667 expression group and low LINC00667 expression group. Then, the results revealed that high expression of LINC00667 was associated with lymph node metastasis and advanced TNM stage, but there was no statistical difference between LINC00667 expression and other clinicopathological parameters of patients with CCA (Table 1). Moreover, we analyzed the survival correlation by Kaplan–Meier method, and the results confirmed that patients with increased LINC00667 expression had poorer overall survival than those with decreased expression of LINC00667 (log rank P < 0.001, Fig. 1d). We also performed univariate and multivariate analyses to evaluate the prognostic value of LINC00667 expression and other clinicopathological features. The univariate analysis showed that the lymph node invasion, advanced TNM stage and high LINC00667 expression were significantly correlated with the overall survival, and the multivariate analysis revealed that high LINC00667 expression was an independent poor prognostic factor for CCA patients (Table 2). Finally, ROC analysis indicated that LINC00667 had great prognostic evaluation ability in CCA when 3-year survival was defined as a threshold for good and poor prognosis. The AUC of LINC00667 expression was 0.830 (95% CI: 0.748–0.911) with 87.5% sensitivity and 72% specificity at the best cut-off value of 6.72 (P < 0.001, Fig. 1e).
The expression level and clinical significance of LINC00667 in CCA. a The prediction of LINC00667 expression in CCA by TCGA database. ***P < 0.001. b Relative expression of LINC00667 in CCA tissues (n = 62) and matched adjacent non-tumor tissues (n = 62). ***P < 0.001. c RT-qPCR analysis of the expression level of LINC00667 in CCA cell lines. Error bars represent the mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001. d Kaplan–Meier curve for overall survival in patients with CCA according to LINC00667 expression (patients grouping according to median expression of LINC00667). Log rank P < 0.001. e ROC curve for prognostic evaluation ability of LINC00667 in predicting prognosis of CCA patients. AUC: 0.830 (95% CI: 0.748–0.911), sensitivity: 87.5%, specificity: 72%. ***P < 0.001
LINC00667 promoted CCA cell proliferation in vitro and in vivo
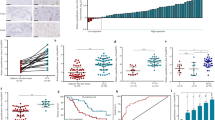
To explore the biological functions of LINC00667 on CCA development, loss-of-function and gain-of-function experiments on CCLP-1 and QBC939 cells were performed by transfection with si-LINC00667 and pcDNA3.1/LINC00667. We found that LINC00667 expression was significantly reduced in CCLP-1 cells after transfection with si-LINC00667-1 and si-LINC00667-2, and its expression was significantly up-regulated in QBC939 cells after transfection with pcDNA3.1/LINC00667 (Fig. 2a). Then, CCK-8 assay revealed that knockdown of LINC00667 suppressed the proliferation of CCLP-1 cells, and up-regulation of LINC00667 promoted QBC939 cell proliferation (Fig. 2b). The proliferative ability of CCA cells was also determined by EdU assays. The results uncovered that knockdown of LINC00667 inhibited CCA cell proliferation, while overexpression of LINC00667 remarkably promoted the proliferation of CCA cells (Fig. 2c). Moreover, the effects of LINC00667 on the clonogenic capabilities of CCA cells were determined by colony formation assays. The results demonstrated that the numbers of cell colonies were markedly decreased after silencing LINC00667, while the numbers of cell colonies were increased after LINC00667 overexpression (Fig. 2d). In addition, we examined the effects of LINC00667 on the proliferation of CCA cells in vivo. As shown in Fig. 2e–g, both volumes and weights of xenograft tumors were significantly restrained in the LINC00667 knockdown group compared with those in the negative control group. Furthermore, the protein expression level of EMT-related markers (E-cadherin, N-cadherin and vimentin) was detected in xenograft tumors by western blot. The results showed that E-cadherin expression was up-regulated after silencing LINC00667, while the expression of N-cadherin and vimentin was down-regulated by knocking down LINC00667 compared with the sh-NC group, respectively (Fig. 2h). These data suggest that LINC00667 as an oncogene promotes CCA proliferation and invasion.
