Abstract
Artificial intelligence (AI) has had a significant impact on human life because of its pervasiveness across industries and its rapid development. Although AI has achieved superior performance in learning and reasoning, it encounters challenges such as substantial computational demands, privacy concerns, communication delays, and high energy consumption associated with cloud-based models. These limitations have facilitated a paradigm change in on-device AI processing, which offers enhanced privacy, reduced latency, and improved power efficiency through the direct execution of computations on devices. With advancements in neuromorphic systems, spiking neural networks (SNNs), often referred to as the next generation of AI, are currently in focus as on-device AI. These technologies aim to mimic the human brain efficiency and provide promising real-time processing with minimal energy. This study reviewed the application of SNNs in the analysis of biomedical signals (electroencephalograms, electrocardiograms, and electromyograms), and consequently, investigated the distinctive attributes and prospective future paths of SNNs models in the field of biomedical signal analysis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Artificial intelligence (AI) has developed rapidly, and it plays a pivotal role in driving innovation in various fields and has become a significant part of our daily lives. AI, which processes data by imitating the neurons of the human brain, exhibits excellent performance in learning and reasoning and is supported by the advancement of hardware and algorithms. Although AI exhibits superior performance in many problems, it incurs considerable computation and resource demands; furthermore, the sizes of the associated models are increasing.
In general, developed AI models are used as a method of delivering model output values for incoming data based on the cloud. Cloud-based AI models require communication with servers for data processing, and they encounter challenges, such as privacy concerns, communication delays, infrastructure costs, and high energy consumption. These limitations have prompted a shift toward on-device AI processing, which minimizes privacy and latency issues via the direct execution of computations on the devices. Lightweight model technologies, such as pruning [1], quantization [2], and knowledge distillation [3], as well as lightweight models, such as DenseNet [4], SqueezeNet [5], and MobileNet [6] have been extensively studied for on-device operation.
On-device processing enables AI applications to operate locally, thereby leveraging the computing power of the device. Moreover, on-device AI can significantly enhance user privacy and data security by processing data locally. In addition, it improves the response time and power efficiency while reducing latency because it preempts the necessity of the data to travel over the network to a server for processing. These advantages have propelled on-device AI to change the paradigm and pave the way for personalized applications.
Research on neuromorphic systems, which differ from the traditional von Neumann-structured computing systems, has been actively conducted along with the advancement of on-device AI. Neuromorphic systems are designed to physically implement neural networks in a way that imitates the operational principles of biological brains. By designing hardware that mimics the structure of biological neurons and synapses, these systems process information asynchronously through discrete spike events. This allows neuromorphic systems to handle event-driven sparse data more efficiently, offering significant improvements in processing speed and power efficiency for tasks. Several neuromorphic hardware systems have been developed over the years such as Intel’s Loihi [7], IBM’s TrueNorth [8], SpiNNaeker [9], and Neurogrid [10], providing real-time processing along with high energy efficiency.
Spiking neural networks (SNNs), which are called the next generation of AI models, are also being studied along with the development of neuromorphic systems. SNNs provide algorithms suitable for neuromorphic hardware and neuromorphic hardware provides a platform to run these networks efficiently. Neuromorphic computing and SNNs represent the forefront of creating computing systems that replicate the functionality of the human brain as well as their energy efficiency. Algorithms for SNNs include input encoding methods, neuron models, network architectures, and training methods. SNNs use biological neuron model and offers a more dynamic and energy-efficient approach compared to deep neural networks. These studies have aimed to overcome the limitations of traditional computing architectures by introducing models capable of real-time processing with minimal energy requirements.
In biomedical engineering, advancements of AI have significantly influenced many research areas, especially changing the analysis of biomedical signals such as electroencephalograms (EEG), electrocardiograms (ECG), and electromyograms (EMG). The transition from manual feature engineering to deep learning-based automatic feature extraction has facilitated significant advancements, consequently enabling more precise and efficient analysis of biological signals. In addition, AI enables the identification of relevant health indicators from complex biological data, thereby offering insights that can drive personalized healthcare solutions.
