Abstract
Popcorn (Zea mays L.) is a special type of maize that pops up when heated leading to high flake volume. It is being used as a popular fiber-rich and nutritious snack all over the world. The genetic control of the popping rate and flake volume is not well understood. The cross made between high popping volume inbred line as the female parent and low popping volume composite as the male parent. The F2 population of 504 plants showed continuous variation for popping volume signifying that the popping volume is a quantitative trait governed by multiple genes. Only a set of 126 out of 313 maize SSR markers were successfully amplified using the standard PCR reaction and 66 among these were found polymorphic between the two parents. Bulk segregant analysis (BSA) of F2 plants based on F2:3 seeds were carried out for mapping popping volume QTL using the 66 polymorphic SSR markers. Only four markers showed an association with popping volume in BSA. Out of the four, three SSR markers (bnlg1331, bnlg1520, bnlg1144) showed high association with the popping volume data after de-bulking of the positive and negative plants used in the bulks. All the 504 F2 plants were genotyped using 3 SSR markers and the F2:3 seeds were phenotyped for popping volume. The single marker analysis of the F2 plants showed that the 3 SSR markers bnlg1331, bnlg1520, and bnlg1836 on chromosome 1, 2, and 5, respectively, were closely associated with the QTL for popping volume covering 78% of total phenotypic variance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maize (Zea mays L.) is one of the important staple crops worldwide as well as in India (Kumar 2016). The most common types of maize are flint, dent, floury, sugary, popcorn, waxy, and multicolored. The kernel types are determined by their physical appearance, the pattern of the endosperm and the amount and quality of the endosperm. Among the different maize types, popcorn is the most popular type of corn snack (Swapna 2017). Maize considered among the top three staple cereal being consumed by the majority of the human population plays a significant role in human nutrition worldwide (Danson et al. 2006; Muthusamy et al. 2014). Maize is being grown in different geographical areas covering 193.7 million hectares globally (Tripathi et al. 2016), with an average production of 1147.62 MT. USA, China, Brazil, Argentina, Mexico, and India are the major producers of maize in which India stands on 6th rank with the production of 27.82 MT (FAOSTAT, 2020; http://www.fao.org/faostat/en/#data/QC). The popcorn plants of most of the cultivars are shorter, especially of inbred lines and open-pollinated varieties and weaker than that of the dent maize. Some popcorn hybrids in tropical environments can be as tall as or taller than those of the dent maize hybrids (Ziegler 2001).
The popped grains of popcorn are the most popular, nutritious, and fiber-rich snack in all parts of the world and are consumed by every age group (Ertas et al. 2009; Karababa 2006). The popcorn production in India is 25–30 q/ha (IASRI 2020) and the global popcorn market was valued at $9060 million in 2016 and is projected to reach $15,098 million by 2023, registering a CAGR of 7.6% from 2017 to 2023 (https://www.alliedmarketresearch.com/). This popular snack production is of ₤785.7 million (AgMRC assessed in 09/03/2020; https://www.agmrc.org/) and its production is increasing steadily in the countries like India, Brazil, and Turkey due to its economic value (Ahmet and Kapar 2011; Vieira et al. 2009; Vijayabharathi et al. 2009). The popcorn kernels are characterized by a high proportion of hard endosperm. Many physical properties of popcorn have been studied including kernel length, width, thickness, diameter, sphericity, volume, surface area and thousand seed weight (Karababa 2006). The popcorn inbred lines that have dent allele may exhibit increased kernel weight, grain yield, reduced stem, and root lodging, and also reduced popping expansion volume (PEV) (Ren et al. 2018). The popping traits associated with PEV are (1) the number of unpopped kernels and (2) the grain moisture content at the time of popping (Ahmet and Kapar 2011; Jele 2012; Mishra et al. 2014; Soylu and Tekkanat 2007). The popping ability of popcorn also depends on the methods used for popping (Dofing et al. 1991). Development of genotypes having high grain yield with high popping volume is an important feature of popcorn breeding (Dhliwayo 2008; Effa et al. 2011; Freitas Júnior et al. 2009; Miranda et al. 2003; Rinaldi et al. 2007). The aim of the breeding programs besides maximizing yielding potential is to improve the popping quality of popped kernels (Gökmen 2004; Soylu and Tekkanat 2007; Srdic and Pajic 2011).
