Abstract
The actin (ACT) family genes are essential for plant growth and development. However, the evolution and function of the ACT family within the Rosaceae species, particularly in pear, remain poorly understood. Here, we identified 41 ACT genes across five Rosaceae species based on phylogenetic and structural features that can be categorized into two primary groups: subfamily I (reproductive) and II (vegetative). Evolutionary analysis suggests that purifying selection played a crucial role in the evolution of the ACT family in Rosaceae, and whole genome duplication (WGD) and dispersed duplication led to the expansion of ACT genes. The pear genome contains twelve ACT genes, which can be classified into two groups based on their phylogeny and expression patterns: reproductive (PbrACT1-5) and vegetative (PbrACT6-12), further validating the reliability of the ACT family classification in Rosaceae. Expression analysis of twelve PbrACT genes across various pear tissues indicated that five genes from subfamily I (PbrACT1-5) were predominantly expressed in pollen tubes, with PbrACT1 exhibiting the highest level of expression. Knockdown of PbrACT1 expression in pear pollen tubes significantly diminished F-actin levels, triggered F-actin depolymerization, and resulted in pollen tube growth inhibition, indicating that PbrACT1 is essential for the formation of the microfilament skeleton during pear pollen tube growth. Overall, this study offers significant insights into the evolution and function of ACT genes in Rosaceae and enhances our understanding of PbrACT in microfilament formation in pear pollen tubes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The actin cytoskeleton, also known as microfilament (MF) or F-actin, is a filamentous polymer built from globular monomers called G-actin. F-actin plays a critical role in plant growth and development. For instance, Arabidopsis seedlings treated with the latrunculin (an inhibitor of actin polymerization) caused a significant reduction in cell expansion, resulting in short plants with distorted velvet hairs and smaller than normal lengths of leaves, roots, embryonic axes, and pollen tubes (Mathur and Hulskamp 2002). Besides, F-actin is also known to be involved in the response to abiotic stresses in plants. For example, treatment of Arabidopsis seedlings with 150 mM NaCl induced F-actin polymerization, whereas exposure to 250 mM NaCl led to F-actin degradation. Concurrently, the salt tolerance of seedlings was significantly diminished by treatment with latrunculin A, whereas treatment with phalloidin clearly increased this salinity tolerance (Wang et al. 2010). Similarly, heat stress has been demonstrated to trigger F-actin depolymerization in tobacco BY-2 cells (Malerba et al. 2010). Further, the actin cytoskeleton has been reported to play a vital role in plant immunity (Li and Staiger 2018). The plant microfilament skeleton can resist fungal invasion through dynamic rearrangement (Opalski et al. 2005). In contrast, the penetration efficiency of the fungus is significantly increased when the plants are treated with cytochalasin E, a drug known to disrupt microfilament rearrangements (Kobayashi and Hakuno 2003; Miklis et al. 2007). Moreover, the F-actin plays a critical function in pollen germination, cytoplasmic circulation, and polar growth (Justus et al. 2004; Li et al. 2001; Gibbon et al. 1999; Qu et al. 2014).
Actin is highly conserved and widespread in eukaryotic cells (McElroy et al. 1990; Chang and Huang 2015). In plants, the actin protein is encoded by multiple genes. For example, the ACT family of Arabidopsis contains eight functionally expressed genes that can be classified into two subfamilies based on their phylogenetic and expression profile: vegetative actins (ACT2, 7, and 8) are strongly expressed mainly in vegetative organs, and reproductive actins (ACT1, 3, 4, 11, and 12) are highly expressed in reproductive tissues (Meagher et al. 1999). The vegetative and reproductive actin are considered functionally redundant due to their strongly conserved amino acid sequences and high identity, such as ACT1 or ACT7 driven by the ACT2 promoter can backfill the act2-1 mutants (Gilliland et al. 2002). Notably, in Arabidopsis, the overexpression of ACT1, which is abundantly expressed in vegetative tissues, caused severe alterations in actin cytoskeletal structure and abnormality in plant tissues and cell types, ultimately leading to severe plant dwarfism. However, overexpression of ACT2 reached similar levels, producing non-significant phenotypic changes (Kandasamy et al. 2007, 2002). The above findings indicated that ACT genes of the same type were partially functionally redundant; however, different types of ACT genes, besides differing in expression patterns, were not functionally equivalent.
Actin, a key component of the cytoskeleton, is highly abundant in pollen, constituting approximately 2–20% of the total soluble proteins in pollen grains (Xu and Huang 2020). Pollen germination and subsequent growth of pollen tubes are critically dependent on the dynamic actin cytoskeleton (Chang and Huang 2015). For example, pharmacological disruption of F-actin organization and dynamics inhibits both pollen germination and pollen tube growth (Justus et al. 2004; Fang et al. 2018; Gibbon et al. 1999). Additionally, numerous actin-binding proteins (ABPs) participate in the modulation of microfilament dynamics (Ren and Xiang 2007; Zhang et al. 2023). For instance, Arabidopsis ADF7 and ADF10 are pollen-specific actin-depolymerizing factors, and their functional loss results in the inhibition of pollen tube growth (Zheng et al. 2013; Jiang et al. 2017). The Arabidopsis actin-interacting protein 1 (AIP1) modulates the dynamics and spatial organization of apical microfilaments within pollen tubes, thereby regulating pollen tube growth (Diao et al. 2020). Furthermore, alterations in actin dynamics induce apoptosis or programmed cell death in multiple organisms (Franklin-Tong and Gourlay 2008; Poulter et al. 2010; Thomas et al. 2006). For instance, S-RNase directly interacts with PbrActin1 in pear, leading to the depolymerization of the actin cytoskeleton and facilitating programmed cell death in self-incompatible pollen tubes (Liu et al. 2007; Chen et al. 2018). Similarly, S-RNase interacts with PavAct1 in sweet cherry pollen to disrupt coordinated actin dynamics in pollen tubes (Matsumoto and Tao 2012). Actin is essential for pollen tube growth; however, the investigation of actin function in Rosaceae is limited.
