Abstract
To assess genetic variation, 33 novel microsatellite markers were identified through nucleotide sequencing of enriched genomic libraries of Pteris cretica. Di- repeats (79.7 %) were found to be most predominant followed by tri (15.8 %), tetra (2.3 %) and hexa (2.3 %) type of repeat motifs. Evaluating these markers in six populations (N = 48) of Western Himalayan range detected average polymorphism information content (PIC) of 0.32. Combined neighbor joining (NJ) and principal coordinate analysis (PCoA) grouped all the populations in two major clusters with high levels of intermixing of accessions in each cluster. This suggests that P. cretica populations of Western Himalaya have broadly been mixed with two sub-populations. High within population variance (98.7 %) and low genetic differentiation (Φst: 0.013), recorded in the analysis of molecular variance (AMOVA). For the first time, highly polymorphic novel genomic microsatellite markers were identified and utilized for revealing genetic diversity of P. cretica in Western Himalayan range in context of established hypothesis of genetic variations based on allozyme markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pteris cretica (family; Pteridaceae) is an ornamental plant with pantropical distribution (Morton 1957). In India, it is found across the Himalayan and Central Mountain regions. It has a remarkable ability to tolerate and accumulate high concentrations of arsenic as well as antimony (Robinson 2009), and therefore also used as a model plant for heavy metal tolerance studies (Raab et al. 2004). It may be proved as promising bio resource for reclaiming barren lands intoxicated with heavy metals.
P. cretica is one of the most ancient fern species often known for apogamous mode of reproduction (Debary 1878; Farlow 1874; Huang et al. 2011). In such reproduction, antheridia produce unreduced spores that give rise to sporophytic plant bypassing fertilization (Ishikawa et al. 2003). Apogamy creates reproductive barriers, thereby facilitating sympatric speciation (Werth and Windham 1991). Earlier studies suggests that more than 75 % of apogamous fern are polyploids (Kanamori 1972; Park and Kato 2003; Walker 1962), and opportunities of polyploidization increase due to a rise in gene dosage or addition of chromosomes (Heilbronn 1932). P. cretica exists as a species complex with its different cyto-reproductive types; diploid, triploid and tetraploid (agamosporous), and diploid sexual types (Kato et al. 1992; Walker 1962). Sexual cytotypes are reported but less frequently in Yunnan, China and Indonesia, Central India and Nepal (Jha and Sinha 1987; Kato et al. 1992; Roy et al. 1971; Walker 1962). Apogamous cytotypes are widely distributed expanding from Europe, Africa, Asia, and North and South America (The Global Biodiversity Information Facility; http://data.gbif.org). Based on the extensive survey and cytological studies done by Verma and Khullar (1965), two apomictic cytotypes namely diploid (n = 2x = 58) and triploid (n = 3x = 87) were found to be the most prevalent groups distributed in Western Himalayan range. Earlier, Suzuki and Iwatsuki (1990) used allozyme based method to reveal genetic variations in apogamous P. cretica in Japan. Nonetheless, different reproductive strategies followed by the species and lack of genomic marker resource makes genetic analysis of P. cretica challenging.
Among various marker systems, microsatellite markers are preferred due to hyper variability, co-dominance, high reproducibility and genome wide distribution (Varshney et al. 2005), and have been utilized for various genetic diversity, mapping and evolutionary studies. However, implementation of such studies in P. cretica is limited largely due to non-availability of microsatellite markers. The purpose of this study is to develop microsatellite markers through nucleotide sequencing of genomic libraries enriched with SSR repeat motifs and their utilization to examine genetic diversity in natural populations of P. cretica existing in Western Himalayan mountain range.