The QTL mapping is one of the approaches to identify loci underlying genetic variation in PEV and other desired traits. The QTL for high PEV are located on different chromosomes (Amaral Junior et al. 2016). Four to six QTL were detected in different studies and several reports have shown that at least one QTL for high PEV was located on chromosome 1 of maize (Babu et al. 2006; Li et al. 2007b, c; Lu et al. 2003). Bulked segregant analysis (BSA) has been considered as a simplified approach for the identification of QTL (Giovannoni et al. 1991; Michelmore et al. 1991). The BSA has been used to identify and map important QTL for different traits in many crops (Trick et al. 2012; Zou et al. 2016). The present study was carried out to map the QTL for popping volume in an F2 mapping population developed from a cross of commercial popping corn inbred line and popcorn composite for use in molecular marker-assisted selection.
Materials and methods
Plant material
The mapping population was developed from a cross between a HPV inbred line introduced from the USA and commercially used in India, as the female parent and a LPV composite (Bajaura Popcorn developed by HPKV, Palampur) as the male parent. The parental lines were maintained through selfing as well as intermating in isolation to avoid contamination. The HPV × LPV cross was made at the experimental farm area, Eternal University, Baru Sahib (H.P.) during Kharif season 2015. A total of 95 F1 hybrid plants along with the parents were selfed and harvested. Only 10 F1:2 cobs were bulked to raise 800 F2 plants. A mapping population of 504 F2 plants was used for genotyping and phenotyping for mapping the QTL for popping volume.
Screening for popping volume
To determine the popping volume of seeds, a commercial popcorn machine (SHAKTI/Model No.VBG-803) was used. The machine was standardized by the use of 15 seeds of HPV at 10–14% moisture content giving around 100 ml popping volume at 220–260 °C. A sample of 15 seeds from each F2:3 cobs was popped and the volume (ml) of the popped seeds was measured in a 250 ml measuring cylinder (Brunson 1937).
Molecular analysis
The genomic DNA from parents and each of the F2 plants was isolated by using the CTAB method (Murray and Thompson 1980) with some modifications. The extracted DNA concentration was visualized on a 1% agarose gel for quality assessment, and quantification of DNA was found to be 20–70 ng/µl with A(260/280) = 1.8 ± 0.5 absorbance. The 20 µl volume PCR amplification reaction mixture contained 50 ng DNA, 2 µl 10× reaction buffer, 1.5 mM MgCl2, 0.75 U Taq polymerase (Promega, WI, USA), 0.2 mM dNTPs, and 0.2 µM of each primer. The protocol of the PCR cycle included an initial denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 55 °C or 58 °C (depending on the SSR primers) for 1 min, and extension at 72 °C for 2 min, and a 10 min final extension at 72 °C. Amplification products were resolved by gel electrophoresis on 2.5–3.0% polyacrylamide gels at 150 V for 3 h. The amplicon band size was determined using “1 kb plus DNA ladder” (Invitrogen Life Technology, Carlsbad, CA, USA). The PCR amplicon size, banding pattern, and length polymorphism were detected and recorded by using gel documentation system (G Box-Sygene, Germany).
Marker-phenotyping using BSA
The molecular mapping of popping volume was carried out using the mapped maize SSR markers. The bin map from MaizeGDB was used for the selection of SSR markers for the parental survey. A total of 313 SSR markers of maize were used representing all the 10 maize linkage groups (Supplementary Table 1). The genetic map of maize has been divided into 100 segments called bins, of approximately 20 cM between two fixed core markers (Gardiner et al. 1993). The SSR markers were selected in such a way that they could cover all the bins of the 10 maize chromosomes. The sequences of the SSR markers were adapted from the maize genome database (MizeGDB) (https://www.maizegdb.org/data_center/ssr). The molecular mapping work was carried out at the School of Agricultural Biotechnology, PAU, Ludhiana. BSA was used in conjunction with SSR analysis (Michelmore et al. 1991) to find markers linked to popping volume QTL. High and low popping volume bulks were prepared from F2 plants based on the popping volume of F2:3 seeds by pooling aliquots containing equivalent amounts of total DNA, approximately 50 ng/µl each from 10 high and 10 low popping volume F2 plants. The SSR molecular markers were then screened for polymorphic survey and linkage using the parents along with the high and low volume bulk DNA samples. The SSR molecular markers revealing polymorphism between parental genotypes as well as the pair of high and low popping volume bulks. The co-dominant SSR markers showing association with popping volume bulks were used for genotyping debulk and all the F2 plants for the identification of QTL for popping volume.