Healthcare AI models are implemented to process data by sending it to a model developed based on a cloud server. However, the server-based approach has several limitations, including the risk of user’s health data privacy and slow response times in emergencies, where rapid reaction is crucial. Additionally, the maintenance costs associated with servers can be substantial. Consequently, a shift towards on-device processing, where data is managed and analyzed directly on the device, can address these issues. The shift towards on-device AI and neuromorphic computing in healthcare underscores the broader trend of migrating towards personalized, efficient, and privacy-preserving technology solutions. This approach addresses the limitations of centralized data processing and paves the way for new possibilities for real-time personalized health monitoring and interventions.
From this perspective, research on SNNs, which offer faster real-time processing speeds and higher energy efficiency on-device compared to traditional deep learning models, is gaining interest in the field of biomedical signal analysis. This article reviews recent studies on SNNs models in the domain of biomedical signals, focusing the following parts: (1) background of SNNs architectures and training approaches; (2) review of SNNs studies in EEG signals; (3) review of SNNs studies in ECG signals; (4) review of SNNs studies in EMG signals; (5) a performance comparison of SNNs models in biomedical signal analysis. By comparing the characteristics of SNNs with those of traditional deep-learning models, this review highlighted the potential of SNNs to revolutionize the analysis and interpretation of biomedical signals in healthcare applications. This review will help researchers to develop SNNs model and provide a future direction for biomedical signals processing systems with power and energy-efficient models.
2 Spiking neural networks
The field of neuromorphic computing in computer science and engineering aims to create more efficient computing systems that mimic the structure and functionality of the human brain. A key aspect of neuromorphic computing is the use of SNNs, which model neuron behavior more closely with biological neurons by representing the information as discrete spikes.
SNNs provide a unique approach to the temporal dynamics of signals by using spikes, and spike neuron models generate spikes in response to input signals. This facilitates SNNs to process information in a time-dependent manner, rendering them particularly suitable for tasks involving temporal sequences or real-time processing. A notable advantage of SNNs is their high energy efficiency, due to the nature of their neurons being selectively activated in response to specific events. This spiking mechanism ensures that not all the neurons are concurrently active, thereby reducing power consumption. This feature is particularly advantageous for on-devices that necessitate energy conservation. By contrast, neurons in deep-learning networks are continuously operational, necessitating higher energy consumption, particularly in large-scale networks engaged in complex computations.
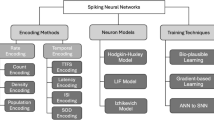
The key elements constituting SNNs include spike neuron models, spike encoding methods, and learning methods for encoded spike trains, as shown in Fig. 1. First of all, there are several models for representing spiking neurons. The Hodgkin-Huxley model [11] is a highly detailed and biophysically accurate representation of the electrical properties of excitable cells like neurons. This model explains the initiation and propagation of action potentials in neurons through the dynamics of ion channels.
The Integrate-and-Fire (IF) model [12] is a simple and idealized model of neuron dynamics, where the membrane potential integrates incoming signals until a threshold is reached, causing the neuron to fire a spike and then reset its potential. The Leaky IF (LIF) model extends the basic IF model by including a “leak” term that models the passive decay of the membrane potential over time. The inclusion of the leak term means that the membrane potential will naturally decay towards the resting potential if no input is present, making it a more realistic representation of biological neurons. The following equation represents the dynamics of the LIF neuron:
Where \(c\) represents the membrane time constant, \(V\left(t\right)\) indicates the membrane potential of the neuron at the time \(t\), \({V}_{reset}\) is the resting membrane potential, \(R\) indicates the membrane resistance, and \(I\left(t\right)\) represents the input to the neuron at the time \(t\). When \(V\left(t\right)\) reaches the threshold \({V}_{th}\), the neuron fires and the membrane potential \(V\left(t\right)\) is reset to \({V}_{reset}\). The Izhikevich model [13] is another neuron model that effectively combines biological plausibility with computational efficiency. By using a set of relatively simple differential equations, it can replicate a wide range of neuronal firing patterns, making it a valuable tool in both theoretical studies and practical applications of neural dynamics.