In this study, we characterized ACT genes across five Rosaceae genomes: Pear (Pyrus bretschneideri), peach (Prunus persica), strawberry (Fragaria vesca), apple (Malus domestica), and plum (Prunus mume). Our analysis encompassed their phylogeny, gene structure, motif identification, and the inference of their expansion history. Additionally, we examined the expression profiles of PbrACTs in various tissues and throughout pollen development. Using antisense oligodeoxynucleotides (as-ODN) to knock down the PbrACT1 expression, we found that PbrACT1 may be required for microfilament skeleton formation and growth in pear pollen tubes. In this work, we conducted a detailed study of ACT genes in five Rosaceae species and provided valuable insights for functional studies of ACT proteins in Rosaceae.
Materials and methods
Identification and physicochemical analysis of ACT proteins
Two methods were employed to identify ACT family genes across five Rosaceae genomes and obtain their sequences. Firstly, a conserved domain (Pfam: PF00022) search was conducted using the Hidden Markov Model with E-values < 1e−5 to detect candidate ACT members. Subsequently, Arabidopsis ACT protein sequences served as queries for BLASTP searches against the genome databases of the five Rosaceae species. Potential ACT genes were further validated by consulting the Pfam (http://pfam.xfam.org) and SMART (http://smart.embl-heidelberg.de/) databases. Finally, 41 ACT genes were characterized across the five Rosaceae genomes. The pear genome (v1.0) sequences were sourced from the Pear Genome Project (Wu et al. 2013), while the genome sequences for apple (v1.1), plum (v1.0), peach (v2.1), and strawberry (v4.0) were downloaded from the GDR (http://www.Rosaceae.org/). The ExPASy (http://web.expasy.org/protparam/) website was utilized to predict the physicochemical properties of ACT proteins (Artimo et al. 2012).
Phylogenetic, gene structure, and motif identification analysis
Phylogenetic trees were constructed using MEGA7.0 with the neighbor-joining method, and their reliability was confirmed through 1000 replicates of bootstrap analysis (Kumar et al. 2016). The gene structure and conserved motifs of ACT genes were identified using the GSDS (http://gsds.gao-lab.org/) and MEME (http://meme-suite.org/tools/meme) websites, respectively (Hu et al. 2015; Bailey et al. 2006). Multiple sequence alignments between ACTs from Arabidopsis and Rosaceae were performed using DNAMAN 6.0 with default settings to elucidate sequence features. Consensus sequence logos were generated with WebLogo (https://weblogo.berkeley.edu/logo.cgi) (Crooks et al. 2004). Identity of CDS sequences of ACT genes was analyzed by the Clustal Omega tool (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Madeira et al. 2022).
Gene duplication modes, syntenic, and Ka/Ks analysis
Initially, syntenic genes were analyzed with methods developed by PGGD (http://chibba.agtec.uga.edu/duplication/) (Lee et al. 2013). Subsequently, various duplication modes within the ACT family of five Rosaceae genomes were characterized with MCScanX (Wang et al. 2012). Ultimately, all identified duplication types and syntenic genes were visualized using Circos (Krzywinski et al. 2009). The calculate_Ka_Ks_pipeline was used to generate Ka and Ks values and Ka/Ks ratio (Qiao et al. 2019).
Expression analysis of ACTs in various pear tissues
Different pear tissues (fruits, leaves, stems, roots, styles, and mature pollen) were collected at the fruit experimental field of Nanjing Agricultural University, China. Total RNA was extracted from these tissues using the RNA isolation Kit (Vazyme, China). The RNA samples were then reverse transcribed following the protocol provided with the HiScript® III RT SuperMix for qPCR kits (Vazyme, China). The PbrACTs-specific primers were designed by Primer 5.0, and their specificity was verified in the pear genome website (Table S1). Due to the 100% sequence identity in the CDS among PbrACT9, 10, 11, and 12, and the 99.21% identity between PbrACT6 and 7, which precluded the design of specific primers, co-primers were employed (Table S2). Quantitative real-time PCR (qRT-PCR) was conducted on a LightCycler® 480 II (Roche, Germany) using SYBR qPCR Master Mix (Vazyme, China), with PbrUBQ genes as internal control. Expression data were analyzed using the 2–ΔΔCt method.
The antisense oligodeoxynucleotide (as‑ODN) assays
The as-ODN assays were conducted as previously described (Chen et al. 2018; Zhang et al. 2024). RNAfold (https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) was utilized to design ODN primers for PbrACT1 (Table S1). The pollen medium was composed of 0.5 mM Ca(NO3)2, 5 mM 1.5 mM H3BO3, 2-(N-morpholino) ethanesulfonic acid hydrate, and 450 mM sucrose, with the pH adjusted to 6.2 using Tris. Primers with a final concentration of 50 µM ODN were mixed with Lipofectamine 2000 (Thermo Fisher Scientific, USA) and incubated for 15 min. The mixture was then added to the pollen medium that had been pre-incubated for 45 min, followed by further incubation for 2 h. Pollen samples were imaged with a Nikon Eclipse E100 microscope (Tokyo, Japan), and the lengths of at least 50 pollen tubes were measured with Image ProPlus 6.0 software (Media Cybernetics, USA).
