Abstract
Although plant actins have highly conserved sequences, they are classified as vegetative or reproductive actin based on genetic relationship and the role of the tissue. To characterize the 1,590 bp actin (ACT) gene cloned from the petals of the Ascocenda Princess Mikasa ‘Blue’ orchid, the encoded protein sequence of Ascocenda ACT was aligned to 31 related proteins from orchids and other plant species. The comparison of the amino acids of the actin proteins showed that Ascocenda ACT1, Cymbidium ACT, Phalaenopsis ACT1, Phalaenopsis ACT2, and Phalaenopsis ACT4 contained 13 positions different in amino acid residues in actin polypeptides. Consequently, Ascocenda ACT, Cymbidium ACT, and Phalaenopsis ACT2 were classified as vegetative actins. The classification of Ascocenda ACT was consistent with the results of qualtitative real-time polymerase chain reaction (qPCR) analysis, which demonstrated that actin is highly expressed in all vegetative tissues of the Ascocenda orchid but not in pollen. The predicted three-dimensional structure of the orchid actins revealed that most of the 13 variable residues were located outside the dynamic functional loops, with the exception of Thr16 and Pro353, which were located in the S-loop and target binding site of Ascocenda ACT, respectively. We suggest that the DNA sequence in actin may play a role in the functions of each actin isoform in a tissue-specific manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant actins, encoded by a multigene family, are fundamental plant proteins, the function of which depends on the type of tissue. They have been identified as isovariant molecules based on their cellular localization and functions, such as in cellular processes (Kandasamy et al. 2009), cell growth (Hussey et al. 2006), defense mechanisms (de Almeida Engler et al. 2010), and transcriptional regulation (Miralles and Visa 2006). Based on their gene expression patterns and phylogenetic analysis, eight different functional ACTs of Arabidopsis thaliana were recognized and divided into two classes: vegetative actins (Arabidopsis ACT2, ACT8, and ACT7) and reproductive actins (Arabidopsis ACT1, ACT3, ACT4, ACT12, and ACT11) (McDowell et al. 1996b; Huang et al. 1996, 1997; An et al. 1999; Kandasamy et al. 1999, 2012; Meagher et al. 1999). Vegetative actins are predominantly expressed in leaves, stems, roots, petals, and sepals, and reproductive actins are expressed in pollinia, ovules, and embryonic tissues (McDowell et al. 1996a; Slajcherova et al. 2012).
A monomeric molecule of actin (42 kDa) forms a globular (G) structure (G-actin) in a dilute salt and polymerizes into a filamentous (F) structure (F-actin), in the form of a double helix, when the ionic strength increases (Reisler 1993; Dominguez and Holmes 2011). G-actin typically consists of a polypeptide chain folded into two major α/β domains, each consisting of two subdomains (Kabsch et al. 1990). The structure of these domains is the same as that of the sugar kinase/ heat shock protein 70/ACT superfamily (Dominguez and Holmes 2011). Although the biological functions of the actin isovariants differ, the conformation of G-actin in the variants is similar because of the highly conserved amino acid sequences in actin polypeptide chains.
Despite recent advances in understanding of actin gene organization based on accessible actins from several plant species, such as Arabidopsis (McKinney and Meagher 1998), rice (Reece et al. 1990), cotton (Li et al. 2005), and soybean (Shah et al. 1982), the role of this gene has not yet been examined in orchid species. Therefore, we report here the molecular cloning and gene expression patterns of the actin gene isolated from Ascocenda Princess Mikasa ‘Blue’ orchid flowers. A phylogenetic analysis and the three-dimensional (3D) structures of Ascocenda actin and four other orchid actins from GenBank were analyzed to compare the genetic variation in the Ascocenda orchid actins with that of the other plant actins.
Materials and methods
Total RNA extraction
Ascocenda Princess Mikasa ‘Blue’ (Ascocenda Royal Sapphire × Vanda coerulea) was purchased from a orchid nursery in Bangkok, Thailand. Five tissues (roots, stems, leaves, petals and pollinia) were immediately collected and subjected to total RNA extraction using the lithium chloride precipitation method (Lievens et al. 1997).