Materials & methods
Plant material and DNA extraction
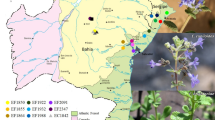
Forty eight accessions of Pteris cretica were sampled across six different locations (Chamba, Kangra, Kullu, Mandi, Shimla and Dehradun) of Western Himalaya (Supplementary Fig. 1). The distance between sampled accessions was kept approximately 15 m (15 m × 15 m quadrat). Minimum number of accessions sampled per population was at least 5. Two accessions collected from Dehradun region (Uttarakhand) were included for comparison purpose to evaluate potential of novel SSR markers for diversity characterization of wider geographical range. Fresh leaves were collected and processed for DNA extraction using CTAB method (Doyle and Doyle 1990). The quantity of DNA was estimated spectrophotometrically using Nanodrop (Thermo Scientific, USA) and quality was assured on 0.8 % agarose gel with respect to λ uncut (Fermentas, USA).
Construction of microsatellite enriched genomic libraries
Di-repeat (GA)n enrichment of genomic DNA of P. cretica was done as described earlier (Bhardwaj et al. 2013; Kijas et al. 1994). Plasmid DNA was isolated using R.E.A.L. prep. plasmid kit (Qiagen, USA) and sequenced directly with M13 universal primers in both forward and reverse directions using ABI 3730 xl DNA Analyzer in 20 μl of sequencing reaction volumes as per the manufacture’s procedure. The base calling and post processing of the sequence data were performed using Sequence Analysis Software (Applied Biosystem, v5.2).
Microsatellite identification and primer designing
Sequenced raw fragments were curated to obtain high quality non-redundant (NR) sequence data using SeqMan (DNAstar Lasergene, version 7.1) as per the parameters reported earlier by Sharma et al. (2009). These NR sequence data were individually processed with SSRIT (http://www.gramene.org/db/searches/ssrtool) to identify SSR containing sequences with a SSRs length of ≥12 bp. Primers were designed with the help of web based Primer3 software (Koressaar and Remm 2007; Untergrasser et al. 2012) with default parameters (GC percentage from 20.0 to 80.0 %; primer length from 18 to 27; primer Tm from 57.0 to 63.0 °C). Considering their derivation from Pteris cretica genome, these microsatellites were prefixed as PCGMS (Pteris Cretica Genomic MicroSatellite) markers.
Marker validation and evaluation of genetic diversity
Designed SSR primers were PCR validated in 48 random collections of P. cretica using 20 ng of template DNA and appropriate annealing temperatures (Table 1) as described by Sharma et al. (2009). Amplified fragments were separated on denaturing sequencing gel and visualized by the procedure described earlier by Sharma et al. (2009).
Considering, earlier reports on distribution of apogamous P. cretica population in targeted Western Himalayan range (Verma and Khullar 1965), various possible ploidy and reproductive modes of different cytotypes, all the SSR marker loci were dominantly scored across all accessions. Null alleles were assigned to accessions with no/ambiguous amplification products under standard conditions. Polymorphic information content (PIC) was calculated for individual marker based on amplification profiles in 48 accessions of P. cretica according to Anderson et al. (1993). Determination of clone mates and genetic diversity among sampled populations was done using Genotype and Genodive software (Meirmans and Van Tienderen 2004). This software package was specially developed for diversity analysis in asexual species of various ploidy states existing within the same population. Although, it can also handle the data of sexually reproducing species. Genotype is useful for identification of clones and duplicate removal for subsequent analyses. Genodive efficiently determines unbiased genetic diversity by normalizing potential biases due to small sample size (Meirmans and Van Tienderen 2004). Infinite mutation model was employed to calculate AMOVA based on distance matrix obtained. Estimates of genetic similarity between the accessions were calculated using Jaccard coefficient and a dendrogram was obtained based on Unweighted Neighbour Joining method using Darwin software version 5.0.158 (Perrier and Jacquemoud-Collet 2006). Principal coordinate analysis was done using the same software.