LINC00667 promoted CCA cell proliferation in vitro and in vivo. a LINC00667 expression with knockdown or overexpression was confirmed by RT-qPCR. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. b CCK8 assay was performed to measure the effect of LINC00667 on CCA cell proliferation. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. c Cell proliferation was further examined in si-LINC00667 and oe-LINC00667 cells by EdU assay. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. d The effect of LINC00667 on colony forming ability was detected by colony formation assay in CCLP-1 and QBC939 cells after transfection. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. e CCLP-1 cells transfected with sh-LINC00667 or sh-NC were subcutaneously injected into the posterior flanks of the mice (n = 6 per group), and the xenograft tumors transfected with sh-LINC00667 were significantly inhibited in comparison with the tumors transfected with sh-NC after 21 days. f The tumor volumes were calculated every 3 days. ***P < 0.001. g The tumor weights were measured after excision. ***P < 0.001. h The effect of LINC00667 on EMT process was detected in xenograft tumors by western blot assays. *P < 0.05, **P < 0.01
LINC00667 promoted CCA cell migration, invasion and EMT process
Next, we explored the role of LINC00667 in the migration and invasion of CCA cells. As shown in Fig. 3a, wound healing assay indicated that the migratory ability of CCLP-1 cells was attenuated by LINC00667 knockdown compared with the si-NC group, while the migratory ability of QBC939 cells was enhanced by LINC00667 overexpression compared with the oe-NC group. In addition, transwell migration and invasion assays demonstrated that knockdown of LINC00667 reduced the numbers of migratory and invasive cells compared with the si-NC group, whereas the numbers of migratory and invasive cells were increased by overexpressing LINC00667 compared with the oe-NC group (Fig. 3b, c). To explore the effect of LINC00667 on EMT, we detected the expression of E-cadherin, N-cadherin, and vimentin in CCA cells with LINC00667 knockdown or overexpression. The results showed that E-cadherin expression was down-regulated by overexpressing LINC00667 but up-regulated by silencing LINC00667 in CCA cells. Conversely, the expression of N-cadherin and vimentin was up-regulated in oe-LINC00667 cells but down-regulated in si-LINC00667 cells (Fig. 3d). These findings suggest that LINC00667 as a cancer-promoting lncRNA contributes to cell migration, invasion and EMT process in CCA.
LINC00667 promoted CCA cell migration, invasion and EMT process. a Wound healing migration assay detected the effects of silencing and overexpressing LINC00667 on cell migration. Error bars represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01. b Transwell without matrigel assay showed that LINC00667 knockdown inhibited cell migration, but LINC00667 overexpression promoted cell migration. Error bars represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01. c The effect of LINC00667 on cell invasion was detected by transwell with matrigel assay. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. d Western blot analysis of the EMT-related markers including E-cadherin, N-cadherin and vimentin in si-LINC00667 and oe-LINC00667 cells, respectively. Error bars represent the mean ± SD of three independent experiments. **P < 0.01
LINC00667 was transcriptionally activated by YY1
Mounting evidence confirmed that transcription factors played vital roles in the modulation of lncRNA expression [17]. Hence, for further finding the regulator of LINC00667, we first performed bioinformatics analysis using JASPAR database (https://jaspar.genereg.net/). As shown in Fig. 4a, transcription factor YY1 was predicted to potentially bind to LINC00667 promoter including three binding sites (E1, E2 and E3). We next successfully synthesized YY1 siRNAs and pcDNA3.1/YY1 overexpressing vector (Fig. 4b). The data demonstrated that knockdown of YY1 remarkably reduced LINC00667 expression, while overexpression of YY1 markedly increased LINC00667 expression in CCA cells (Fig. 4c). The binding relation between YY1 and LINC00667 promoter was verified by ChIP assay and luciferase reporter assay. ChIP assay demonstrated that the binding site E1 of LINC00667 promoter was responsive to YY1-induced transcription, while no evident changes were found in the binding sites E2 and E3 (Fig. 4d). Furthermore, luciferase reporter assay confirmed that overexpression of YY1 enhanced luciferase activities of CCA cells transfected with binding site E1 wild type, but not binding site E1 mutant type (Fig. 4e). These data suggest that YY1 functions as a transcription regulator of LINC00667 expression by directly binding to LINC00667 promoter.