F-actin visualization and relative content assay in pollen tubes
F-actin imaging and relative content assays in pear pollen tubes were performed as previously described (Chen et al. 2018). Method for microfilament imaging is summarized as follows: Firstly, pollen tubes were fixed in 4% paraformaldehyde for 1 h following 10 min treatment with 200 µM MES-buffered saline. Subsequently, samples were washed three times with PBS (pH = 6.8). Samples were then stained with 5 µM FITC-phalloidin overnight, followed by three washed with PBS. Finally, samples were anti-fluorescently sealed and observed by LSM800. For F-actin relative content assays, the procedure is as follows: pollen samples were fixed for 1 h and washed three times with PBS. Samples were then stained with 5 µM FITC-phalloidin and 5 µM ethidium bromide (EB) after overnight incubation. Following three washes with PBS, the excess fluorescence was eliminated with methanol. The fluorescence intensity was measured by fluorescence spectrometry. F-actin content was calculated as the ratio of FITC-phalloidin fluorescence (excitation wavelength: 492 nm; emission wavelengths: 514 nm) to EB fluorescence (excitation wavelength: 513 nm; emission wavelengths: 615 nm).
Statistical analysis
Experiments were conducted with at least three replicates, and data were analyzed using GraphPad Prism 6.01. Statistical differences between the two groups were calculated using Student’s t-test.
Results
Identification and phylogenetic analysis of ACTs
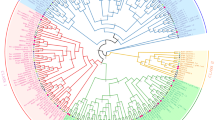
A total of 41 ACTs were characterized in five Rosaceae species (Fig. 1 and Table S3). Specifically, 12 ACTs were identified in pear (PbrACTs), 6 in strawberry (FveACTs), 6 in peach (PpeACTs), 11 in apple (MdoACTs), and 6 in plum (PmuACTs). Notably, the PbrACTs exhibited uneven distribution across the pear chromosome, with five genes located on chromosome 15. Additionally, PbrACT9 (Chr15: 33,484,562–33,486,017), PbrACT10 (Chr15: 33,428,764–33,430,219), PbrACT11 (Chr15: 33,284,053–33285508), and PbrACT12 (Chr15: 33,340,016–33341471) were found to comprise 377 amino acids (aa), possess an isoelectric point (PI) of 5.31, and a molecular weight (MW) of 41.74 kDa (Table 1, S3). It was further determined that these four genes are identical and are located in neighboring regions of the same chromosome.
Phylogenetic analysis of the ACT family proteins in Rosaceae and Arabidopsis. The phylogenetic tree was constructed based on full-length protein sequences using the neighbor-joining method in MEGA 7.0, with bootstrap analysis assessed through 1000 replicates. The red and blue backgrounds denote the two subfamilies (I and II) of ACT proteins. The species abbreviations and their corresponding colors are as follows: At for Arabidopsis thaliana (black), Pbr for Pyrus bretschneideri (red), Mdo for Malus domestica (green), Ppe for Prunus persica (yellow), Fve for Fragaria vesca (blue), and Pmu for Prunus mume (pink)
Based on the neighbor-joining evolutionary tree, the ACT family members were classified into two subfamilies (subfamily I and II). Notably, subfamily I contains the largest number of ACT genes, with only five ACTs in pear (Fig. 1). Conversely, subfamily II formed a smaller clade, comprising 16 members, including seven ACTs from pear (Fig. 1).
Conserved motif and exon/intron analysis
To investigate the structural diversity of ACT genes in Rosaceae, an exon/intron analysis of the gene sequences was performed. The results indicated that most ACT genes possess four exons, with the exception of nine apple genes (MdoACT1/3/5–11), which contain three exons (Fig. 2a). MEME website was utilized to characterize conserved motifs within ACT proteins. The analysis predicted five motifs for all ACT proteins, except for MdoACT1, MdoACT3, and MdoACT5-11, which exhibited four motifs (Fig. 2b, Table S4), underscoring the relative conservation of ACT proteins during evolution.
Exon–intron (a) and conserved motifs (b) analysis of ACT proteins in five Rosaceae species and Arabidopsis according to phylogenetic relationships. Neighbor-joining tree was constructed using MEGA 7.0, with bootstrap analysis performed on 1000 replicates and a scale bar representing 0.01 substitutions per site. Exons, introns, and untranslated regions are represented by red boxes, black lines, and blue boxes, respectively. Five conserved motifs were identified by MEME, with boxes of different colors representing distinct motifs. The red and blue backgrounds denote the two subfamilies (I and II) of ACT proteins in Arabidopsis and Rosaceae, respectively.
To understand the structural characteristics of ACT proteins, we conducted an identity analysis on the amino acid sequence of ACT proteins in Arabidopsis and Rosaceae. Our findings revealed a high degree of conservation in ACT protein sequences, with identity percentages ranging from 91.25 to 100% compared to the amino acid sequence of Arabidopsis (Table S5). Similarly, the amino acid sequence identity of ACT proteins within pear, apple, peach, plum, and strawberry exhibited high conservation, with identity percentages ranging from 94.96 to 100%, 94.72 to 100%, 96.02 to 99.2%, 95.76 to 99.2%, and 95.76 to 99.47%, respectively (Table S5). Furthermore, we performed multiple sequence alignments for all ACT proteins under investigation. The alignments demonstrated a striking consistency in the amino acid sites between ACT proteins in Rosaceae and Arabidopsis, with the notable exception of the N-terminal region in nine apple proteins (MdoACT1, 3, 5–11), which exhibited a deletion of 17 amino acids (Fig. S1).
Physicochemical analysis of ACT proteins
To investigate the function of ACT proteins in Rosaceae, a comprehensive analysis of their physicochemical properties was performed (Table 1). We found that the sequence length of ACT proteins varied from 360 to 377 aa, and most ACTs contain 377 aa. The PI of all ACT proteins are acidic, suggesting that ACT proteins from the Rosaceae family are rich in acidic amino acids. Additionally, the MW of all ACT proteins ranges from 39.92 to 41.81 kDa (Table 1). The negative and positive scores of the grand average of hydropathicity (GRAVY) score correspond to hydrophilicity and hydrophobicity, respectively. The findings revealed that all ACT proteins exhibited negative GRAVY score, suggesting that these proteins possess hydrophilic characteristics. Furthermore, the aliphatic indexes of all ACT proteins ranged from 83.05 to 86.42, which indicates that they are thermally stable (Table 1).