Reverse transcriptase-polymerase chain reaction
Total RNA (1 μg) extracted from Ascocenda petals was reverse transcribed to obtain complementary DNA (cDNA) using Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Buckinghamshire, UK) according to the manufacturer’s instructions and a primer specific to the polyA tail of mRNA (RACE-T, 5′-GAC TCG AGT CGA CAT CG (T)17-3′). The cDNA was amplified in an RT-PCR reaction mixture including i-Taq ™ Polymerase (iNtRON Biotechnology, Inc., Seongnam-si, Korea) according to the manufacturer’s recommendations and the MikAct-F(1) (5′-AAA CTA TGG CTG AAG CAG AGG A-3′) and RACE (5′-GACTCGAGTCGACATCG-3′) primers under the following thermal cycling conditions: initial denaturation for 3 min at 94 °C, 30 amplification cycles of denaturation for 30 s at 94 °C, annealing for 40 s at 60 °C, and extension for 1 min at 72 °C, and a final extension for 5 min at 72 °C. An aliquot of the PCR product was resolved on a 0.8 % agarose gel. Subsequently, the 1,400 bp PCR product was extracted from the agarose gel utilizing a HiYield Gel/PCR DNA Fragment Extraction kit (RBC Bioscience, NewTaipei City, Taiwan), cloned into pGEM®-T Easy (Promega Co., Wisconsin, USA) and transformed into XL1-blue Escherichia coli. The positive clones were confirmed to be the actin gene by Automated DNA Sequencing (Macrogen, Seoul, Korea).
Actin gene sequence analysis
The DNA sequence of the cloned gene was analyzed using Blastn (Altschul et al. 1997) and Blastp (Altschul et al. 2005) in the non-redundant protein sequence database (http://www.blast.ncbi.nlm.nih.gov). The physicochemical parameters of the protein sequence were calculated using ProtParam (http://au.expasy.org/tools/protparam.html) (Artimo et al. 2012). The inverted repeat in the act sequences was analyzed using DNAman version 7.0 (Lynnon Corporation, Canada). The subdomain annotation was adopted from the interaction between actin cytoskeleton and heat shock proteins (Mounier and Arrigo 2002) by the alignment method based on rabbit actin (1atn chain A) (Kabsch et al. 1990) and the Conserved Domain Database (CDD) (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) (Marchler-Bauer et al. 2011).
Primary structure analysis
Thirty-two total actin sequences (Ascocenda ACT and actins from twelve monocots, eighteen dicots, and an abalone) were retrieved from the NCBI protein database (http://www.ncbi.nlm.nih.gov/) to identify a genetic relationship. The evolutionary relationships between these actins, based on their amino acid sequences, were illustrated as phylogenetic trees using MEGA package version 5.1 (Tamura et al. 2011) and the Maximum Likelihood method, a JTT matrix-based model (Jones et al. 1992) and MUSCLE alignment (Edgar 2004). A bootstrap analysis with 1,000 replicates was performed to assess the statistical reliability of the tree topology.
3D structure homology modeling
Identification of regions of homology between the amino acid sequences of actins of the five orchid species, rice and Arabidopsis with the actin of Drosophila melanogaser (3eksA) (Nair et al. 2008) using the structural alignment tool of the SWISS-MODEL workspace (Bordoli and Schwede 2012). The four main steps in this approach are following: finding the most homologous amino acid sequences between the query and the 3eksA template; aligning query amino acid and template; generating a 3D structure based on the 3eksA template; and then selecting for the most native conformations. The percentage sequence identity and three-dimensional structures were determined using QMEAN4 (Benkert et al. 2011) employing the most homologous amino acid sequence resulting from the 3eksA sequences as standard. Figure constructions were superimposed and performed with the Accelrys DS visualizer 4.0 (Accelrys, Inc., San Diego, CA USA).
Gene expression analysis by qualitative real-time polymerase chain reaction (qPCR)
To quantitate the actin transcripts in the various orchid tissues, qPCR was performed, and 5.8S rRNA was selected as the reference gene. One microgram of total RNA from the root, stem, leaf, petal, or pollen was treated with RNase-free DNase-I (Promega, USA). The total RNA was reverse transcribed into cDNA using MikAct1-R (1401) (5′-GCT TGT GAA GCA CAA TAA C-3′) and 5.8S rRNA (R) (5′-GCT TGA AGC CCA GGC AGA CG-3′) with Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, UK). The 342 base pairs (bp) corresponding to the ACT gene were amplified using the following primers: MikAct1-F (1060) (5′-CAT TCC AGC AGA TGT GGA T-3′) and MikAct1-R (1401). In addition, 5.8S rRNA (198 bp) was amplified with 5.8S rRNA (F) (5′-ATG ACT CTC GAC AAT GGA TTT-3′) and 5.8S rRNA (R) using KAPA SYBR® FAST qPCR Kits (Kapa Biosystems, USA). Each reaction containing 0.2 μl of cDNA template and 0.625 μM of each primer in a final reaction volume of 10 μl was performed in triplicate to ensure reproducibility. qPCR was performed on an Eppendorf Mastercycler® ep realplex4 S real-time PCR instrument (Eppendorf, USA) using the following conditions: denaturation for 3 min at 95 ° C, followed by 38 amplification cycles of denaturation for 10 s at 95 ° C and annealing and extension for 20 s at 60 ° C. At the end of the PCR program, a melt curve was generated to verify the homogeneity of the PCR product using the following program: denaturation for 15 s at 95 ° C, 15 s at 60 ° C, ramping to 95 ° C over 20 min, and 15 s at 95 ° C. The CT mean and standard deviation for SYBR were calculated throughout. The results were analyzed by the 2-∆∆CT method (Livak and Schmittgen 2001).