Spike-encoding methods are crucial for translating data into spike patterns that can be processed using SNNs. Spike encoding methods capture complex temporal relationships while reducing computational resources by encoding information using data patterns and timing. The representative spike-encoding methods include rate, temporal coding, delta modulation, and encoding layer. The rate encoding [14] is among the simplest and most intuitive methods, wherein the frequency of spikes represents the input signal magnitude. In this encoding method, an increased input value leads to a higher spike firing rate, while a lower input value results in a decreased firing rate. Temporal coding [15] represents information through the exact timing of spikes rather than their firing rate, relying on the relative timing of spikes to transmit information. This approach depends on the relative timing of spikes to transmit information. For instance, in a time-to-first spike mechanism, bright pixels are encoded as initial spikes, whereas dark inputs either spike later or not at all. The temporal encoding mechanisms allocate much more meaning to each individual spike compared to rate encoding.
The spike-timing-dependent plasticity (STDP) algorithm [16, 17] serves as a representative example, functioning as a learning rule in SNNs based on biological mechanisms seen in the brain. Synaptic weights are adjusted by the STDP mechanism based on the precise timing of incoming spikes. The STDP adjusts the synaptic weights based on the timing of the incoming spikes. This contrasts with the backpropagation algorithm utilized in deep learning, wherein the error gradients are calculated to optimize the network weights. Backpropagation provides a powerful and generalizable framework for deep learning; however, it is difficult to use for the training of spiking neurons. Spiking neurons communicate through discrete spikes or impulses, which are essentially binary events (neurons either fire or do not fire at a given time). This spiking mechanism is inherently nondifferentiable, rendering its direct application to the backpropagation algorithm challenging. Therefore, the STDP algorithm and various methods for spike learning have been proposed.
The fundamental principle of STDP is that changes in synaptic strength (synaptic weight) are determined by the time difference between presynaptic and postsynaptic spikes. When a presynaptic neuron fires just prior to a postsynaptic neuron within a specific time window, the synaptic strength is enhanced, referred to as long-term potentiation (LTP). When a presynaptic neuron fires just after a postsynaptic neuron, the synaptic strength is reduced, referred to as long-term depression (LTD). The change of synaptic weight is represented by the following equation:
Here, \(\varDelta w\) represents the change in synaptic weight, \({A}^{+}\)and \({A}^{-}\)are learning rate constants for potentiation and depression, respectively, \({\tau }^{+}\)and \({\tau }^{-}\) are the time constants for the decay of the learning window.
3 SNNs in biomedical signals
A type of artificial neural network, SNNs mimics the way biological neurons communicate, using discrete events referred to as spikes. Time series data inherently contains temporal patterns, and SNNs can encode information in the timing of spikes, thus having strength in capturing and processing these time patterns. The temporal and event-driven processing capabilities of SNNs have been studied in various fields of biosignals; in particular, many studies have been conducted based on EEG signals, which are closely related to spiking neurons in the brain. This section reviews research related to the application of SNNs to EEG, ECG, and EMG biosignals.
3.1 Electroencephalogram signal
The electrical activity of the brain is represented by the EEG signal, which is measured with electrodes positioned on the scalp and EEG signals reflect the aggregated synchronous activities of numerous neurons that have similar spatial orientations [18]. When neurons in the brain fire electrical impulses, they generate electrical fields that can be detected and recorded using these electrodes. EEG is used in numerous applications, from diagnosing neurological conditions like epilepsy and sleep disorders to cognitive neuroscience research and control signals in brain-computer interfaces (BCIs).