Evolutionary pattern analysis of ACT genes
Several types of gene duplication contribute to the expansion of gene families (Qiao et al. 2019). Our analysis revealed that five duplication types drive the expansion of the ACT family: 59% WGD, 20% dispersal, 12% transposition, 7% proximal, and 2% tandem (Fig. 3a). Notably, WGD was identified in five Rosaceae species. Specifically, 90.9% of ACT genes in apple, 80% in pear, 50% in peach, and 50% in plum were replicated and retained from WGD, while only 33.3% in strawberry (Fig. 3b, Table S6). Furthermore, the percentage of dispersed duplication (DSD) in pear (8.3%), strawberry (50%), peach (33.3%), and plum (33.3%) was assessed (Fig. 3b, Table S6). Transposed duplication was identified at rates of 16.7% in peach, 16.7% in strawberry, 16.7% in plum, and 8.3% in pear (Fig. 3b, Table S6). These findings indicate that WGD significantly affects the expansion of the ACT family in apple, pear, peach, and plum, while DSD plays a key role in the expansion of the strawberry ACT family.
Statistical analysis of duplication types within the ACT family members in Rosaceae species. a Statistics of ACT genes from different duplication types in Rosaceae genomes. b Statistics of ACT genes from different duplication types in each Rosaceae genomes. The MCScanX software was utilized to analyze the various duplication patterns within the ACT family
In this study, collinearity maps were generated for each Rosaceae species to investigate the evolution of the ACT genes. The analysis revealed that PbrACTs are distributed across seven chromosomes in pear, with eleven syntenic pairs identified (Fig. 4). Among them, eight, six, and seven homozygous gene pairs were detected in PpeACT, FveACT, and PmuACT, respectively (Fig. 4). Furthermore, the distribution of eleven MdoACTs was mapped onto eight chromosomes in apple, with twelve syntenic pairs identified (Fig. 4).
Intragenic collinearity and duplication type analysis of the ACT family in Rosaceae species. a Chromosomal localization and intra-genomic collinearity of ACT genes in each Rosaceae genome. Gene pairs connected by lines of different colors represent different duplication types (WGD, whole genome duplication; TD, tandem duplication; TRD, transposed duplication; PD, proximal duplication; DSD, dispersed duplication). b Statistical analysis of various duplication types in five species, including P. bretschneideri, M. domestica, P. persica, P. mume, and F. vesca. The five colored rectangles represent different types of gene duplication: WGD (red), DSD (black), PD (yellow), TD (blue), and TRD (green). The Y-axis indicates the number of duplicated gene pairs
To elucidate the selective pressures shaping the evolution of the ACT family, we computed Ks values and Ka/Ks ratios for ACT gene pairs. The results show that all Ka/Ks ratios are less than 1 (Fig. 5a, Table S7), indicating that the ACT family has undergone purifying selection during evolution. Furthermore, Ks values were utilized to infer the evolutionary history of duplication events. The results show that Ks values for all duplicated gene pairs range from 0.0343 to 4.1280 (Table S7). As shown in Fig. 5b and Table S7, the Ks values for gene pairs in pear and apple predominantly corresponded to recent (Ks ~ 0.15–0.3) and ancient (Ks ~ 1.5–1.8) WGD events, whereas peach, plum, and strawberry are mainly associated with ancient WGD. The findings align with previous studies that pear and apple have also experienced recent WGD events compared to strawberry, peach, and plum. Notably, gene pairs with higher Ks values, such as PbrACT5-PbrACT3 (Ks = 3.96689), MdoACT2-MdoACT8 (Ks = 3.91788), and MdoACT7-MdoACT2 (Ks = 3.9353), suggest that they may have originated from more ancient duplication event (Fig. 5b, Table S7).
Ka/Ks ratios (a) and Ks values (b) for duplicated gene pairs within the ACT family in five Rosaceae species, including P. bretschneideri (PbrACTs), M. domestica (MdoACTs), P. persica (PpeACTs), P. mume (PmuACTs), and F. vesca (FveACTs). The X-axis represents five different species, while the Y-axis denotes the Ka/Ks ratio or Ks values
Expression patterns of PbrACT genes
The qRT-PCR method was performed to assess the expression profiles of PbrACTs in six pear tissues. The findings revealed that five PbrACT genes (PbrACT1-5) were strongly expressed in pollen tubes (Fig. 6a). Additionally, the transcriptome data and qRT-PCR results indicated that PbrACT1 exhibited the highest expression level in pollen tubes compared to other PbrACT genes (Fig. 6b, c). Based on previous studies, Arabidopsis ACT1 and ACT3 exhibit a preferential accumulation at high levels in mature pollen (An et al. 1996). In this study, the significant expression of PbrACT1 in pollen underscores its pivotal role in pear reproductive development.
Expression patterns of twelve PbrACT genes. a Expression analysis of PbrACTs in different pear tissues by qRT-PCR. b Expression heatmap of PbrACT genes during pollen growth. Red and blue indicate high and low expression, respectively. The three stages of pear pollen growth include mature pollen grains (MP), hydrated pollen (HP), and pollen tubes growing 6 h after hydration (PT). c Expression of five PbrACTs in pollen tubes analyzed by qRT-PCR
PbrACT1 controlling pollen tube growth
F-actin is crucial for pollen tube growth (Qu et al. 2014; Zhang et al. 2018; Susaki et al. 2023). To elucidate the role of PbrACT1, we utilized the as-ODN assay, a gene knockdown technique, to silence the expression of PbrACT1 in pear pollen tubes. When the expression of PbrACT1 was dramatically reduced after as-ODN treatment, pollen tube growth was markedly inhibited (Fig. 7a–c), indicating that PbrACT1 contributes to pollen tube growth. Subsequently, we observed a significant increase in the rate of F-actin depolymerization in pollen tubes after as-ODN treatment (Fig. 7d, e), along with a significant decrease in F-actin levels (Fig. 7f). These results indicate that the knockdown of PbrACT1 expression reduces the F-actin content within pollen tubes, thereby inhibiting pollen tube growth. In conclusion, the microfilament skeleton plays a critical role in the growth of pear pollen tubes, and PbrACT1 contributes importantly to the formation of the microfilament skeleton.