Results and discussion
Primary structure of Ascocenda ACT
The full-length cDNA corresponding to the Ascocenda act gene was 1,590 bp, encoding one open reading frame of 377 residues, with a calculated molecular weight of 41.65 kDa and a theoretical isoelectric point of 5.39. The length of the encoded Ascocenda actin was similar to the average length of that of the other actin proteins, which were 374–376 amino acids long (Hennessey et al. 1993), and to the molecular mass of the crystal structure of G-actin (Dos Remedios et al. 2003). Fig. 1 shows that the predicted Ascocenda actin polypeptide contained four major putative domains: a functional nucleotide binding domain of the sugar kinase/heat shock protein 70/actin superfamily located at the N-terminus (Leu10 to Ala183) and lined with 11 conserved residues (Asp13, Asn14, Gly15, Thr16, Met18, Lys20, Gln139, Asp156, Ser157, Gly158, and Asp159); an ATP binding site located along the entire length of the polypeptide, with 6 conserved residues (Thr16, Asp156, His163, Glu216, Gly304, and Lys338); a gelsolin binding site located from Gly148 to Pro353 with 9 conserved residues (Tyr145, Gly148, Glu169, Gly344, Ile347, Leu348, Leu351, Ser352, and Pro353); and a profilin binding site from Tyr168 to Lys375 at the C-terminus with 11 conserved residues (Tyr168, Glu169, Tyr171, Leu173, His175, Asp288, Asp290, Pro353, Glu363, Arg374, and Lys375).
3D structure of the actin protein isolated from Ascocenda Princess Mikasa ‘Blue’. a The four subdomains, predicted functional regions, binding clefts, and 13 variable residues of the orchids are illustrated. b A diagram of the predicted conserved domains in the Ascocenda actin protein. The black bar represents the polypeptide chain. The boxes refer to one conserved domain of sugar kinase/HSP70/actin. The binding sites for sugar kinase, ATP, gelsolin, and profilin are indicated with a line, and the locations of the conserved residues are depicted with the following symbols: sugar/kinase (▼), ATP (∆), gelsolin (▲), and profilin binding sites (▲). The four subdomains of the folded ACT polypeptide structure are marked with boxes over the polypeptide chains
The flanking 3′-UTR sequence of Ascocenda mRNA was 375 bp in length. It contained a putative uracil (U)-rich polyadenylation signal of UUUGUA. The 3′-UTR of the Ascocenda ACT gene lacks the classical poly (A) signal (AAAUAA) commonly found in numerous yeast and plant actin genes (Rothnie et al. 1994). Alternative cis-elements, such as UGUA, UAUA, and U-rich elements, have been identified in the former (Hu et al. 2005; Ji et al. 2007). One of these elements, UGUA, was shown to be an important functional signal in far-upstream elements in the rice actin gene (Shen et al. 2008), an additional accessory element in the cauliflower mosaic virus polyadenylation signal (Rothnie et al. 1994), and the sequence-specific for the heterologous polyadenylation site in yeasts and plants (Rothnie et al. 1994). According to some studies, the UUUGUA element in all Arabidopsis vegetative actins is associated with tissue-specific polyadenylation (Liu et al. 2010; Di Giammartino et al. 2011). The sequence data reported herein for the Ascocenda act gene was deposited in GenBank under the accession number HQ596370.