EEG-based SNNs algorithms are being studied in various fields such as epileptic detection, emotional recognition, auditory attention detection, and BCI. Table 1 summarizes the EEG-based SNNs studies. In studies related to the epileptic seizure detection research, Zhang et al. [19] introduced an EEG-based SSN (EESNN), which features a recurrent spiking convolution structure. This model was designed to capture the inherent temporal and biological characteristics of EEG signals, aiming to enhance the detection of epileptic seizures in different patients. EESNN achieved comparable or even better performance in cross-patient epileptic seizure detection than traditional artificial neural networks (ANNs), while also significantly reducing energy consumption. Burelo et al. [20] introduced a custom-designed SNNs for the detection of high-frequency oscillations (HFO) in scalp EEG recordings. The SNNs model features a two-layer network utilizing LIF neurons and synapses, employing delta modulator-based spike encoding method. The proposed SNNs achieved an 80% accuracy in associating HFO occurrences with active epilepsy and found a strong correlation between the HFO rate detected by the SNNs and seizure frequency. These results demonstrated the potential of SNNs for non-invasive epilepsy monitoring using compact, energy-efficient devices.
. Li et al. [21] introduced a fractal-SNNs scheme for recognizing emotions from EEG data, incorporating multi-scale temporal, spectral, and spatial (TSS) information. Their key innovation, the Fractal-SNN block, mimics biological neural structures with spiking neurons and a novel fractal rule, efficiently extracting distinctive TSS features. They also proposed the inverted drop-path training technique, enhancing generalization by allowing the network to learn from various sub-networks. Extensive experiments on public benchmark databases showed that their model outperforms other advanced methods in EEG-based emotion recognition, particularly under subject-dependent protocols. Xu et al. [22] proposed the Emo-EEGSpikeConvNet (EESCN) model featuring neuromorphic data generation and a neurospiking framework to classify EEG-based emotions. EEG signals are converted into 2D frames by the neuromorphic data generation module, which captures spatial and temporal information and reduces computational costs. Within the neurospiking framework, there is an encoding layer to convert EEG data into spikes, a spike convolution layer to extract features, and a fully connected layer to perform classification. The EESCN demonstrated high accuracy on the DEAP and SEED-IV datasets, outperforming existing SNNs methods. This model also achieved faster computation and lower memory usage, making it suitable for practical applications. Xu et al. [23] demonstrated that SNNs can effectively decode emotional brain networks from EEG data related to the viewing of emotional videos. The study found high decoding accuracies for these emotions using the SNNs with self-backpropagation, highlighting specific frequency bands as biological markers. By analyzing EEG signals while participants watched emotional videos, it identified distinct brain network activations for fear, sadness, and happiness. These findings support the potential of SNNs for effective emotion recognition and BCI applications. Luo et al. [24] proposed a novel approach for identifying emotional states based on EEG signals. EEG signal features were extracted using discrete wavelet transform, variance, and fast fourier transform algorithms, and extracted features were then used to train NeuCube [25]-based SNNs model. NeuCube, an architecture for spatio- and spectro-temporal brain data, includes a 3D SNN reservoir module, an input data module, and an output function module. When compared to existing benchmark methods, the proposed method achieved high accuracy in emotion recognition classification.
Studies have also been conducted on SNNs for detecting auditory attention. Cai et al. [26] introduced BSAnet, which is an innovative end-to-end modular architecture designed to decode auditory attention using EEG signals. BSAnet consists of biologically plausible LIF neurons along with an event-driven neural encoder. In addition, it incorporates a temporal attention mechanism and a recurrent spiking layer, which enhance its ability to represent detailed temporal patterns in brain signals. This architecture achieved improved performance in auditory spatial attention detection tasks compared to other state-of-the-art systems, confirming its efficacy in realistic brain-like information processing. Faghihi et al. [27] presented a neuroscience-inspired SNNs for detecting auditory spatial attention from EEG data. The model utilized biologically realistic IF neurons and employed sparse coding through synaptic pruning to enhance classification accuracy. Remarkably, the proposed SNNs achieved an average accuracy of 90% with only 10% of the EEG signals used as training data, demonstrating its efficiency and robustness with minimal data.