PbrACT1 promotes pear pollen tube growth. a Pollen tube growth was inhibited by as-ODN-PbrACT1 treatment. Bar = 100 µm. b Expression level of PbrACT1 was decreased by as-ODN treatment. c Measurement of pollen tube length. d Phenotypes of FITC-phalloidin staining in pollen tubes under as-ODN treatment. Bars = 10 μm. e Statistics of F-actin depolymerization rate. f Measurement of relative F-actin levels. Significant differences (p < 0.01) by Student’s t-test indicated as “**”
Discussion
F-actin is an important component of the cytoskeleton and plays critical roles in various aspects of plant growth, including root growth, pollen germination and growth, and immune responses (Nishimura et al. 2003; Justus et al. 2004; Li et al. 2001; Li and Staiger 2018; Zepeda et al. 2014; Zhang et al. 2023). ACT is essential for F-actin formation, and the ACT gene family members have been characterized and analyzed in many species (Meagher et al. 1999; Zhang et al. 2010; Li et al. 2005; McElroy et al. 1990). However, the characterization and functional analysis of the ACT family in Rosaceae remain limited. In this study, we characterized 41 ACT genes within five Rosaceae species (Fig. 1 and Table S3) and investigated that PbrACT1 affects the growth of pear pollen tubes by regulating F-actin levels.
Phylogenetic relationships provided new insights into the evolution and genetic diversity of different gene family members (Smith et al. 2008; Kou et al. 2020). Phylogenetic analysis revealed that ACT genes in Rosaceae can be divided into two subgroups (Fig. 1), which classification is consistent with that reported on Arabidopsis (Kandasamy et al. 2007, 2009). Phylogenetic analysis also demonstrated that ACTs with similar gene structures and conserved motifs cluster together (Fig. 2). Furthermore, amino acid sequence alignment and identity analysis indicated that the Rosaceae ACT protein sequences are highly conserved (Fig. S1), with identity ranging from 94.72 to 100% (Table S5). Similarly, the divergence between vegetative and reproductive actins at the amino acid sequence level in Arabidopsis ranged from 4 to 7% (Kandasamy et al. 2007). The findings suggest that the ACT proteins exhibit a high degree of conservation.
Different patterns of gene duplication within plant genomes play distinct roles in the expansion of gene families (Qiao et al. 2019). Duplication pattern analysis indicates that the expansion of the ACT family in apple, peach, pear, and plum is primarily attributed to WGD, whereas DSD mainly contributes to the expansion of the ACT family in strawberry (Fig. 3 and Table S6). In summary, the expansion of the ACT family in Rosaceae has undergone various duplication types, with WGD and DSD collectively accounting for approximately 79% of the ACT family genes and serving as the predominant forces in its expansion (Fig. 3a). Previous studies have demonstrated that pear and apple have experienced both recent WGD (30–45 MYA, Ks ~ 0.15–0.3) and ancient WGD (~ 140 MYA, Ks ~ 1.5–1.8) events (Wu et al. 2013; Velasco et al. 2010). However, peach, plum, and strawberry have not undergone a recent WGD event. Ks value analysis revealed that most Ks values in pear and apple are predominantly associated with the two peaks corresponding to WGD events, while the Ks values for peach, plum, and strawberry are mainly distributed around the Ks values of ancient WGD events (Fig. 5b and Table S7). Consequently, the recent WGD events have contributed to the expansion of the ACT family in pear and apple. These findings elucidate why the number of ACT genes in pear (12) or apple (11) is nearly double that observed in peach (6), strawberry (6), and plum (6).
Gene expression patterns are often correlated with gene function (Kou et al. 2020). For instance, Arabidopsis ACT1 and ACT3 are strongly expressed mainly in pollen, with evidence indicating their distinct and crucial roles in the plant cytoskeleton (An et al. 1996; Vitale et al. 2003). The PtrACT1 gene is mainly expressed in mature xylem fiber cells, suggesting that it may regulate the formation of mature xylem in the trunk (Zhang et al. 2010). Similarly, the GhACT1 gene is mainly expressed in fibroblasts and played an essential function in fiber elongation (Li et al. 2005). In this study, the pear genome contains twelve ACT genes, which were categorized into two main groups based on their expression patterns: reproductive (PbrACT1-5) and vegetative (PbrACT6-12) (Fig. 6). Phylogenetic and expression analyses of PbrACT genes have further validated the classification of the ACT family in Rosaceae (Fig. 1). This study focused on PbrACT genes that are pivotal in pear reproduction. Transcriptome data and qRT-PCR results indicated that five PbrACTs (PbrACT1-5) exhibit low expression levels in various tissues but demonstrate strong expression in pollen tubes, with PbrACT1 showing the highest expression level (Fig. 6). These findings suggest that PbrACT1 plays an essential role in pear pollen tube growth.