LINC00667 was transcriptionally activated by YY1. a YY1 binding motif and the prediction of YY1 binding sites in the promoter region of LINC00667 provided by the JASPAR database. b The knockdown and amplification efficiencies of YY1 in CCA cells were determined by RT-qPCR and western blot. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. c RT-qPCR analysis of LINC00667 expression in CCA cells after transfection with si-YY1 and pcDNA3.1/YY1. Error bars represent the mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001. d RT-qPCR analysis of the ChIP products validated the binding capacity of YY1 to the LINC00667 promoter. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. e Luciferase reporter assay was further performed to confirm the direct binding of YY1 to LINC00667 promoter. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001
LINC00667 was involved in CCA progression by sponging miR-200c-3p
To further investigate the mechanism by which LINC00667 promoted the proliferation and invasion of CCA cells, we analyzed LINC00667 localization in CCA cells by subcellular fractionation assay. The results confirmed that LINC00667 was widely distributed in cytoplasm (Fig. 5a), suggesting LINC00667-induced mechanism mainly at post-transcriptional level. It is known that lncRNAs can function as a ceRNA to indirectly enhance protein expression by competing for shared miRNAs. Thus, we predicted the potential miRNA targets of LINC00667 using the bioinformatics database StarBase v3.0 (https://starbase.sysu.edu.cn/). The miR-200c-3p was speculated as a target gene of LINC00667 in CCA on the basis of the predicted binding site between them.
LINC00667 contributed to CCA progression by sponging miR-200c-3p. a Relative LINC00667 expression in the cytoplasm and nuclei of CCLP-1 and QBC939 cells was determined by RT-qPCR. b The miR-200c-3p expression was measured in CCA cells with LINC00667 overexpression or knockdown. Error bars represent the mean ± SD of three independent experiments. **P < 0.01, ***P < 0.001. c Relative expression of miR-200c-3p in CCA tissues (n = 62) and matched adjacent non-tumor tissues (n = 62). ***P < 0.001. d Correlation between LINC00667 and miR-200c-3p expression in CCA specimens (n = 62) detected by RT-qPCR. r = − 0.5168, ***P < 0.001. e RT-qPCR analysis of the expression level of miR-200c-3p in CCA cell lines. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. f, g The miR-200c-3p expression with knockdown or overexpression was confirmed by RT-qPCR. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. h Luciferase activity in CCA cells was detected by co-transfection with miR-200c-3p mimics and luciferase reporter plasmids containing LINC00667 wild type or mutant type. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. i Ago2 RIP assay was performed in CCLP-1 and QBC939 cells. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. j–l LINC00667 siRNA and miR-200c-3p inhibitor were co-transfected into CCLP-1 cells, and cellular proliferation and invasion were detected by CCK8, EdU and transwell assays. Error bars represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01
As shown in Fig. 5b, knocking down LINC00667 was found to observably elevate miR-200c-3p expression in CCA cells, whereas overexpressing LINC00667 suppressed the expression of miR-200c-3p compared with controls. The expression of miR-200c-3p in both tumor tissues and paired adjacent tumor-free tissues was detected via RT-qPCR. The data revealed that miR-200c-3p expression was remarkably decreased in tumor tissues in comparison with that in paired adjacent tumor-free tissues (Fig. 5c). Moreover, miR-200c-3p expression displayed a negative correlation with the LINC00667 expression in CCA tissues (r = − 0.5168, P < 0.001, Fig. 5d). In addition, miR-200c-3p expression was significantly down-regulated in CCA cells compared with controls (Fig. 5e). To explore whether LINC00667 could interact with miR-200c-3p, we constructed the luciferase reporter plasmids containing wild-type or mutant binding sites of LINC00667. The knockdown and overexpression efficiencies of miR-200c-3p were showed in Fig. 5f, g. The results of luciferase reporter assays demonstrated that the luciferase activity in wild-type LINC00667 group was decreased after co-transfection with miR-200c-3p mimics, but the luciferase activity in mutant LINC00667 group was not changed (Fig. 5h). Subsequently, to validate whether LINC00667 could sponge miR-200c-3p, Ago2 RIP assay was further conducted. As shown in Fig. 5i, LINC00667 was more abundant in anti-Ago2 + miR-200c-3p mimics group. These results suggest that LINC00667 acts as a miR-200c-3p sponge in CCA cells.