Genetic relationships and phylogenetic tree for actins
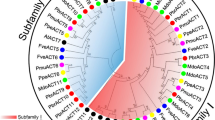
A pair-wise comparison revealed that the amino acid sequences of Ascocenda and 30 actins isolated from the orchids and various other plant species were highly conserved, with 93–96 % amino acid identity, whereas actin from abalone (Haliotis diversicolor Reeve) shared 89 % identity with Ascocenda actin (Supplementary Table 1). These 32 actins were grouped into two clusters based on their genetic relationships, amino acid composition, and multiple alignment scores (Fig. 2). Some orchid actins exhibited a distinct genetic relationship, thereby placing them in different clusters from the other orchid actins. In Cluster I, Phalaenopsis True Lady ACT1, Phalaenopsis Tropicana ACT4, and 10 other actins from both monocots and dicots were genetically closely related to Arabidopsis ACT1 and ACT11, which served as representatives of reproductive actins. In contrast, Ascocenda ACT, Cymbidium ensifolium ACT, and the Phalaenopsis hybrid ACT2 were grouped in Cluster II, together with the representative of a vegetative actin, Arabidopsis ACT7. As shown in Fig. 2, the actin members in Cluster II were further divided into two subclusters: subcluster IIA (actins from dicots) and subcluster IIB (actins from monocots). The Ascocenda ACT, Cymbidium ensifolium ACT, and Phalaenopsis hybrid ACT2 were grouped in subcluster IIB, together with the vegetative actin Arabidopsis ACT7, with 95–96 % amino acid identity. However, the 1 % amino acid difference between Ascocenda ACT, Phalaenopsis True Lady ACT1, and Phalaenopsis Tropicana ACT4 led to these orchid actins being grouped in different clusters. The vegetative actins in Cluster II with 95 % amino acid identity, on average, are more closely related than the reproductive actins in Cluster I (93.8 % identity). These results are consistent with the classification of eight known functional actins isolated from A. thaliana, which were classified as vegetative or reproductive classes based on their amino acid sequences, with 0.3–8.8 % amino acid diversity (McDowell et al. 1996b; An et al. 1999; Bhattacharya et al. 2000; Slajcherova et al. 2012).
Phylogenetic analysis of the 32 actin polypeptides. The tree with the highest log likelihood (−2184.3915) is presented. The percentage of trees in which the associated taxa clustered together is indicated next to the branches. The tree is drawn to scale, with the branch length based on the number of substitutions per site
Amino acid variations in the actins
The multiple alignments of the 32 actins revealed that Ser159, Tyr281, and Gly297 were the most conserved amino acid residues (Supplementary Table 2). With regard to specific conserved residues in each cluster, four conserved residues (Thr131/Ala131, Ile221, Thr234, and Ala360) were consistently present in Cluster I (the reproductive actins), and Val131, Val221, Ser234, and Ser360 were consistently present in Cluster II (the vegetative actins). Furthermore, Met201, Asn241, and Gly362 were present in actins from dicots in subcluster IIA, and Ile214 and Ala370 were restricted to actins from monocots in subcluster IIB. It can be assumed that the conserved residues can be used to group the 32 actins into distinct clusters in the phylogenetic tree. The identification of two conserved residues, Val221 and Ser360, in Cluster II of the vegetative actins was consistent with previous reports of vegetative actions in ferns and algae (An et al. 1999). However, residues Ala7 and Ser14, which are conserved in nonmuscle β-actin in humans and most other eukaryotes (Sheterline et al. 1995), were not very common in these 32 actins. Nevertheless, the protein alignment was useful in determining the genetic relationships between these polypeptides, and their predicted protein structures provided an alternative approach for gene characterization.
There were 13 variable positions in the actins of the five orchid species (Glu3-Asp3, Gly4-Pro4-Val4, Glu5-Asp5, Glu6-Asp6, Ala190-Ser190, Ala131-Val131, Ile214-Met214, Val221-Ile221, Ser234-Thr234, Ser241-Thr241, Leu268-Met268, Ala360-Phe360-Ser360, and Ala370-Ser370) (Fig. 3a). The amino acid at position 4 in the N-terminus of the 32 actins was the most divergent residue. The residues at this position (Ala4, Phe4, Gly4, Pro4, Ser4, Thr4 and Val4) exhibited different chemical properties. Three different amino acids, Val4, Pro4, and Gly4, were present in this position in the orchid actins. The Phalaenopsis True Lady ACT1 and Phalaenopsis Tropicana ACT4 reproductive actins (Fig. 3b) contained the same amino acid, Gly4, whereas variable residues, including Gly4, Pro4, and Val4, were identified in the Phalaenopsis ACT2, Ascocenda ACT, and Cymbidium ACT vegetative actins (Fig. 3c). Mutating the amino acid at this position in chicken β-actin (Aspenstrom et al. 1992), Drosophila ACT88F (Reedy et al. 1991), or yeast ACT1 (Johannes and Gallwitz 1991; Wertman et al. 1992) affected the activity, growth, and structure of ATPase, respectively, in previous studies (Hennessey et al. 1993).