Finally, research on SNNs has also seen in the field of BCI. Liao et al. [28] proposed SCNet, a novel SNNs with convolutional neural networks (CNN) for motor imagery (MI) classification. SCNet used an adaptive coding mechanism that reduces information loss during the encoding process. This mechanism improves the model’s capacity to learn from the data, enhancing both the accuracy and efficiency of processing EEG signals. SCNet showed classification performance improvements compared to several state-of-the-art SNNs and traditional machine learning models in motor imagery tasks. Gong et al. [29] introduced SGLNet, an innovative SNNs that combines adaptive graph convolution with Long Short-Term Memory (LSTM) units. Proposed model includes a direct learnable spike encoding mechanism that converts EEG signals into spike trains effectively. In addition, proposed architecture effectively captures both the spatial topological relationships and temporal relationships in EEG signals, which are crucial for accurate BCI applications. Consequently, SGLNet enhanced the classification performance in emotion recognition and MI tasks compared to representative baseline models. Tan et al. [30] introduced SNNs framework for short-term emotion recognition modeling of spatiotemporal EEG patterns without relying on handcrafted features. In this study, model was proposed that combined the SNN reservoir module (SNNr) based on the NeuCube framework with the deSNN representation, which allows the spatio-temporal activation pattern generated through the trained SNNr to generate output neurons. In this study, when the deSNN representation was added to the basic SNNs model, improved results were obtained in arousal and valence classification of the emotion classification dataset.
Wu et al. [31] enhanced EEG-based patter recognition in NeuCube [32] SNNs model, by addressing limited labeled data and distribution variability through transfer learning. To optimize the hyperparameters of the NeuCube reservoir, an improved cuckoo search algorithm [33] was developed aiming to extract the best spatio-temporal features from EEG data. NeuCube’s output classifier was improved by transferring weights to the support vector machine model, and as a result, performance in MI recognition was siginificantly improved compared to other deep learning models. Singanamalla et al. [34] introduced SNNs that is adaptable to different EEG signal modalities, such as MI and Steady-State Visually Evoked Potentials (SSVEP), without needing extensive adjustments in hyperparameters. In addition, they proposed a method for generating synthetic EEG data using neural perturbation and synaptic filter in SNNs, which can enhance classifier performance with only a limited number of original data samples. Antelis et al. [35] proposed SNNs model for MI tasks recognition. SNNs model consists of an Izhikevich neuron model [13] that is biologically realistic and requires little computation, and was trained using the Particle Swarm Optimization algorithm [32], which optimizes a problem by iteratively enhancing a candidate solution according to a defined quality metric. They conducted a comparative analysis demonstrating the advantages of SNNs over traditional neural network models, particularly in terms of handling dynamic, time-sensitive data inherent to EEG signals used for BCI.
3.2 Electrocardiogram signal
An ECG signal represents the heart’s electrical activity over time in a graphical format, recorded and displayed through a medical device known as an electrocardiograph. This signal is acquired by placing electrodes on the patient’s body, which detect tiny electrical changes on the skin arising from the electrophysiological pattern of depolarization and repolarization during each heartbeat [18]. ECG signals are essential for diagnosing and monitoring a range of cardiac conditions, including arrhythmias, heart attacks, and other heart diseases.
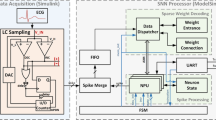
Most SNNs research based on ECG signals has focused on arrhythmia detection. Table 2 summarizes the ECG-based SNNs studies. Chu et al. [46] introduced a neuromorphic processing system using a spike-driven SNNs processor for ECG classification. Proposed neuromorphic system includes a novel Level-crossing sampling methodto encode temporal ECG signals with a single-bit representation, an enhanced hardware-aware spatio-temporal backpropagation training method to minimize firing rates, and a specialized SNNs processor design for energy-efficient ECG classification. The proposed SNNs processor, validated on field programmable gate array (FPGA) and implemented to an application-specific integrated circuit, achieved high classification accuracy with low power operation in MIT-BIH database. Xing et al. [47] introduced ECG classification approach using SNNs integrated with a channel-wise attention mechanism specifically designed for real-time applications on personal portable devices. The proposed model positioned the channel attention mechanism between SNN layers to incorporate channel information, enhancing the model’s ability to capture relevant features. Additionally, wavelet threshold denoising method was employed to reduce ECG signal noise. The proposed algorithm was implemented on FPGA and achieved improved results in terms of arrhythmia automatic classification accuracy, energy consumption, and real-time functionality compared to state-of-the-art methods. Feng et al. [48] introduced a deep SNNs for ECG classification comprising a 14-layer deep layer using ANN-to-SNNs conversion. By transferring the trained parameters from an ANN to SNNs and employing ReLU activation functions during the ANN-to-SNN transformation, proposed model achieved the highest accuracy on ECG classification tasks. The efficiency and effectiveness of this method were demonstrated by experimental results, showing that the transformed SNNs outperformed the original ANN.