Next, we intended to investigate whether LINC00667 promoted the proliferation and invasion of CCA cells through sponging miR‐200c-3p. The CCK-8 and EdU rescue assays showed that the inhibition of cell proliferation caused by si-LINC00667 was partly rescued by miR‐200c-3p inhibitor in CCLP-1 cells (Fig. 5j, k). LINC00667 knockdown obviously repressed CCA cell invasion confirmed by transwell assays; nevertheless, the inhibitory effect was attenuated by co-transfection with miR-200c-3p inhibitor (Fig. 5l). These data suggest that LINC00667 suppresses miR-200c-3p expression to facilitate the proliferation, migration and invasion of CCA.
LINC00667 up-regulated PDK1 by competitively binding miR-200c-3p to accelerate CCA development
The predicted binding site of miR-200c-3p at the 3′-UTR of PDK1 mRNA was identified using StarBase v3.0 database. Interestingly, PDK1 mRNA and LINC00667 shared the overlapped binding sequences of miR-200c-3p. Accordingly, we tried to verify the correlation between miR-200c-3p and PDK1. RT-qPCR and western blot assays showed that overexpression of miR-200c-3p suppressed the expression of PDK1 mRNA and protein, and inhibition of miR-200c-3p expression promoted the expression of PDK1 mRNA and protein (Fig. 6a). The expression of PDK1 mRNA in both tumor tissues and paired adjacent tumor-free tissues was detected via RT-qPCR. The data revealed that PDK1 mRNA expression was remarkably increased in tumor tissues in comparison with that in paired adjacent tumor-free tissues (Fig. 6b). Moreover, PDK1 mRNA expression displayed a negative correlation with the miR-200c-3p expression in CCA tissues (r = − 0.5074, P < 0.001, Fig. 6c). In addition, the expression of PDK1 was higher in CCA cells than normal HIBEC cells at both mRNA and protein levels (Fig. 6d). Luciferase reporter plasmids of PDK1 wild type and mutant type were constructed, and only co-transfection of the PDK1 wild-type plasmids and miR-200c-3p mimics significantly decreased the luciferase activity of CCA cells (Fig. 6e). Consistently, RIP assay displayed that PDK1 mRNA was significantly enriched by anti-Ago2 antibodies with miR-200c-3p mimics in CCA cells (Fig. 6f). These results suggest that PDK1 is a direct target of miR-200c-3p in CCA cells.