3D structure of Ascocenda actin constructed using the Accelrys DS Visualizer. a The 13 variable residues identified in the orchids, together with their predicted functional regions and binding clefts are illustrated. The variable residues in the reproductive and vegetative actins are marked in the green boxes with white and black, respectively. All the mutated residues in Ascocenda actin are in red. The remaining variable residues in the orchids are shown in black in the yellow boxes. The structures of the actins from the b reproductive cluster and c vegetative cluster include the specific conserved amino acids
In the 32 actins, another four positions containing variable residues (Thr131-Ala131, Ile221, Thr234, and Ala360) in the reproductive actins in Cluster I were replaced with Val131, Val221, Ser234, and Ser360, respectively, in the vegetative actins in Cluster II . These variable residues were consistently identified in 73 actins when the actins from 41 dicots and 22 monocots were aligned (Supplementary Fig. 1). Surprisingly, all 73 plant actin polypeptides were two residues longer at the N-terminus than actins from human, yeast, and fungi. Furthermore, two residues (Met201 and Gly362) were present in the vegetative actins from dicots in subcluster IIA, and one residue, Ala370, was present in the monocot actins in subcluster IIB containing three orchid actins, with 96 % amino acid identity. Therefore, the actins in the five orchid species placed in Cluster I and subcluster IIB were subjected to 3D structure analysis by comparing their 3D structures with those of actins from rice and Arabidopsis.
3D structural analyses of orchid actins
The sequence identity between each orchid actin and that of the 3eksA template varied approximately from 89 to 90 % after folding into 3D structures, with a model reliability score of 0.69–0.7 (Supplemental Table 3). The 3D structure of Ascocenda ACT contained four domains. These four subdomains were annotated as subdomain 1 (residues 3–34, 72–146, and 340–377), subdomain 2 (residues 35–71), and subdomain 3 (residues 147–182 and 272–339, with the most variable sequence region located at amino acids 312–323 and subdomain 4 (residues 183–271) (Fig. 3a, 4). The reshuffling of subdomains 1 and 3 was structurally related to gene duplication, whereas subdomains 2 and 4 can be viewed as large insertions into the subdomains 1 and 3 (Dominguez and Holmes 2011). It was not surprising that the 3D structures of the eight plant actin samples were nearly the same in the four subdomain structures, with the exception that the folded structure of subdomain 2 was slightly shorter at the helix DNase I-binding loop (D-loop) region of Phalaenopsis ACT2 and Arabidopsis ACT1 (Fig. 3b, c, 4). Two binding pockets in the ATP binding site, the nucleotide binding cleft (upper) associated with a divalent cation, (Mg2+), in cells and the target binding cleft (lower), formed within the linkage regions between domains 1 and 3 (Fig. 3a, b, 4). The CDD search revealed that the 3D domains of Ascocenda actin were closed related to many other domains, including functional domains in the 1ATN chain A of rabbit actin (Kabsch et al. 1990; Dominguez and Holmes 2011), the ATP binding site (1DGA chain A) in Dictyostelium discoideum (Vorobiev et al. 2003), cofilin binding site (1COF chain A) in Saccharomyces cerevisiae (Fedorov et al. 1997), and profilin binding site (1HLU) in Bos taurus (Chik et al. 1996).