Results
Enrichment of microsatellites
Three hundred fifty (GA)n enriched genomic positive clones were selected for nucleotide sequencing in both forward and reverse directions. Non-redundant (NR) nucleotide sequences with size range of 200–800 bp were utilized for identification of PCGMS markers. Analysis of sequence data with SSRIT identified 133 clones, which contained one or more targeted or non-targeted microsatellite repeat motifs (37 perfect and 49 compound excluding mononucleotides) suggesting 39.4 % enrichment. Among 133 SSRs, di- repeats (106, 79.7 %) were most predominant followed by tri (21, 15.8 %), tetra (3, 2.3 %) and hexa (3, 2.3 %) type of repeat motifs. GA/TC type of repeat motif formed a major portion (78, 73.6 %) of di-repeats followed by CA/TG (28, 26.4 %) type of repeat motif. Among tri repeats, ATG/CAT motifs were most common (7, 33 %) followed by CAG/CTG type (5, 24 %) of SSR motifs (Figs. 1a and 2b).
Marker development and evaluation of polymorphism
Eighty six primers flanking the microsatellite repeat could be designed. Of these, thirty seven markers produced successful amplicon. However, four PCGMS markers showed no genetic variation (monomorphic) among the accessions, and hence were excluded from the study. A total of thirty microsatellite sequences (deposited under Gen Bank IDs: KJ000492 to KJ000521) containing thirty three polymorphic PCGMS markers produced a total of 194 amplicons ranging from 2 (PCGMS27.1) to 12 (PCGMS1, PCGMS13, PCGMS17) with an average of 6 amplicons per SSR locus (Table 1; Fig. 2). Polymorphic information content (PIC) ranged from 0.15 (PCGMS27.1) to 0.43 (PCGMS1) with a mean value of 0.32.
Genetic diversity and cluster analysis
Based on Genotype software, we found none of the sampled accessions were clones (at threshold value = 10). Further, choosing an appropriate threshold value is essential for assigning clones to a particular lineage. However, in case of arbitrary populations like this, determining threshold value was difficult. When the threshold value was set 15 based on distance matrix, 48 accessions assigned to 43 genotypes (Table 2). Based on genotypic frequency, total diversity was found to be 0.96 with high level of within population diversity (d = 0.87) and low level of among populations diversity (d = 0.1) (Supplementary Table 1). Further, AMOVA analysis revealed high level of variance partitioned within populations (98.7 %) and low variance partitioned among population (1.3 %). Φst values (0.013) suggest very low level of genetic differentiation among populations (Supplementary Table 2). However, pair wise population differentiation suggested Shimla and Chamba populations were most diverse followed by Dehradun and Kullu (Supplementary Table 3).
Combined Neighbor Joining (Fig. 3a) and principal coordinate analysis (Fig. 3b) detected two major clusters with high level of intermixing among 6 populations of Western Himalaya. Majority of accessions (33) were grouped in cluster 2 followed by fifteen accessions in cluster 1.
Dendrogram showing genetic relationships among 48 accessions of P. cretica based on 194 alleles produced by 33 PCGMS markers. Scale represents Jaccard’s similarity coefficient. Tree branches with bootstrap values greater ≥50 % are indicated (a). Principal coordinate analysis revealing clustering pattern of 48 accessions of P. cretica (b)
Discussion
Genomic microsatellites
Microsatellite enrichment technique based on pre-cloning, selective hybridization are shown to be a robust, reproducible and cost effective approach for identification of large numbers of SSR markers in diverse plant species (Parida et al. 2010). Efficiency of enrichment obtained in present study was 39.4 % of the NR cloned sequences, which was comparatively higher than that reported in apple (Guilford et al. 1997), Eucalyptus (Brondani et al. 1998) and tea (Bhardwaj et al. 2013). Among di- repeats (106, 79.7 %), GA/TC type was most predominant (58.6 %) showing successful enrichment of desirable repeat motifs.