PDK1 was a direct target of miR-200c-3p in CCA. a The PDK1 expression was measured in CCA cells with miR-200c-3p overexpression or knockdown. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. b Relative expression of PDK1 mRNA in CCA tissues (n = 62) and matched adjacent non-tumor tissues (n = 62). ***P < 0.001. c Correlation between miR-200c-3p and PDK1 mRNA expression in CCA specimens (n = 62) detected by RT-qPCR. r = − 0.5074, ***P < 0.001. d Relative expression of PDK1 mRNA and protein in CCA cells. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. e Luciferase activity in CCA cells was detected by co-transfection with miR-200c-3p mimics and luciferase reporter plasmids containing PDK1 wild type or mutant type. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. f Ago2 RIP assay further verified the direct interaction between miR-200c-3p and PDK1. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001. g The knockdown efficiency of PDK1 in CCA cells were determined by RT-qPCR and western blot. Error bars represent the mean ± SD of three independent experiments. ***P < 0.001
To investigate whether LINC00667 promoted the proliferation and invasion of CCA cells via miR-200c-3p and PDK1, rescue assays were performed. PDK1 expression was significantly decreased by transfection with PDK1 siRNA in CCA cells (Fig. 6g). The restoration of miR-200c-3p by transfection with miR-200c-3p inhibitor reversed the reduced mRNA and protein levels of PDK1 induced by LINC00667 knockdown. On the contrary, the restoration of miR-200c-3p by transfection with miR-200c-3p mimics rescued the increased mRNA and protein levels of PDK1 induced by overexpressing LINC00667 (Fig. 7a). The promotive effects of cell proliferation and invasion induced by LINC00667 overexpression in CCK8, EdU and transwell assays were partly restored by co-transfection with si-PDK1 (Fig. 7b–d). Finally, we confirmed that down-expression of miR-200c-3p obviously promoted CCA cell proliferation and invasion; however, these promotive effects were attenuated by PDK1 knockdown (Fig. 7e–g). Taken together, our study confirmed that LINC00667 was a pivotal cancer-promoting lncRNA in CCA, and YY1/LINC00667/miR-200c-3p/PDK1 pathway exhibited crucial functions in CCA development.
LINC00667 up-regulated PDK1 by competitively binding miR-200c-3p to accelerate CCA development. a Relative expression level of PDK1 was assessed by RT-qPCR and western blot after co-transfection with LINC00667 siRNA and miR-200c-3p inhibitor, or LINC00667-overexpressing plasmid and miR-200c-3p mimics. Error bars represent the mean ± SD of three independent experiments. **P < 0.01. b–d CCK8, EdU and transwell assays indicated that the promotive effects of proliferation and invasion induced by LINC00667 overexpression were partly reversed by silencing PDK1. Error bars represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01. e–g The promotive effects of proliferation and invasion induced by miR-200c-3p inhibitor were partly reversed by silencing PDK1 in CCK8, EdU and transwell assays. Error bars represent the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01
Discussion
The crucial roles of lncRNAs in the occurrence and development of tumors have become a focus of cancer basic research. Numerous lncRNAs have been identified as oncogenes or tumor-suppressive genes involved in the development of cancers [18]. Shen et al. [19] confirmed that lncRNA KCNQ1OT1 was up-regulated in osteosarcoma and promoted osteosarcoma growth. Wu et al. [20] testified that lncRNA MEG3 served as a tumor-suppressive gene in prostate cancer and inhibited the progression of prostate cancer. Interestingly, we observed that LINC00667 was highly expressed in CCA tissues according to TCGA database analysis. Moreover, LINC00667 has been confirmed as a cancer-promoting lncRNA in various cancers including glioma, non-small cell lung cancer, and Wilms’ tumor. Therefore, we decided to investigate whether LINC00667 played a biological role in CCA.
We explored the clinical significance of LINC00667 in patients with CCA, and found that its overexpression was correlated with lymph nodes metastasis, advanced TNM stage, and poor 5-year overall survival. To further determine the potential of LINC00667 as a valuable biomarker for patients with CCA, we performed univariate and multivariate analyses. And our results demonstrated that increased LINC00667 expression was an independent poor prognostic marker of overall survival of CCA patients. In addition, the ROC analysis indicated that the expression detection of LINC00667 had an acceptable prognostic efficacy in predicting the overall survival of CCA patients. LncRNAs were involved in tumor progression by regulating cellular biological behaviors, such as proliferation, apoptosis, invasion, stemness maintenance, and chemotherapy resistance [21]. Shen et al. [22] confirmed that lncRNA AGAP2-AS1 accelerated esophageal cancer development by restraining cellular apoptosis and promoting tumor cell proliferation, migration and invasion. In our study, CCK-8, EdU, and colony formation assays were conducted to detect CCA cell viability, and wound healing and transwell assays were performed to measure cellular mobility. The data demonstrated that knockdown of LINC00667 inhibited cell proliferation, migration and invasion, whereas overexpression of LINC00667 promoted cell proliferation, migration and invasion. Moreover, tumor xenograft assay confirmed that knockdown of LINC00667 inhibited tumor growth in vivo. In addition, the effect of LINC00667 on the EMT process was detected by western blot assays. The results demonstrated that LINC00667 promoted the EMT process of CCA by suppressing epithelial marker E-cadherin expression and promoting the expression of mesenchymal marker N-cadherin and vimentin.