To map the conserved and divergent residues in the G-actin structures, the 13 divergent residues were labeled on the 3D structures of the orchid actins and the Arabidopsis and rice actins, the representatives of the reproductive and vegetative actin classes (Fig. 3a). Fig. 3a and 4 show that the two most divergent residues, Pro4 in domain 1 and Ala190 in domain 4, are located at the center of the nucleotide binding site in the upper cleft. None of the 13 divergent residues were located in the following dynamic functional regions: the DNase-I binding loop (D-loop, residues 39–51), G-loop (Gly158), hinge helix region (starting at Gln137), histidine-loop (H-loop; methylated His73; residues 70–78), serine-loop (S-loop; Ser14; residues 11–16), TrpHis2-loop (WH2-loop; W-loop; residues 165–172), or the C-terminus (residue 377) based on atomic crystal structure analysis of G-actin (Zheng et al. 2007; Kudryashov et al. 2010; Oztug Durer et al. 2010; Dominguez and Holmes 2011). Interestingly, Ser14 in the S-loop of animal actins was replaced with Thr16 in all the plant actins. Ser14 plays an important role at the start of the opening and closing of the nucleotide cleft processes by phosphorylation, which may strengthen the traction of filaments (Dominguez and Holmes 2011). Serine is commonly replaced by threonine (Pearlman et al. 2011) because both amino acids can perform a similar role in the phosphorylation mechanism (Betts and Russell 2003). As mentioned above, this mutation did not affect the 3D structure of actin. The activity of these actin isoforms could be the subject of a future study of plant/animal actin Ser14/Thr16 substitution, as the protein activity may vary after phosphorylation of these substituted amino acids (Kisselev et al. 2000).
Ten conserved residues (Tyr145, Ala146, Gly148, Thr150, Gly170, Ile343, Ile347, Leu348, Leu351, and Met357) were located in the target binding cleft. These conserved residues, which are predominantly hydrophobic, form the major binding site of most reported actin-binding proteins (Novick and Botstein 1985; Hennessey et al. 1992; Dominguez and Holmes 2011). As clearly presented in Fig. 3, more variable residues were identified in the reproductive actins (Ile7, Val131, IIe214, Val221, Ser234, Ser237, Gly297, Ser305, and Ala362) than in the vegetative actins (Ser201, Leu268, Ala274 and Ala370). In addition, in the target binding cleft, one residue, the conserved Thr353 was replaced with Pro353 in Ascocenda ACT.
The subdomain 2 structure generated from Arg41-Asp53 was identical to the D-loop in the 1ATN_A template (Kabsch et al. 1990). However, the folding of the helix loop in subdomain 2 of the orchid actins showed small differences (Fig. 3b, c). The D-loop plays a potential role in binding by forming over 80 complex structures with DNase I and small molecules, adopting an extended open loop conformation between subdomains 1 and 3 (Kabsch et al. 1990; Dominguez and Holmes 2011). Thus, the different D-loop structures may affect the binding activity and polymerization of F-actin (Chik et al. 1996; Oztug Durer et al. 2010; Dominguez and Holmes 2011). The DNase I binding loop is directly involved in conformational changes (Otterbein et al. 2001), and it is likely that this non-synonymous substitution will affect the structure and function of Gossypium hirsutum actin (Li et al. 2005).
ACT gene expression pattern in Ascocenda Princess Mikasa ‘Blue’
Real-time quantitative PCR was performed to quantify ACT gene expression in the vegetative (root, stem, leaf and petal) and reproductive tissues (pollen) of Ascocenda. As illustrated in Fig. 5, the expression of ACT was substantially different in the reproductive tissue compared to the vegetative tissue, with the latter showing 15 to 20-fold higher expression.
The cloned ACT gene from the Ascocenda flower was ubiquitously expressed in all the vegetative tissues, especially in the petals. However, the expression of the ACT gene transcript was very low in pollen. This observation is in accordance with ACT expression data for vegetative actins (Arabidopsis ACT2, ACT7, and ACT8) in Arabidopsis (Meagher et al. 1999). It is further supported by the expression patterns of Arabidopsis ACT1, ACT3, ACT4, and ACT12, which are strongly expressed in mature pollen but present at low levels in the other major organs (root, stem, and flower) (An et al. 1996; Huang et al. 1996). In addition, Arabidopsis ACT11 was more highly expressed in ovules and, to a certain extent, in embryos and young meristematic tissues (Huang et al. 1997). The specialized functional expression of actin in plants is cell- and tissue-specific for appropriate plant development (Li et al. 2005). In addition, the gene product contains conserved residues that are common to vegetative actins, including Val136, Val226, Ser239, and Ser366 (An et al. 1999).
We identified and characterized a full-length cDNA of a putative ACT gene isolated from the flowers of Ascocenda Princess Mikasa ‘Blue’. The conserved sequences and expression patterns confirmed that the cloned ACT gene from the petals of the orchid was a vegetative actin based on the classification of Arabidopsis actin. The discovery of differences in the amino acid sequences of the five orchid species may affect current vegetative and reproductive classifications. For the most part, the variability in the amino acid sequences of the orchid actins had no effect on the structure of actin. However, it may affect the activity of actin, including tissue-specific functions, in particular the Ser14 /Thr16 substitution in the S-loop of Ascocenda ACT and plant actins. Therefore, it is important to identify the role of each actin isoform.