Single marker analysis for QTL mapping
The analysis of main QTL effects with Single-marker analysis (SMA) and recombination frequency was performed by using the method of Liu (1998). A significant association of a potential marker with a QTL for popping volume was detected by mapdisto v 2.1.1. (Lorieux 2012).
Results
The popping volume of HPV and LPV parental lines were 100 ml and 30 ml, respectively. The F1 seeds showed negligible popping (42 ml) indicating that the high popping volume was recessive. Since the HPV was used as the female parent, the low popping volume of the F1 hybrid seeds further indicated that the popping volume was not under the seed maternal genotypic control like many other quality traits but the endosperm genotype control due to metaxenia like many endosperm traits in maize.
Screening of F2 population for popping volume
The popping volumes of 15 dried seeds from each HPV and LPV were 100 ± 1.49 ml and 30 ± 2.00 ml, respectively with a very little season-to-season variation for popping volume within the parental cobs indicating that they were genetically uniform. A total of 95 F1 plants were selfed carefully and harvested cob-wise. A large variation was observed in F1:2 cobs (Table 1) for popping volume confirming that the popping volume is under endosperm genotypic control. The seeds of ten F1:2 cobs were bulked and space planted to raise about 800 F2 plants for genotypic and phenotypic characterization. Out of 800 F2 plants, 504 healthy plants could be selfed to develop F2:3 seeds for phenotyping for popping volume and to develop F3 lines. The popping volume data of 504 F2:3 seeds showed continuous normal distribution of popping volume ranging from 20 to 140 ml indicating it as a quantitative trait (Table 2, Fig. 1) controlled by multiple genes.
Bulk segregant analysis
Bulk segregation analysis was carried out to identify the SSR markers linked to the popping volume. Ten high popping volume F2 plants (Plant number 28, 63, 183, 234, 272, 304, 326, 333, 449, and 661) were selected to make the high volume bulk (HVB) and similarly, ten low popping volume plants (Plant number 34, 112, 121 144, 153, 171, 180, 200, 367, and 403) were selected as the low volume bulk (LVB) (Table 3). The equal amount of genomic DNA (50 µg) from the high and low popping volume plants was bulked to make HVB and LVB.
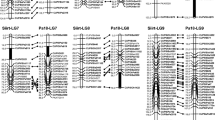
Only 126 out of 313 SSR markers were successfully amplified using the standard PCR reaction and 66/126 were found polymorphic. All the polymorphic SSR molecular markers were amplified among HPV, LPV, HVB, and LVB (Fig. 2). Out of 66 polymorphic SSR molecular markers, nine markers viz. umc1160, umc1169, bnlg1331, umc1184, bnlg1520, bnlg1144, bnlg1836, umc1632, and bnlg1583 present on chromosome number 1, 2, 3, 5, 7, and 9, respectively (Supplementary Table 2 and 3) showed association in the bulk segregant analysis.
Debulking of high and low volume bulks
The nine SSR markers showing association in the bulk segregant analysis were used to confirm their association in individual plants of high and low volume bulks. Out of nine, four SSR molecular markers confirmed the association with the popping volume data of these high and low volume F2 plants used in bulk segregant analysis. There was only one heterozygote for HVB and LVB plants during de-bulk analysis with SSR marker bnlg1331, none with bnlg1520, two in HVB with bnlg1836 indicating that these SSR markers are tightly linked with QTL for popping volume with 0–20% recombination. There were many heterozygotes in both debulks with SSR marker bnlg1144 mapped on chromosome 3 suggesting that it may not be tightly linked with the QTL for popping volume of maize (Fig. 3). The four SSR molecular markers found associated with the popping volume were genotyped in 504 F2 plants.