It has been previously demonstrated that transcription factors like STAT1, AR, and SP1 may act as positive regulators involved in the transcription of downstream factors including lncRNAs [23,24,25]. In this study, to explore the mechanism involved in the dysregulation of LINC00667, we used JASPAR online database to predict the possible transcription factors. The data found that transcription factor YY1 bound to the LINC00667 promoter region with high scores. YY1 has been confirmed to be involved in various tumor progression and associated with poor clinical outcome of tumor patients [26]. For example, YY1 activated LINC00673 transcription to promote the growth of breast cancer [27]. In our study, we confirmed that knockdown of YY1 markedly inhibited LINC00667 expression, whereas overexpression of YY1 promoted the expression of LINC00667. Moreover, we performed ChIP and luciferase reporter assays, and these assays demonstrated the direct binding between YY1 and LINC00667 promoter region. To sum up, these results revealed that the up-regulation of LINC00667 in CCA was partly induced by YY1.
It has been confirmed that lncRNAs can act as endogenous molecular sponges to regulate target mRNAs expression by competitively binding to their shared binding sites on miRNAs [28]. By bioinformatics analysis, we found that LINC00667 and PDK1 had the overlapped binding site on miR-200c-3p, suggesting the possible ceRNA relation between them. RT-qPCR assays confirmed that LINC00667 and PDK1 were significantly up-regulated in CCA tissues and cells, while miR-200c-3p was down-expressed in CCA tissues and cells compared with controls. Pearson correlation analysis confirmed that the expression of LINC00667 and miR-200c-3p in CCA tissues possessed inverse correlation, and PDK1 expression was also negatively related to miR-200c-3p expression in CCA tissues. Furthermore, the loss-of-function and gain-of-function experiments demonstrated that LINC00667 inhibited miR-200c-3p expression, and miR-200c-3p inhibited PDK1 expression in CCA cells. More importantly, LINC00667 overexpression promoted PDK1 expression, which was partly rescued by overexpressing miR-200c-3p. The luciferase reporter and Ago2 RIP assays further confirmed the direct binding abilities of LINC00667 to miR-200c-3p and miR-200c-3p to PDK1 mRNA. Finally, the rescue assays indicated that LINC00667 promoted the proliferation and invasion of CCA cells by regulating miR-200c-3p/PDK1.
In conclusion, we first reported that LINC00667 exerted an oncogenic function in CCA. LINC00667 promoted the proliferation, migration, invasion and EMT of CCA cells by the regulation axis of YY1/LINC00667/miR-200c-3p/PDK1. In addition, LINC00667 overexpression represented a poor prognosis of CCA patients. Consequently, LINC00667 is expected to be a valuable target for diagnosis and therapy of CCA.
References
Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383(9935):2168–79. https://doi.org/10.1016/S0140-6736(13)61903-0.
Hoyos S, Navas MC, Restrepo JC, Botero RC. Current controversies in cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864(4 Pt B):1461–7. https://doi.org/10.1016/j.bbadis.2017.07.027.
Wang L, Cao L, Wen C, Li J, Yu G, Liu C. LncRNA LINC00857 regulates lung adenocarcinoma progression, apoptosis and glycolysis by targeting miR-1179/SPAG5 axis. Hum Cell. 2020;33(1):195–204. https://doi.org/10.1007/s13577-019-00296-8.