Abbreviations
- ACT:
-
Actin
- D-loop:
-
DNase-I binding loop
- G-actin:
-
Globular actin
- H-loop:
-
Histidine-loop
- S-loop:
-
Serine-loop
- W-loop:
-
Tryptophan-loop
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109
An YQ, Huang S, McDowell JM, McKinney EC, Meagher RB (1996) Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell 8:15–30
An SS, Mopps B, Weber K, Bhattacharya D (1999) The origin and evolution of green algal and plant actins. Mol Biol Evol 16:275–285
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucl Acids Res 40:W597–W603
Aspenstrom P, Lindberg U, Karlsson R (1992) Site-specific amino-terminal mutants of yeast-expressed beta-actin. Characterization of the interaction with myosin and tropomyosin. FEBS Lett 303:59–63
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350
Betts MJ, Russell RB (2003) Amino acid properties and consequences of subsitutions. In: Barnes MR, Gray IC (eds) Bioinformatics for Geneticists. Wiley, United Kingdom
Bhattacharya D, Aubry J, Twait EC, Jurk S (2000) Actin gene duplication and the evolution of morphorlogical complexity in land plants. J Phycol 36:813–820
Bordoli L, Schwede T (2012) Automated protein structure modeling with SWISS-MODEL workspace and the protein model portal. Methods Mol Biol 857:107–136
Chik JK, Lindberg U, Schutt CE (1996) The structure of an open state of β-actin at 2.65 Å resolution. J Mol Biol 263:607–623
de Almeida Engler J, Rodiuc N, Smertenko A, Abad P (2010) Plant actin cytoskeleton re-modeling by plant parasitic nematodes. Plant Signaling Behav 5:213–217
Di Giammartino DC, Nishida K, Manley JL (2011) Mechanisms and consequences of alternative polyadenylation. Mol Cell 43:853–866
Dominguez R, Holmes KC (2011) Actin structure and function. Annu Rev Biophys 40:169–186
Dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ (2003) Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 83:433–473
Edgar RC (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res 32:1792–1797
Fedorov AA, Lappalainen P, Fedorov EV, Drubin DG, Almo SC (1997) Structure determination of yeast cofilin. Nat Struct Biol 4:366–369
Hennessey ES, Harrison A, Drummond DR, Sparrow JC (1992) Mutant actin: a dead end? J Muscle Res Cell Motil 13:127–131
Hennessey ES, Drummond DR, Sparrow JC (1993) Molecular genetics of actin function. Biochem J 282:657–671
Hu J, Lutz CS, Wilusz J, Tian B (2005) Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11:1485–1493
Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB (1996) The Arabidopsis thaliana ACT4/ACT12 actin gene subclass is strongly expressed throughout pollen development. Plant J 10:189–202
Huang S, An YQ, McDowell JM, McKinney EC, Meagher RB (1997) The Arabidopsis ACT11 actin gene is strongly expressed in tissues of the emerging inflorescence, pollen, and developing ovules. Plant Mol Biol 33:125–139
Hussey PJ, Ketelaar T, Deeks MJ (2006) Control of the actin cytoskeleton in plant cell growth. Annu Rev Plant Biol 57:109–125
Ji G, Zheng J, Shen Y, Wu X, Jiang R, Lin Y, Loke JC, Davis KM, Reese GJ, Li QQ (2007) Predictive modeling of plant messenger RNA polyadenylation sites. BMC Bioinformatics 8:43
Johannes FJ, Gallwitz D (1991) Site-directed mutagenesis of the yeast actin gene: a test for actin function in vivo. EMBO J 10:3951–3958
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. CABIOS 8:275–282
Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC (1990) Atomic structure of the actin:DNase I complex. Nature 347:37–44
Kandasamy MK, McKinney EC, Meagher RB (1999) The late pollen-specific actins in angiosperms. Plant J 18:681–691
Kandasamy MK, McKinney EC, Meagher RB (2009) A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21:701–718
Kandasamy MK, McKinney EC, Roy E, Meagher RB (2012) Plant vegetative and animal cytoplasmic actins share functional competence for spatial development with protists. Plant Cell 24:2041–2057
Kisselev AF, Songyang Z, Goldberg AL (2000) Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J Biol Chem 275:14831–14837
Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E (2010) A nucleotide state-sensing region on actin. J Biol Chem 285:25591–25601