F-actin plays an essential function in plant polar growth (Qu et al. 2020, 2014; Zhang et al. 2018; Kandasamy et al. 2009; Numata et al. 2022; Zhou et al. 2010). For instance, the Arabidopsis mutant act2-1 exhibits reduced root hair length and markedly enlarged root hair bases compared to the wild type (Gilliland et al. 2002). Similarly, the mutant der1, which is associated with the microfilament backbone, has been found to display an abnormally enlarged midsection of the root hairs (Ringli et al. 2002). Besides, much pharmacological evidence indicates that disruption of actin structure inhibits pollen germination and pollen tube growth, underscoring the necessity of an intact and dynamic actin cytoskeleton for proper pollen development (Gibbon et al. 1999; Xu and Huang 2020; Gossot and Geitmann 2007). For example, treatment with cytochalasin B, an inhibitor of actin polymerization, inhibits apple and pear pollen germination and pollen tube growth (Fang et al. 2018; Liu et al. 2008). Further, genetic evidence indicates that individual knockdown of the four ACT genes (ACT1, 3, 4, and 12) did not result in any apparent sterility phenotypes, but simultaneous silencing of these four ACTs through RNA interference led to significant reproductive defects (Pawloski et al. 2006). Surprisingly, the loss of ACT11 function promotes pollen tube growth, possibly as a compensatory response to reduced F-actin levels (Chang and Huang 2015). Consequently, the F-actin is essential for pollen tube growth. In this study, our findings indicate that PbrACT1 exerts a promotional influence on the growth of pollen tubes. When the expression of PbrACT1 in pollen tubes was reduced by as-ODN treatment, the growth of pollen tubes was inhibited (Fig. 7a–c). Furthermore, the knockdown of PbrACT1 expression led to a significant enhancement of F-actin depolymerization and a decrease in F-actin levels in pear pollen tubes (Fig. 7d–f). Consequently, PbrACT1 is critical for F-actin formation during pear pollen tube growth. Overall, our results contribute to a comprehensive understanding of the ACT family in Rosaceae, elucidating the pivotal role of PbrACT1 in pollen tube growth and offering a valuable foundation for future research on the functions of other actin proteins within the Rosaceae.
Conclusion
In this study, we characterized 41 ACT genes in five Rosaceae genomes, which were divided into two subfamilies. Subsequent evolutionary analyses revealed that purifying selection played a crucial role in the evolution of the ACT family in Rosaceae, with whole genome duplication and dispersed duplication leading to the expansion of ACT genes. Furthermore, PbrACT genes were classified into two major groups based on phylogenetic relationships and expression patterns: reproductive (PbrACT1-5) and vegetative (PbrACT6-12). Notably, PbrACT1 exhibits strong expression in pollen tubes and confirms its essential role in F-actin formation during pear pollen tube growth. In summary, these findings contribute to a comprehensive understanding of the Rosaceae ACT gene family and offer valuable insights for the functional studies of ACT proteins in Rosaceae.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
An YQ, Huang S, McDowell JM, McKinney EC, Meagher RB (1996) Conserved expression of the Arabidopsis ACT1 and ACT3 actin subclass in organ primordia and mature pollen. Plant Cell 8(1):15–30. https://doi.org/10.1105/tpc.8.1.15
Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:W597-603. https://doi.org/10.1093/nar/gks400
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369-373. https://doi.org/10.1093/nar/gkl198
Chang M, Huang S (2015) Arabidopsis ACT11 modifies actin turnover to promote pollen germination and maintain the normal rate of tube growth. Plant J 83(3):515–527. https://doi.org/10.1111/tpj.12910
Chen J, Wang P, de Graaf BHJ, Zhang H, Jiao H, Tang C, Zhang S, Wu J (2018) Phosphatidic acid counteracts S-RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell 30(5):1023–1039. https://doi.org/10.1105/tpc.18.00021
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190. https://doi.org/10.1101/gr.849004
Diao M, Li X, Huang S (2020) Arabidopsis AIP1-1 regulates the organization of apical actin filaments by promoting their turnover in pollen tubes. Sci China Life Sci 63(2):239–250. https://doi.org/10.1007/s11427-019-9532-0
Fang K, Zhang Q, Yang R, Cao Q, Qin L (2018) Cytochalasin B treatment of apple (Malus pumila mill.) pollen tubes alters the cytoplasmic calcium gradient and causes major changes in the cell wall components. Russ J Plant Physiol 65(3):384–393. https://doi.org/10.1134/S1021443718030111
Franklin-Tong VE, Gourlay CW (2008) A role for actin in regulating apoptosis/programmed cell death: evidence spanning yeast, plants and animals. Biochem J 413(3):389–404. https://doi.org/10.1042/BJ20080320
Gibbon BC, Kovar DR, Staiger CJ (1999) Latrunculin B has different effects on pollen germination and tube growth. Plant Cell 11(12):2349–2363. https://doi.org/10.1105/tpc.11.12.2349
Gilliland LU, Kandasamy MK, Pawloski LC, Meagher RB (2002) Both vegetative and reproductive actin isovariants complement the stunted root hair phenotype of the Arabidopsis act2-1 mutation. Plant Physiol 130(4):2199–2209. https://doi.org/10.1104/pp.014068
Gossot O, Geitmann A (2007) Pollen tube growth: coping with mechanical obstacles involves the cytoskeleton. Planta 226(2):405–416. https://doi.org/10.1007/s00425-007-0491-5