Evaluation of genetic diversity and population structure
Several marker tools have been used previously for determining the level of genetic variation in P. cretica including allozymes (Suzuki and Iwatsuki 1990). However, these markers suffered from one or more limitation in revealing the actual level of variation. For the first time, we developed novel SSR markers (which exploits DNA polymorphism) from enriched genomic libraries and marked their utilization in revealing the genetic diversity P. cretica from Western Himalaya region of India. Thirty three SSR markers outperformed limited allozyme markers (Suzuki and Iwatsuki 1990), with a total of 194 allelic variants (average: 6 alleles per SSR locus). Further, the genomic SSR markers (PCGMS) revealed higher allelic variants as compared to EST SSR markers (<5 alleles per SSR locus) in Alpine lady fern (Woodhead et al. 2003). This study reveals potential of newly developed SSR markers for genetic diversity and evolutionary studies in P. cretica complex. Previous cytological and phylogenetic analyses on P. cretica suggest different cytotypes and reproductive strategies are common in different parts of the world including India (Grusz et al. 2009; Jaruwattanaphan et al. 2013; Jha and Sinha 1987; Roy et al. 1971; Suzuki and Iwatsuki 1990; Verma and Khullar 1965; Walker 1962). Further, apogamictic cyotypes has been reported in many fern species including P. cretica (Debary 1878; Farlow 1874; Huang et al. 2011). Apogamy must give rise to clonal populations (Chao et al. 2012). However, analyses of clone mates and genetic diversity using Genotype Genodive software suggests different genotypes of P. cretica do exist in Western Himalaya region contributing to high level of within population genetic variance (98.7 %) and diversity (d = 0.87). Various hypotheses have been proposed supporting our observations of genetic variation in apogamous species: (1) hybridization of apoamous species with sexual diploid cytotype or allied species like P. kidoi (Grusz et al. 2009; Jaruwattanaphan et al. 2013; Suzuki and Iwatsuki 1990; Walker 1962), (2) Unequal meiosis (Lin et al. 1992), (3) genetic segregation by homoeologous pairing of chromosome (Ootsuki et al. 2012; Ishikawa et al. 2003; Klekowski and Edward 1973), (4) Watano and Iwatsuki (1988) and Manton (1950) suggested recurrent origin of apogamous species from ancestral sexual species due to translocations, chromosomal changes and natural selection. Theory of multiple origins (Ellstrand and Roose 1987; Parker 1979) which suggests the existing diversity in populations is due to diversity already existing long back to their time of origin. Moreover, rare occurrence of sexual cytotypes of P. cretica in Yunnan, China and Indonesia, Central India and Nepal regions cannot be overlooked. Nevertheless, on the basis of genetic fingerprinting data alone, it would be too early to conclude which reproductive strategy (ies) is (are) being adopted by the species that has led to high genetic diversity.
Low level of genetic differentiation among population and high level of intermixing of accessions of distant geographic locations as revealed by AMOVA, NJ and PCoA signifies long range spore dispersal (up to hundreds of kilo meters with wind and/or water currents prevailing in the regions) (Soltis and Soltis 1990) and establishment to new locations. Furthermore, due to ornamental importance of the species, anthropogenic activities might have resulted in migration of genotypes to distant locations. Clustering pattern based on combined NJ and PCoA suggests basically two genetic populations contributing to overall diversity of the species in the region. This also suggests accessions from same geographic locations might not exhibit similar individual genotype, and that natural selection has not played any significant role in genetic differentiation and diversification of the species. Majority of accessions (33) grouped in cluster 2, suggesting that the genetic pool is more successful and invaded larger locality in Western Himalayan range as reported earlier by Schneller et al. (1998). Our results suggest that the whole species complex functions as a single large population with intermixing of sub-populations of similar genetic backgrounds as previously reported (Bucharova and Munzbergova 2012; Schneller et al. 1998).
Current research presents the first study to assess genetic diversity and differentiation in Western Himalaya populations of Pteris cretica using hyper variable genomic SSR markers. Population genetic analysis using SSR markers is a landmark for population biology studies of Indian ferns. Novel microsatellite markers with high level of polymorphic potential suggests that PCGMS markers are capable of revealing substantial variation in natural populations of P. cretica, thus, can be extrapolated for future population biology and evolutionary studies. Our study provides a new outlook to the existing allozyme based genetic variability of P. cretica complex across Western Himalayan range. High level of genetic diversity in this apomictic fern allows the plant to adapt and withstand various environmental stress. In future, these markers can assist cytological studies to decipher reproductive strategies and evolutionary pattern of this fern.