Single marker analysis of F2 plants using SSR markers
To confirm the marker-trait association, SMA was used to confirm the extent of association of the SSR markers with the QTL for popping volume and their contribution to the phenotypic variance. The popping volume was recorded in 504 F2:3 seeds of F2 cobs (Supplementary Table 2). All the 504 F2 individual plants were genotyped using four SSR markers which were found associated in BSA (Supplementary Fig. 1–4). These data were subjected to single marker analysis using the software mapdisto v2.1.1. These four SSR molecular markers bnlg1331, bnlg1520, bnlg1144, and bnlg1836 located on chromosome numbers 1, 2, 3, and 5 were confirmed to be associated with popping volume. Out of the four, two SSR molecular markers bnlg1331 (bin 1.09) and bnlg1836 (bin 5.01) were highly robust each covering 32% phenotypic variation for popping volume. The SSR molecular marker bnlg1520 (bin 2.09) showed only 14% phenotypic variation. The SSR marker bnlg1144 (bin 3.02) covered only 4% phenotypic variation (Table 4). The QTL mapping indicated that the major QTL for popping volume are present on chromosomes 1, 2, and 5 linked to the SSR molecular markers bnlg1331 (bin 1.09), bnlg1520 (bin 2.09) and bnlg1836 (bin 5.01), respectively. These three SSR markers on chromosome1, 2 and 5 contributing 78% of phenotypic variance (R2) for popping volume can be used for validation of popping volume QTL in subsequent generations and other populations. The QTL for popping volume mapped on chromosomes 1, 2, and 5 have been designated as qPVEU-1, qPVEU-2, and qPVEU-5, respectively.
Discussion
Popcorn is a widely preferred snack of maize worldwide. Popping characteristics of maize were originally selected by American Indians in early western civilizations (Carter et al. 1989; Sweley et al. 2013). However, the popcorn cultivars have lower yields as compared to the dent and flint corn hybrids and hence the improvement of popcorn cultivars has been undertaken by many researchers by using conventional breeding programs (Jele et al. 2014; Li et al. 2007a; Lu et al. 2003). High popping rate but the negligible popping volume of F1 hybrid seeds from a cross between HPV with high popping volume and popping rate as the female parent and LPV as the male parent indicated that the high popping volume is controlled by recessive endosperm genotype. The popping volume of F1:2 cobs ranging from 10 to 138 ml with a mean value of 54.12 ml confirms that the popping volume is under the control of the endosperm genotype and not the maternal genotype and the low popping volume is partially dominant. Teosinte, the wild progenitor of maize also had no popping volume and popping rate further confirming that the genetic control of high popping volume is under the spontaneously induced mutations which are usually recessive. The F1 hybrid seeds in a cross of popcorn A-1-6 and flint V273 showed partial dominance of low popping rate of V273 over the high popping rate of popcorn and no to partial dominance of high flake volume of popcorn (Babu et al. 2006). Crumbaker et al. (1949), crossed dent maize × popcorn lines and found that the low popping volume and dent-type kernels were partially dominant over high popping volume and flint-type kernels. Their data also suggested that two backcrosses to the popcorn recurrent parent were sufficient to recover popping volume equal to the popcorn parent. Both the additive and dominant genetic effects play a very important role in the inheritance of popping characteristics of maize (Dofing et al. 1991; Li et al. 2003).
Frequency distribution of popping volume of 15 seeds of each of the large number of F2:3 cobs of HPV × LPV population ranging from 20 to 140 ml was nearly normal indicating that the popping volume is a quantitative trait controlled by multiple genes with transgressive segregation further suggesting the presence of QTL for popping volume in both the parents. In several studies, the popping characteristic of popcorn maize was found quantitatively inherited and controlled by multiple genes (Babu et al. 2006; Li et al. 2007c, 2009; Lu et al. 2003; Ziegler 2001). Babu et al. (2006), in a mapping population of 194 F2 plants developed from a cross of popcorn (A-1-6) × flint corn V273 identified four QTL on chromosome 1, 3, 8, and 10 for high flake volume covering 62% of the phenotypic variance. Li et al. (2007c) in 259 F2 plants from Dan232 (dent corn) × N04 (popcorn) cross also identified six QTL for popping volume, three on chromosome 1 and one each on chromosome 6, 7, and 8 with 54% of phenotypic variance.