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Lievens S, Goormachtig S, Holsters M (1997) Identification of differentially expressed mRNAs using the differential display technique, workshop on genome diversity and genome expression in plants EMBO course. Ghent, Belgium, pp 1–17
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3’ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327:94–97
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔC T Method. Methods 25:402–408
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucl Acids Res 39:D225–D229
McDowell JM, An YQ, Huang S, McKinney EC, Meagher RB (1996a) The Arabidopsis ACT7 actin gene is expressed in rapidly developing tissues and responds to several external stimuli. Plant Physiol 111:699–711
McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB (1996b) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142:587–602
McKinney EC, Meagher RB (1998) Members of the Arabidopsis actin gene family are widely dispersed in the genome. Genetics 149:663–675
Meagher RB, McKinney EC, Vitale AV (1999) The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet 15:278–284
Miralles F, Visa N (2006) Actin in transcription and transcription regulation. Curr Opin Cell Biol 18:261–266
Mounier N, Arrigo AP (2002) Actin cytoskeleton and small heat shock proteins: How do they interact. Cell Stress Chaperones 7:167–176
Nair UB, Joel PB, Wan Q, Lowey S, Rould MA, Trybus KM (2008) Crystal structures of monomeric actin bound to cytochalasin D. J Mol Biol 384:848–864
Novick P, Botstein D (1985) Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell 40:405–416
Otterbein LR, Graceffa P, Dominguez R (2001) The crystal structure of uncomplexed actin in the ADP state. Science 293:708–711
Oztug Durer ZA, Diraviyam K, Sept D, Kudryashov DS, Reisler E (2010) F-actin structure destabilization and DNase I binding loop: Fluctuations mutational cross-linking and electron microscopy analysis of loop states and effects on F-actin. J Mol Biol 395:544–557
Pearlman SM, Serber Z, Ferrell JE Jr (2011) A mechanism for the evolution of phosphorylation sites. Cell 147:934–946
Reece KS, McElroy D, Wu R (1990) Genomic nucleotide sequence of four rice (Oryza sativa) actin genes. Plant Mol Biol 14:621–624
Reedy M, Beall C, Fyrberg E (1991) Do variant residues among the six actin isoforms of Drosophila reflect functional specialization? Biophys J. 59:187(abstract).
Reisler E (1993) Actin molecular structure and function. Curr Opin Cell Biol 5:41–47
Rothnie HM, Reid J, Hohn T (1994) The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3’-end formation in plants. EMBO J 13:2200–2210
Shah DM, Hightower RC, Meagher RB (1982) Complete nucleotide sequence of a soybean actin gene. Proc Natl Acad Sci U S A 79:1022–1026
Shen Y, Ji G, Haas BJ, Wu X, Zheng J, Reese GJ, Li QQ (2008) Genome level analysis of rice mRNA 3’-end processing signals and alternative polyadenylation. Nucl Acids Res 36:3150–3161
Sheterline P, Clayton J, Sparrow J (1995) Actin. Protein Profile 2:1–103
Slajcherova K, Fiserova J, Fischer L, Schwarzerova K (2012) Multiple actin isotypes in plants: diverse genes for diverse roles. Front Plant Sci. doi:10.3389/fpls.2012.00226
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Vorobiev S, Strokopytov B, Drubin DG, Frieden C, Ono S, Condeelis J, Rubenstein PA, Almo SC (2003) The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci U S A 100:5760–5765
Wertman KF, Drubin DG, Botstein D (1992) Systematic mutational analysis of the yeast ACT1 gene. Genetics 132:337–350
Zheng X, Diraviyam K, Sept D (2007) Nucleotide effects on the structure and dynamics of actin. Biophys J 93:1277–1283
Acknowledgments
This project was supported by the Science Achievement Scholarship of Thailand (SAST) contract no.02/2551, Chulabhorn Research Institute (CRI) and Kasetsart University Research and Development Institute (KURDI), Thailand.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
(PPTX 286 kb)
Supplementary Table 1
(DOCX 37 kb)
Supplementary Table 2
(DOCX 546 kb)
Supplementary Table 3
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Pangjai, W., Huehne, P.S. Variable amino acid sequences in the S-loop and target binding site of vegetative actin in flowers of the Ascocenda orchid. J. Plant Biochem. Biotechnol. 24, 408–416 (2015). https://doi.org/10.1007/s13562-014-0291-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-014-0291-5