Abbreviations
- SSRs:
-
Simple sequence repeats
- ESTs:
-
Expressed sequence tags
- PIC:
-
Polymorphic information content
- PCR:
-
Polymerase chain reaction
References
Anderson JA, Churchill GA, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Bhardwaj P, Kumar R, Sharma H, Tewari R, Ahuja PS, Sharma RK (2013) Development and utilization of genomic and genic microsatellite markers in Assam tea (Camellia assamica ssp. assamica) and related Camellia species. Plant Breed 132:748–763
Brondani RPV, Brondani C, Tarchini R, Grattapaglia D (1998) Development, characterization and mapping of microsatellite markers in Eucalyptus grandis and E. urophylla. Theor Appl Genet 97:816–827
Bucharova A, Munzbergova Z (2012) Gene flow among populations of two rare co-occurring fern species differing in ploidy level. PLoS ONE 7:e45855
Chao YS, Liu HY, Chiang YC, Chiou WL (2012) Polyploidy and speciation in Pteris (Pteridaceae). J Bot 2012: 7, doi:10.1155/2012/817920. http://dx.doi.org/10.1155/2012/817920
DeBary A (1878) Uber apogamie farne und die erscheinung der apogamieim allgemeinen. Bot Zeit 36:450–495
Doyle JJ, Doyle JE (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ellstrand NC, Roose ML (1987) Patterns of genotypic diversity of clonal plant species. Am J Bot 74:123–131
Farlow WG (1874) Memoirs: an asexual growth from the prothallus of Pteris cretica. Q J Microsc Sci 2:266–272
Grusz AL, Windham MD, Pryer KM (2009) Deciphering the origins of apomictic polyploids in the Cheilanthesyavapensis complex (Pteridaceae). Am J Bot 96:1636–1645
Guilford P, Prakash S, Zhu JM, Rikkerink E, Gardiner S, Bassett H, Forster R (1997) Microsatellites in Malus x domestica (apple): abundance, polymorphism and cultivar identification. Theor Appl Genet 94:249–254
Heilbronn A (1932) Polyploidy and generational change. Ber Dtsch Bot Ges 50:289–289
Huang YM, Hsu SM, Hsieh TS, Chou HU, Chiou WL (2011) Three Pteris species (pteridacae, pteridophyta) reprodcue by apogamy. Bot Stud 52:79–87
Ishikawa H, Ito M, Watano Y, Kurita S (2003) Extensive electrophoretic variation in apogamous fern species. Dryopterisnipponensis (Dryopteridaceae) Actaphytotaxgeobot 54:59–68
Jaruwattanaphan T, Matsumoto S, Watano Y (2013) Reconstructing hybrid speciation events in the Pteris cretica group (pteridaceae) in Japan and adjacent regions. Syst Bot 38:15–27
Jha J, Sinha BM (1987) Cytomorphological variability in apogamous populations of Pteris cretica L. Caryologia 40:71–78
Kanamori K (1972) Apogamy in ferns with special reference to the apogamous embryogenesis. Sci Rep TKD Sec B 15:111–131
Kato M, Nakato N, Cheng X, Iwatsuki K (1992) Cytotaxonomic study of ferns of Yunnan, southwestern China. Bot Mag Tokyo 105:105–124
Kijas JMH, Fowler JCS, Garbett CA, Thomas MR (1994) Enrichment of microsatellites from the citrus genome using biotinylated oligonucleotide sequences bound to streptavidin coated magnetic particles. Biotechniques 16:656–662
Klekowski JR, Edward J (1973) Sexual and subsexual systems in homosporous pteridophytes: a new hypothesis. Am J Bot 60:535–544
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291
Lin SJ, Kato M, Iwatsuki K (1992) Diploid and triploid offspring of triploid agamosporous fern Dryopteris pacifica. Bot Mag Shokubutsu-Gaku-Zasshi 105:443–452
Manton I (1950) Problems of cytology and evolution in the Pteridophyta. Cambridge University Press
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Morton CV (1957) Observations on cultivated ferns. I. Am Fern J 47:7–14
Ootsuki R, Sato H, Nakato N, Murakami N (2012) Evidence of genetic segregation in the apogamous fern species Cyrtomium fortunei (Dryopteridaceae). J Plant Res 125(5):605–612