Jiang et al. [49] proposed the MSPAN system, a memristive spike-based computing engine with adaptive neurons for the purpose of edge arrhythmia detection. They introduced a deep integrative SNN architecture comprising spike-based convolution layers and fully connected layers with an integrate unit and an adaptive unit. Additionally, the study proposed a memristor-based computing-in-memory architecture, eliminating the necessity for an analog-to-digital converter. A threshold-adaptive LIF neuron module mimicking human brain function is included in the architecture, which improves energy efficiency and processing speed. The MSPAN system achieved a significant reduction in computational complexity relative to traditional CNN-based methods, without compromising on performance. Yan et al. [50] introduced a two-stage CNN and SNN framework for energy-efficient ECG classification. Initially, a two-step CNN workflow classifies ECG beats as normal or abnormal, then further categorizes abnormal beats into specific types. They proposed a convolutional SNN with an identical structure as the CNN, transferring network weights from CNN to SNN. This approach allows the SNN to inherit the learned features from the CNN, achieving high accuracy with reduced computational complexity. They achieved significant energy savings, with the two-class SNN system consuming only 0.077 W and maintaining 90% accuracy. Shekhawat et al. [51] proposed a Binarized Spiking Neural Network (BSNN) optimized using a momentum search algorithm to improve the detection of fetal arrhythmia detection from ECG signals. By optimizing the network’s weight parameters and employing hexadecimal local adaptive binary pattern for feature extraction, the proposed method tried to overcome the challenges posed by noisy ECG signals and the complexity of fetal arrhythmias. BSNN achieved significant enhancement of fetal arrhythmia detection, thereby supporting better prenatal care and clinical outcomes.
Yin et al. [52] introduced spiking recurrent neural networks (SRNNs) that leverage novel surrogate gradients and adaptive spiking neurons for improved time-domain classification tasks, including ECG wave-pattern classification, speech and gesture recognition. This study proposed the use of adaptive LIF trained with a novel surrogate gradient, the multi-gaussian, which significantly outperforms other surrogate gradients. This training method optimized the SRNNs for sequential and temporal tasks, resulting in significantly less energy consumption and computational efficiency than traditional networks.
3.3 Electromyogram signal
EMG is a signal of electrical activity generated by skeletal muscles and is recorded through electrodes placed on the surface of the skin over the muscles or inserted directly into muscle tissue. The presence of an electrical potential, produced by muscle cells upon electrical or neurological activation, is reflected in this signal [18]. EMG is used for various clinical and research purposes. In clinical practice, EMG testing can assist in diagnosing diseases and disorders that impact muscles and their controlling nerves.
Research on SNNs based on EMG signals has primarily focused on the detection of hand gestures. Table 3 summarizes the EMG-based SNNs studies. Xu et al. [56] introduced an innovative event-driven spiking CNN (SCNN) for EMG pattern recognition. They designed a SCNN structure that integrates a fully connected module with a cyclic CNN and improved LIF neurons to enhance learning efficiency and reduce information loss. Furthermore, they employed an event-driven differential coding approach for effective EMG signal encoding and spatio-temporal backpropagation for model training. The research, centered on high-density surface EMG signals across six gestures and various electrode positions, demonstrated high energy efficiency and high performance in small-sample training experiments.