During BSA for popping volume QTL, ten high popping volume (> 100 ml) F2 plants were used as the positive bulk and ten low popping volume (< 30 ml) F2 plants as the negative bulk. Four SSR markers (bnlg1331, bnlg1520, bnlg1144, and bnlg1836) on chromosome 1, 2, 3, and 5, respectively were found to be linked to high popping volume bulk. Screening of debulks using these SSR markers, bnlg1331, bnlg1520, and bnlg1836 showed only 0–20% recombination from the QTL for popping volume while contributing 32, 14, and 32% phenotypic variance, respectively. This is the first report on the use of BSA for the identification and mapping of markers linked to QTL for popping volume. The SSR markers linked to the QTL for popping volume have been validated and used for the development of popcorn lines with high popping volume and resistance to southern corn leaf blight and sheath blight diseases of maize. The BSA is mostly used for preliminary identification of molecular markers linked to major genes controlling a trait. During the use of BSA for mapping molecular markers linked to a quantitative trait, only the QTL with the additive gene action can be identified more effectively. As expected all the three QTL for popping volume had additive gene action with the positive alleles for popping volume contributed by HPV. Four QTL have also been identified by Lu et al. (2003) on chromosomes 1, 3, 5S, and 5L for popping expansion volume, which together explained 45% of the phenotypic variation for popping expansion volume in a popcorn × dent corn cross. Jain-Poster and Woodford-Thomas (2015) identified QTL for popping expansion volume in a popcorn × dent corn cross covering 49.8% of the total phenotypic variance on chromosomes 1 and 3. The region on chromosome 1 appears to have a robust QTL for popping volume which may be a common QTL identified in various mapping populations (Da Silva et al. 1993; Hoseney et al. 1983; Matz 1984) including the one identified here. A QTL reported on chromosome 3, also identified here linked with bnlg1144 SSR marker, had only 4% coverage of phenotypic variance. There were only two polymorphic markers for chromosome 3. The use of more polymorphic markers of the bin may also help to identify the marker closely linked to the QTL for its use in MAS. The QTL for popping volume mapped on chromosomes 1, 2, and 5 identified in this investigation have been designated as qPVEU-1, qPVEU-2, and qPVEU-5, respectively, explaining 78% of the total phenotypic variance. The SSR markers linked to the QTL for popping volume have been validated and could be used for the development of popcorn lines with high popping volume and resistance to southern corn leaf blight (SCLB) and banded leaf sheath blight (BLSB) introgressed from teosinte (Zea mexicana L.).
Conclusion
The frequency distribution of the popping volume of 504 F2:3 cobs ranging from 20 to 140 ml was nearly normal suggesting that the popping volume is a quantitative trait controlled by multiple genes with additive effect indicating the presence of genes for popping volume in both the parents. Only four SSR markers bnlg1331, bnlg1520, bnlg1144, and bnlg1836 on chromosome numbers 1, 2, 3, and 5, respectively were found to be linked to popping volume or flake volume and popping rate. Screening of de-bulk using 4 SSR markers, 3 markers showed 0–20% recombination with the QTL for popping volume contributing 32, 14, and 32% phenotypic variance, respectively. The three QTL for popping volume mapped on chromosomes 1, 2 and 5 contributing 78% of the total phenotypic variance have been designated as qPVEU-1, qPVEU-2 and qPVEU-5, respectively. The three QTL qPVEU-1, qPVEU-2 and qPVEU-5 could be used in maker assistant selection for high popping volume breeding in maize. We have also used and validated these markers to select SCLB and BLSB resistant lines with high popping volume.