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Jiang Y, Wang J, Xie Y, Chen N, Huang S (2017) ADF10 shapes the overall organization of apical actin filaments by promoting their turnover and ordering in pollen tubes. J Cell Sci 130(23):3988–4001. https://doi.org/10.1242/jcs.207738
Justus CD, Anderhag P, Goins JL, Lazzaro MD (2004) Microtubules and microfilaments coordinate to direct a fountain streaming pattern in elongating conifer pollen tube tips. Planta 219(1):103–109. https://doi.org/10.1007/s00425-003-1193-2
Kandasamy MK, McKinney EC, Meagher RB (2002) Functional nonequivalency of actin isovariants in Arabidopsis. Mol Biol Cell 13(1):251–261. https://doi.org/10.1091/mbc.01-07-0342
Kandasamy MK, Burgos-Rivera B, McKinney EC, Ruzicka DR, Meagher RB (2007) Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19(10):3111–3126. https://doi.org/10.1105/tpc.107.052621
Kandasamy MK, McKinney EC, Meagher RB (2009) A single vegetative actin isovariant overexpressed under the control of multiple regulatory sequences is sufficient for normal Arabidopsis development. Plant Cell 21(3):701–718. https://doi.org/10.1105/tpc.108.061960
Kobayashi I, Hakuno H (2003) Actin-related defense mechanism to reject penetration attempt by a non-pathogen is maintained in tobacco BY-2 cells. Planta 217(2):340–345. https://doi.org/10.1007/s00425-003-1042-3
Kou X, Liu Q, Sun Y, Wang P, Zhang S, Wu J (2020) The peptide PbrPSK2 from phytosulfokine family induces reactive oxygen species (ROS) production to regulate pear pollen tube growth. Front Plant Sci 11:601993. https://doi.org/10.3389/fpls.2020.601993
Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645. https://doi.org/10.1101/gr.092759.109
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lee TH, Tang H, Wang X, Paterson AH (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41:D1152-1158. https://doi.org/10.1093/nar/gks1104
Li J, Staiger CJ (2018) Understanding cytoskeletal dynamics during the plant immune response. Annu Rev Phytopathol 56:513–533. https://doi.org/10.1146/annurev-phyto-080516-035632
Li Y, Zee SY, Liu YM, Huang BQ, Yen LF (2001) Circular F-actin bundles and a G-actin gradient in pollen and pollen tubes of Lilium davidii. Planta 213(5):722–730. https://doi.org/10.1007/s004250100543
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17(3):859–875. https://doi.org/10.1105/tpc.104.029629
Liu ZQ, Xu GH, Zhang SL (2007) Pyrus pyrifolia stylar S-RNase induces alterations in the actin cytoskeleton in self-pollen and tubes in vitro. Protoplasma 232(1–2):61–67. https://doi.org/10.1007/s00709-007-0269-4
Liu Z, Jiang X, Xu G, Zhang S (2008) Effects of cytochalasin B and phalloidin on Pyrus pyrifolia self-incompatible pollen germination and tube growth. Acta Botan Boreali-Occiden Sin 28(12):2393–2399
Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50(W1):W276–W279. https://doi.org/10.1093/nar/gkac240
Malerba M, Crosti P, Cerana R (2010) Effect of heat stress on actin cytoskeleton and endoplasmic reticulum of tobacco BY-2 cultured cells and its inhibition by Co2+. Protoplasma 239(1–4):23–30. https://doi.org/10.1007/s00709-009-0078-z
Mathur J, Hulskamp M (2002) Microtubules and microfilaments in cell morphogenesis in higher plants. Curr Biol 12(19):R669-676. https://doi.org/10.1016/s0960-9822(02)01164-8
Matsumoto D, Tao R (2012) Isolation of pollen-expressed actin as a candidate protein interacting with S-RNase in Prunus avium L. J Jpn Soc Hortic Sci 81(1):41–47. https://doi.org/10.2503/jjshs1.81.41
McElroy D, Rothenberg M, Reece KS, Wu R (1990) Characterization of the rice (Oryza sativa) actin gene family. Plant Mol Biol 15(2):257–268. https://doi.org/10.1007/BF00036912
Meagher RB, McKinney EC, Vitale AV (1999) The evolution of new structures: clues from plant cytoskeletal genes. Trends Genet 15(7):278–284. https://doi.org/10.1016/s0168-9525(99)01759-x
Miklis M, Consonni C, Bhat R, Lipka V, Schulze-Lefert P, Panstruga R (2007) Barley MLO modulates actin-dependent and actin-independent antifungal defense pathways at the cell periphery. Plant Physiol 144(2):1132–1143. https://doi.org/10.1104/pp.107.098897
Nishimura T, Yokota E, Wada T, Shimmen T, Okada K (2003) An Arabidopsis ACT2 dominant-negative mutation, which disturbs F-actin polymerization, reveals its distinctive function in root development. Plant Cell Physiol 44(11):1131–1140. https://doi.org/10.1093/pcp/pcg158
Numata T, Sugita K, Ahamed Rahman A, Rahman A (2022) Actin isovariant ACT7 controls root meristem development in Arabidopsis through modulating auxin and ethylene responses. J Exp Bot 73(18):6255–6271. https://doi.org/10.1093/jxb/erac280
Opalski K, Schultheiss H, Kogel K, Hückelhoven R (2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41(2):291–303. https://doi.org/10.1111/j.1365-313X.2004.02292.x
Pawloski L, Kandasamy M, Meagher R (2006) The late pollen actins are essential for normal male and female development in Arabidopsis. Plant Mol Biol 62(6):881–896. https://doi.org/10.1007/s11103-006-9063-5
Poulter NS, Staiger CJ, Rappoport JZ, Franklin-Tong VE (2010) Actin-binding proteins implicated in the formation of the punctate actin foci stimulated by the self-incompatibility response in Papaver. Plant Physiol 152(3):1274–1283. https://doi.org/10.1104/pp.109.152066
Qiao X, Li Q, Yin H, Qi K, Li L, Wang R, Zhang S, Paterson AH (2019) Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol 20(1):38. https://doi.org/10.1186/s13059-019-1650-2
Qu X, Jiang Y, Chang M, Liu X, Zhang R, Huang S (2014) Organization and regulation of the actin cytoskeleton in the pollen tube. Front Plant Sci 5:786. https://doi.org/10.3389/fpls.2014.00786