Molist C, Navarro N, Giralt I, et al. miRNA-7 and miRNA-324-5p regulate alpha9-Integrin expression and exert anti-oncogenic effects in rhabdomyosarcoma. Cancer Lett. 2020;477:49–59. https://doi.org/10.1016/j.canlet.2020.02.035.
Li J, Huang L, Li Z, et al. Functions and roles of long noncoding RNA in cholangiocarcinoma. J Cell Physiol. 2019;234(10):17113–26. https://doi.org/10.1002/jcp.28470.
Ma Q, Qi X, Lin X, Li L, Chen L, Hu W. LncRNA SNHG3 promotes cell proliferation and invasion through the miR-384/hepatoma-derived growth factor axis in breast cancer. Hum Cell. 2020;33(1):232–42. https://doi.org/10.1007/s13577-019-00287-9.
Chen W, Zhou ZQ, Ren YQ, et al. Effects of long non-coding RNA LINC00667 on renal tubular epithelial cell proliferation, apoptosis and renal fibrosis via the miR-19b-3p/LINC00667/CTGF signaling pathway in chronic renal failure. Cell Signal. 2019;54:102–14. https://doi.org/10.1016/j.cellsig.2018.10.016.
Wang D, Zheng J, Liu X, et al. Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212-3p and linc00667/miR-429 axis. Mol Ther Nucleic Acids. 2019;14:465–82. https://doi.org/10.1016/j.omtn.2018.12.017.
Yang Y, Li S, Cao J, Li Y, Hu H, Wu Z. RRM2 regulated by LINC00667/miR-143-3p signal is responsible for non-small cell lung cancer cell progression. Onco Targets Ther. 2019;12:9927–39. https://doi.org/10.2147/OTT.S221339.
Liu P, Chen S, Huang Y, et al. LINC00667 promotes Wilms’ tumor metastasis and stemness by sponging miR-200b/c/429 family to regulate IKK-β. Cell Biol Int. 2020;44(6):1382–93. https://doi.org/10.1002/cbin.11334.
Jiang Y, Yang Y, Wang H, Darko GM, Sun D, Gao Y. Identification of miR-200c-3p as a major regulator of SaoS2 cells activation induced by fluoride. Chemosphere. 2018;199:694–701. https://doi.org/10.1016/j.chemosphere.2018.01.095.
Maolakuerban N, Azhati B, Tusong H, Abula A, Yasheng A, Xireyazidan A. MiR-200c-3p inhibits cell migration and invasion of clear cell renal cell carcinoma via regulating SLC6A1. Cancer Biol Ther. 2018;19(4):282–91. https://doi.org/10.1080/15384047.2017.1394551.
Danarto R, Astuti I, Umbas R, Haryana SM. Urine miR-21-5p and miR-200c-3p as potential non-invasive biomarkers in patients with prostate cancer. Turk J Urol. 2019;46(1):26–30. https://doi.org/10.5152/tud.2019.19163.
Dupuy F, Tabariès S, Andrzejewski S, et al. PDK1-dependent metabolic reprogramming dictates metastatic potential in breast cancer. Cell Metab. 2015;22(4):577–89. https://doi.org/10.1016/j.cmet.2015.08.007.
Siu MKY, Jiang YX, Wang JJ, et al. PDK1 promotes ovarian cancer metastasis by modulating tumor-mesothelial adhesion, invasion, and angiogenesis via α5β1 integrin and JNK/IL-8 signaling. Oncogenesis. 2020;9(2):24. https://doi.org/10.1038/s41389-020-0209-0.
Hsu JY, Chang JY, Chang KY, Chang WC, Chen BK. Epidermal growth factor-induced pyruvate dehydrogenase kinase 1 expression enhances head and neck squamous cell carcinoma metastasis via up-regulation of fibronectin. FASEB J. 2017;31(10):4265–76. https://doi.org/10.1096/fj.201700156R.