Parida SK, Pandit A, Gaikwad K, Sharma TR, Srivastava PS, Singh NK, Mohapatra T (2010) Functionally relevant microsatellites in sugarcane unigenes. BMC Plant Biol 10:251
Park CH, Kato M (2003) Apomixis in the interspecific triploid hybrid fern Cornopteris christenseniana (Woodsiaceae). J Plant Res 116:93–103
Parker ED (1979) Ecological implications of clonal diversity in parthenogeneticmorpho species. Am Zool 19:753–762
Perrier X, Jacquemoud-Collet JP (2006) Darwin software. http://darwin.cirad.fr/darwin
Raab A, Feldmann J, Meharg AA (2004) The nature of arsenic-phytochelatin complexes in Holcusla natus and Pteris cretica. Plant Physiol 134:1113–1122
Robinson RC (2009) Invasive and problem ferns: a European perspective. Int Urban Ecol Rev 4:83–91
Roy RP, Sinha BMB, Sakya AR (1971) Cytology of some ferns of Kathmandu valley. Br Fern Gaz 10:193–199
Schneller J, Holderegger R, Gugerli F, Eichenberger K, Lutz E (1998) Patterns of genetic variation detected by RAPDs suggest a single origin with subsequent mutations and long -distance dispersal in the apomictic fern Dryopteris remota (Dryopteridaceae). Am J Bot 85:1038–1038
Sharma RK, Bhardwaj P, Negi R, Mohapatra T, Ahuja PS (2009) Identification, characterization and utilization of unigene derived microsatellite markers in tea (Camellia sinensis L.). BMC Plant Biol 9:53
Soltis PS, Soltis DE (1990) Genetic variation within and among populations of ferns. Am Fern J 80:161–172
Suzuki T, Iwatsuki K (1990) Genetic variation in agamosporous fern Pteris cretica L. in Japan. Heredity 65:221–227
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3 - new capabilities and interfaces. Nucleic Acids Res 40:115
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: feature sand applications. Trends Biotechnol 23:48–55
Verma SC, Khullar SP (1965) Cytogenetics of the western Himalayan Pteris cretica complex. Ann Bot 29:673–681
Walker TG (1962) Cytology and evolution in the fern genus Pteris L. Evolution 16:27–43
Watano Y, Iwatsuki K (1988) Genetic variation in the ‘Japanese apogamousform’of the fern Aspleniumun ilaterale Lam. Bot Mag Shokubutsu-Gaku-Zasshi 101:213–222
Werth CR, Windham MD (1991) A model for divergent, allopatric speciation of polyploidy pteridophytes resulting from silencing of duplicate-gene expression. Am Nat 137:515–526
Woodhead M, Russell J, Squirrell J, Hollingsworth PM, Cardle L, Ramsay L, Gibby M, Powell W (2003) Development of EST‐SSRs from the alpine lady‐fern, Athyrium distentifolium. Mol Ecol Notes 3:287–290
Acknowledgments
The research work presented in the manuscript was funded by Department of Biotechnology (DBT) and Council of Scientific and Industrial Research (CSIR), Government of India. This is IHBT communication number 3624.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Vishal Kumar and Abhishek Bhandawat contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Topographical location of forty eight P. cretica accessions sampled from Western Himalayan range (PPTX 324 kb)
Supplementary Table 1
Partitioning of diversity of 48 accessions. (DOCX 11 kb)
Supplementary Table 2
Analysis of molecular variance (AMOVA) of 48 P. cretica accessions (DOCX 12 kb)
Supplementary Table 3
Test for population differentiation. (DOCX 11 kb)
Rights and permissions
About this article
Cite this article
Kumar, V., Bhandawat, A., Sharma, H. et al. Novel microsatellite markers identification and diversity characterization in Pteris cretica L.. J. Plant Biochem. Biotechnol. 25, 104–110 (2016). https://doi.org/10.1007/s13562-015-0316-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-015-0316-8