Abbreviations
- BSA:
-
Bulk segregant analysis
- SMA:
-
Single marker analysis
- HPV:
-
High popping volume
- LPV:
-
Low popping volume
References
Ahmet Ö, Kapar H (2011) Determination of grain yield, some yield and quality traits of promising hybrid popcorn genotypes. Turk J Field Crops 16(2):233–238
Amaral Junior AT, Freitas ILJ, Guimarães AG, Maldonado C, Arriagada O, Mora F (2016) Bayesian analysis of quantitative traits in popcorn (Zea mays L.) through four cycles of recurrent selection. Plant Prod Sci 19(4):574–578
Babu R, Nair S, Kumar A, Rao H, Verma P, Gahalain A, Singh I, Gupta HS (2006) Mapping QTLs for popping ability in a popcorn × flint corn cross. Theor Appl Genet 112(7):1392–1399
Brunson A (1937) Popcorn breeding (USDA Yearbook). US Government Printing Office, Washington, DC
Carter P, Hicks D, Doll J, Schulte E, Schuler R, Holmes B (1989) Popcorn. Alternative field crops manual. University of Wisconsin-Extension, Madison
Crumbaker DE, Johnson I, Eldredge JC (1949) Inheritance of popping volume and associated characters in crosses between popcorn and dent corn 1. Agron J 41(5):207–212
Da Silva W, Vidal B, Martins M, Vargas H, Pereira C, Zerbetto M, Miranda LC (1993) What makes popcorn pop. Nature 362(6419):417
Danson JW, Mbogori M, Kimani M, Lagat M, Kuria A, Diallo A (2006) Marker assisted introgression of opaque2 gene into herbicide resistant elite maize inbred lines. Afr J Biotechnol 5(24):2417–2422
Dhliwayo T (2008) Genetic mapping and analysis of traits related to improvement of popcorn. Ph D thesis, Iowa State university, Ames, Iowa
Dofing S, ĎCroz-Mason N, Thomas-Compton M (1991) Inheritance of expansion volume and yield in two popcorn × dent corn crosses. Crop Sci 31(3):715–718
Effa E, Uwah D, Ukeh D (2011) Yield response of popcorn (Zea mays L. var. everta) to nitrogen and lime amendment in a South Eastern rainforest environment of Nigeria. Am J Plant Physiol 6(6):304–311
Ertas N, Soylu S, Bilgicli N (2009) Effects of kernel properties and popping methods on popcorn quality of different corn cultivars. J Food Process Eng 32(4):478–496
Freitas Junior SP, Amaral Junior AT, Rangel RM, Viana AP (2009) Genetic gains in popcorn by full-sib recurrent selection. Crop Breed Appl Biotechnol 9(1):1–7
Gardiner J, Coe E, Melia-Hancock S, Hoisington D, Chao S (1993) Development of a core RFLP map in maize using an immortalized F2 population. Genetics 134(3):917–930
Giovannoni JJ, Wing RA, Ganal MW, Tanksley SD (1991) Isolation of molecular markers from specific chromosomal intervals using DNA pools from existing mapping populations. Nucleic Acids Res 19(23):6553–6568
Gökmen S (2004) Effects of moisture content and popping method on popping characteristics of popcorn. J Food Eng 65(3):357–362
Hoseney R, Zeleznak K, Abdelrahman A (1983) Mechanism of popcorn popping. J Cereal Sci 1(1):43–52
Jain-Poster K, Woodford-Thomas T (2015) Mapping QTLs for popping ability in a popcorn × dent maize genetic cross. J Emerg Invest 1:1–8
Jele CP (2012) Genetic analysis of agronomic and quality traits in popcorn hybrids. M Sc thesis, University of KwaZulu-Natal, Republic of South Africa
Jele P, Derera J, Siwela M (2014) Assessment of popping ability of new tropical popcorn hybrids. Aust J Crop Sci 8(6):831–839
Karababa E (2006) Physical properties of popcorn kernels. J Food Eng 72(1):100–107
Li Y, Wu S, Niu S, Li Z (2003) Analysis of combining ability of popcorn with normal corn inbreds belonging to different heterotic groups. J Henan Agric Univ 37:1–5
Li Y, Dong Y, Niu S, Cui D (2007a) The genetic relationship among plant-height traits found using multiple-trait QTL mapping of a dent corn and popcorn cross. Genome 50(4):357–364
Li Y, Dong Y, Niu S, Cui D (2007b) QTL for popping characteristics in popcorn. Plant Breed 126(5):509–514
Li Y, Niu S, Dong Y, Cui D, Wang Y, Liu Y, Wei M (2007c) Identification of trait-improving quantitative trait loci for grain yield components from a dent corn inbred line in an advanced backcross BC2F2 population and comparison with its F2:3 population in popcorn. Theor Appl Genet 115(1):129–140