Chen X, Zeng K, Xu M, et al. SP1-induced lncRNA-ZFAS1 contributes to colorectal cancer progression via the miR-150-5p/VEGFA axis. Cell Death Dis. 2018;9(10):982. https://doi.org/10.1038/s41419-018-0962-6.
Li J, Jiang X, Li C, et al. LncRNA-MEG3 inhibits cell proliferation and invasion by modulating Bmi1/RNF2 in cholangiocarcinoma. J Cell Physiol. 2019;234(12):22947–59. https://doi.org/10.1002/jcp.28856.
Shen Y, Xu J, Pan X, et al. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. 2020;11(4):278. https://doi.org/10.1038/s41419-020-2485-1.
Wu M, Huang Y, Chen T, et al. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 2019;23(1):29–38. https://doi.org/10.1111/jcmm.13658.
Bai J, Xu J, Zhao J, Zhang R. LncRNA NBR2 suppresses migration and invasion of colorectal cancer cells by downregulating miRNA-21. Hum Cell. 2020;33(1):98–103. https://doi.org/10.1007/s13577-019-00265-1.
Shen S, Li K, Liu Y, et al. Silencing lncRNA AGAP2-AS1 upregulates miR-195-5p to repress migration and invasion of EC cells via the decrease of FOSL1 expression. Mol Ther Nucleic Acids. 2020;20:331–44. https://doi.org/10.1016/j.omtn.2019.12.036.
Shen Y, Gao X, Tan W, Xu T. STAT1-mediated upregulation of lncRNA LINC00174 functions a ceRNA for miR-1910-3p to facilitate colorectal carcinoma progression through regulation of TAZ. Gene. 2018;666:64–71. https://doi.org/10.1016/j.gene.2018.05.001.
Yang F, Shen Y, Zhang W, et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018;25(12):2209–20. https://doi.org/10.1038/s41418-018-0123-6.
Li C, Liu H, Yang J, et al. Long noncoding RNA LINC00511 induced by SP1 accelerates the glioma progression through targeting miR-124-3p/CCND2 axis. J Cell Mol Med. 2019;23(6):4386–94. https://doi.org/10.1111/jcmm.14331.
Tang W, Zhou W, Xiang L, et al. The p300/YY1/miR-500a-5p/HDAC2 signalling axis regulates cell proliferation in human colorectal cancer. Nat Commun. 2019;10(1):663. https://doi.org/10.1038/s41467-018-08225-3.
Qiao K, Ning S, Wan L, et al. LINC00673 is activated by YY1 and promotes the proliferation of breast cancer cells via the miR-515-5p/MARK4/Hippo signaling pathway. J Exp Clin Cancer Res. 2019;38(1):418. https://doi.org/10.1186/s13046-019-1421-7.
Li P, Zhou B, Lv Y, Qian Q. LncRNA HEIH regulates cell proliferation and apoptosis through miR-4458/SOCS1 axis in triple-negative breast cancer. Hum Cell. 2019;32(4):522–8. https://doi.org/10.1007/s13577-019-00273-1.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81602088).
Funding
This work was supported by the National Natural Science Foundation of China (No. 81602088).
Author information
Authors and Affiliations
Contributions
JL: conceptualization; JL and XJ: experimental design and execution; ZH and LL: data curation and analysis; YC: experimental resources; CG and ZS: writing-original draft; PK: writing-review and editing; YC: supervision. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of The 2nd Affiliated Hospital of Harbin Medical University (KY2018-301), and patient consent was acquired prior to the initiation of experiment. All procedures performed in studies involving animals were in accordance with the ethical standards of the Animal Care and Use Committee of The 2nd Affiliated Hospital of Harbin Medical University (SYDW2018-114).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, J., Guan, C., Hu, Z. et al. Yin Yang 1-induced LINC00667 up-regulates pyruvate dehydrogenase kinase 1 to promote proliferation, migration and invasion of cholangiocarcinoma cells by sponging miR-200c-3p. Human Cell 34, 187–200 (2021). https://doi.org/10.1007/s13577-020-00448-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-020-00448-1