Qu X, Wang Q, Wang H, Huang S (2020) Visualization of actin organization and quantification in fixed Arabidopsis pollen grains and tubes. Bio Protoc 10(1):e3509. https://doi.org/10.21769/BioProtoc.3509
Ren H, Xiang Y (2007) The function of actin-binding proteins in pollen tube growth. Protoplasma 230:171–182. https://doi.org/10.1007/s00709-006-0231-x
Ringli C, Baumberger N, Diet A, Frey B, Keller B (2002) ACTIN2 is essential for bulge site selection and tip growth during root hair development of Arabidopsis. Plant Physiol 129(4):1464–1472. https://doi.org/10.1104/pp.005777
Smith LL, Fessler JL, Alfaro ME, Streelman JT, Westneat MW (2008) Phylogenetic relationships and the evolution of regulatory gene sequences in the parrotfishes. Mol Phylogenet Evol 49(1):136–152. https://doi.org/10.1016/j.ympev.2008.06.008
Susaki D, Izumi R, Oi T, Takeuchi H, Shin JM, Sugi N, Kinoshita T, Higashiyama T, Kawashima T, Maruyama D (2023) F-actin regulates the polarized secretion of pollen tube attractants in Arabidopsis synergid cells. Plant Cell 35(4):1222–1240. https://doi.org/10.1093/plcell/koac371
Thomas SG, Huang S, Li S, Staiger CJ, Franklin-Tong VE (2006) Actin depolymerization is sufficient to induce programmed cell death in self-incompatible pollen. J Cell Biol 174(2):221–229. https://doi.org/10.1083/jcb.200604011
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagne D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R (2010) The genome of the domesticated apple (Malus x domestica Borkh.). Nat Genet 42(10):833–839. https://doi.org/10.1038/ng.654
Vitale A, Wu RJ, Cheng Z, Meagher RB (2003) Multiple conserved 5’ elements are required for high-level pollen expression of the Arabidopsis reproductive actin ACT1. Plant Mol Biol 52(6):1135–1151. https://doi.org/10.1023/b:plan.0000004309.06973.16
Wang C, Zhang L, Yuan M, Ge Y, Liu Y, Fan J, Ruan Y, Cui Z, Tong S, Zhang S (2010) The microfilament cytoskeleton plays a vital role in salt and osmotic stress tolerance in Arabidopsis. Plant Biol 12(1):70–78. https://doi.org/10.1111/j.1438-8677.2009.00201.x
Wang Y, Tang H, Debarry J, Tan X, Li J, Wang X, Lee T, Jin H, Marler B, Guo H, Kissinger J, Paterson A (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49. https://doi.org/10.1093/nar/gkr1293
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd). Genome Res 23(2):396–408. https://doi.org/10.1101/gr.144311.112
Xu Y, Huang S (2020) Control of the actin cytoskeleton within apical and subapical regions of pollen tubes. Front Cell Dev Biol 8:614821. https://doi.org/10.3389/fcell.2020.614821
Zepeda I, Sanchez-Lopez R, Kunkel JG, Banuelos LA, Hernandez-Barrera A, Sanchez F, Quinto C, Cardenas L (2014) Visualization of highly dynamic F-actin plus ends in growing Phaseolus vulgaris root hair cells and their responses to Rhizobium etli nod factors. Plant Cell Physiol 55(3):580–592. https://doi.org/10.1093/pcp/pct202
Zhang D, Du Q, Xu B, Zhang Z, Li B (2010) The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284(2):105–119. https://doi.org/10.1007/s00438-010-0552-5
Zhang R, Xu Y, Yi R, Shen J, Huang S (2023) Actin cytoskeleton in the control of vesicle transport, cytoplasmic organization, and pollen tube tip growth. Plant Physiol 193(1):9–25. https://doi.org/10.1093/plphys/kiad203
Zhang H, Liu X, Tang C, Lv S, Zhang S, Wu J, Wang P (2024) PbRbohH/J mediates ROS generation to regulate the growth of pollen tube in pear. Plant Physiol Bioch 207:108342. https://doi.org/10.1016/j.plaphy.2024.108342
Zhang S, Wang C, Xie M, Liu J, Kong Z, Su H (2018) Actin bundles in the pollen tube. Int J Mol Sci 19 (12). https://doi.org/10.3390/ijms19123710
Zheng Y, Xie Y, Jiang Y, Qu X, Huang S (2013) Arabidopsis actin-depolymerizing factor7 severs actin filaments and regulates actin cable turnover to promote normal pollen tube growth. Plant Cell 25(9):3405–3423. https://doi.org/10.1105/tpc.113.117820
Zhou Y, Yang Z, Guo G, Guo Y (2010) Microfilament dynamics is required for root growth under alkaline stress in Arabidopsis. J Integr Plant Biol 52(11):952–958. https://doi.org/10.1111/j.1744-7909.2010.00981.x
Acknowledgements
We thank Dr. Ma Yuehua for technical assistance in using confocal microscopy. Bioinformatic analysis was supported by the High-performance Computing Platform of the Bioinformatics Center of Nanjing Agricultural University.
Funding
This work was financially supported by the Jiangsu Agricultural Science and Technology Innovation Fund (CX(22)3044), the open funds of the National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops (Horti-KF-2023-05), Ningbo Key Laboratory of Characteristic Horticultural Crops in Quality Adjustment and Resistance Breeding (NBYYL2023001), Xinjiang Forestry and Fruit Industry Research System (XJLGCYJSTX05-2024-07), the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Tianshan Talents Program of Xinjiang Uygur Autonomous Region and the Earmarked Fund for China Agriculture Research System (CARS-28).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Data archiving statement
All PbrACT-related sequences were available from the Pear Genome Project (http://peargenome.njau.edu.cn), and the protein sequences of ACT members including apple, peach, strawberry, and plum were downloaded from the GDR (http://www.rosaceae.org/) database.
Additional information
Communicated by A.M. Dandekar.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Zhang, H., Tang, C. et al. Characterization of the actin (ACT) family in Rosaceae and role of PbrACT1 in pear pollen tube growth. Tree Genetics & Genomes 20, 12 (2024). https://doi.org/10.1007/s11295-024-01647-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-024-01647-9