Li YL, Dong YB, Niu SZ, Cui DQ (2009) Identification of QTL for popping characteristics using a BC2F2 population and comparison with its F2:3 population in popcorn. Agric Sci China 8(2):137–143
Liu B (1998) Statistical genomics. CRC Press, Boca Raton
Lorieux M (2012) MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed 30(2):1231–1235
Lu HJ, Bernardo R, Ohm H (2003) Mapping QTL for popping expansion volume in popcorn with simple sequence repeat markers. Theor Appl Genet 106(3):423–427
Matz SA (1984) Miscellaneous equipment. In: Matz SA (ed) Snack food technology. Springer, Dordrecht, pp 308–333
Michelmore RW, Paran I, Kesseli R (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci 88(21):9828–9832
Miranda GV, Coimbra RR, Godoy CL, Souza LV, Guimarães LJM, Melo AVD (2003) Potencial de melhoramento e divergência genética de cultivares de milho-pipoca. Pesquisa agropecuária brasileira 38(6):681–688
Mishra G, Joshi D, Panda BK (2014) Popping and puffing of cereal grains: a review. J Grain Process Storage 1(2):34–46
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326
Muthusamy V, Hossain F, Thirunavukkarasu N, Choudhary M, Saha S, Bhat JS, Prasanna BM, Gupta HS (2014) Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9(12):1–22
Ren Y, Yobi A, Marshall L, Angelovici R, Rodriguez O, Holding DR (2018) Generation and evaluation of modified opaque-2 popcorn suggests a route to quality protein popcorn. Front Plant Sci 9:1803
Rinaldi DA, Pípolo VC, Gerage AC, Ruas CdF, Júnior F, da Silva N, Souza Ad, Souza SGHd, Garbuglio DD (2007) Correlação entre heterose e divergência genética estimadas por cruzamentos dialélicos e marcadores moleculares RAPD em populações de milho-pipoca. Bragantia 66(2):183–192
Soylu S, Tekkanat A (2007) Interactions amongst kernel properties and expansion volume in various popcorn genotypes. J Food Eng 80(1):336–341
Srdić J, Pajić Z (2011) Variability of yield and kernel quality parameters of popcorn hybrids (Zea mays L. everta). J Process Energy Agric 15(2):84–86
Swapna G (2017) Popcorn—“A healthy nutritional speciality corn value added product”. Int J Food Nutr Sci 6(2):1
Sweley JC, Rose DJ, Jackson DS (2013) Quality traits and popping performance considerations for popcorn (Zea mays L. Everta). Food Rev Int 29(2):157–177
Trick M, Adamski NM, Mugford SG, Jiang CC, Febrer M, Uauy C (2012) Combining SNP discovery from next-generation sequencing data with bulked segregant analysis (BSA) to fine-map genes in polyploid wheat. BMC Plant Biol 12(1):14
Tripathi A, Joshi N, Kumar A (2016) Maize production technologies in India-a review. Octa J Environ Res 4(3):243–251
Vieira R, Scapim C, Moterle L, Tessmann D, Conrado T, Amaral Júnior Ad (2009) Diallel analysis of leaf disease resistance in inbred Brazilian popcorn cultivars. Genet Mol Res 8(4):1427–1436
Vijayabharathi A, Anandakumar C, Gnanamalar R (2009) Combining ability analysis for yield and its components in popcorn (Zea mays var. everta Sturt.). Electron J Plant Breed 1(1):28–32
Ziegler KE (2001) Popcorn. In: Hallauer AR (ed) Specialty Corn, 2nd edn. CRC Press, Boca Raton FL, pp 199–234
Zou C, Wang P, Xu Y (2016) Bulked sample analysis in genetics, genomics and crop improvement. Plant Biotechnol J 14(10):1941–1955
Acknowledgements
The authors are highly thankful to the Department of Genetics, Plant Breeding and Biotechnology, Dr. Khem Singh Gill Akal College of Agriculture, Eternal University for providing the required infrastructure and research facilities to undertake this study. HPKV, Palampur is also acknowledged for providing the seed material used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human and animal rights
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thakur, S., Kumar, R., Vikal, Y. et al. Molecular mapping of popping volume QTL in popcorn (Zea maize L.). J. Plant Biochem. Biotechnol. 30, 496–503 (2021). https://doi.org/10.1007/s13562-020-00636-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-020-00636-y