Vitale et al. [57] demonstrated the potential of neuromorphic computing to enhance edge-computing wearable devices for biomedical applications, focusing on EMG-based gesture recognition. They proposed two SNNs—a spiking convolutional neural network and a spiking fully connected network—and implemented on Intel’s Loihi neuromorphic processor to evaluate gesture recognition. The proposed system demonstrated high accuracy, energy efficiency, and low latency compared to traditional machine learning methods, highlighting its suitability for real-time wearable applications. Ma et al. [58] developed a neuromorphic processing system incorporating mixed signals and employing a spiking recurrent neural network (SRNN) to efficiently and accurately classify EMG-based gesture. They introduced an adaptive delta-encoding method to equalize input firing rates across channels and subjects, incorporated a soft Winner-Take-All network with STDP learning to enhance classification accuracy, and used a sparse representation to reduce connections in SRNN. The proposed system implemented on the DYNAP neuromorphic chip and achieved competitive performance with lower power consumption. Ceolini et al. [59] developed a neuromorphic computing benchmark for hand gesture recognition by integrating EMG and event-based camera data through a sensor-fusion approach. Their framework evaluated the performance of two different neuromorphic processors, Loihi and ODIN + MorphIC, in processing spike-encoded sensory data. They implemented Spiking CNN and Spiking MLP architectures on Intel’s Loihi and ODIN + MorphIC processors, respectively. The system achieved competitive accuracy with significantly lower energy consumption, providing a benchmark for neuromorphic computing for real-time low-power applications.
Garg et al. [60] proposed a new approach to optimize the hyperparameters of the spike encoding algorithm based on the readout layer concept of storage computing. This study used a regulated reservoir computing approach that operates at the edge of chaos. CRITICAL auto-regulation algorithm [61] was used to adjust the dynamics within the reservoir, enhancing the network’s sensitivity and performance. The proposed method was evaluated for hand gesture recognition performance on two open-source datasets and demonstrated improved results compared to existing state-of-the-art methods. Sun et al. [62] explored the feasibility of applying SNNs to myoelectric control systems, addressing challenges such as extensive training demands, low robustness, and excessive energy consumption. This paper introduced an adaptive threshold-based temporal encoding method and used improved LIF neuron that incorporates voltage-current effects to enhance EMG pattern recognition. Proposed SNNs model significantly reduced training repetitions, lower power consumption, and improving robustness compared to conventional methods. Tanzarella et al. [63] proposed a neuromorphic framework for movement intention recognition of human spinal motor neurons. This study introduced SNNs model features a convolutional architecture optimized for decoding spinal motor neuron activity, utilizing LIF neurons and local learning rules. The proposed method showed high hand gesture classification accuracy for all muscles. In addition, the power consumption for the system was efficiently managed, highlighting its suitability for real-time applications on neuromorphic hardware.
4 Conclusion
This paper provides a detailed review of the latest applications of SNNs in the analysis of biomedical signals, particularly focusing on EEG, ECG, and EMG signals. Based on the characteristics of SNNs that mimic the human brain, researchers have explored various applications using EEG signals, with most studies comparing the performance of SNNs to traditional deep learning and machine learning. Meanwhile, studies on ECG and EMG signals have mainly utilized SNN algorithms for classification tasks, and when integrated with neuromorphic systems and wearable devices, both classification performance and energy consumption were analyzed, demonstrating that it can be applied to on-device.
The advance of neuromorphic systems changes from traditional cloud-based AI systems to on-device AI platforms. This transition is driven by the demand for efficient, private, and personalized computing solutions that can function independently of centralized data centers. These systems protect private information and reduce response latency, which are critical in healthcare area. From this perspective, SNNs, which have a structure closer to the neural architecture of the human brain compared to traditional deep learning methods, play an important role and offer a promising approach for real-time processing on-devices. SNN not only demonstrated comparable performance to deep learning models but also exhibited high energy efficiency and fast inference times, indicating the potential for real-time processing. Future research on SNN is expected to focus on reducing computational complexity and improving network scalability to facilitate broader adoption in clinical and real-world environments. For example, studies are likely to explore methods to integrate SNNs with existing medical devices and Internet of Things platforms to enable seamless, real-time health monitoring. Additionally, it is anticipated that personalized healthcare based on online learning from on-device data will become feasible. As SNNs technology continues to evolve, its applications in healthcare will lead to a new era of fast, accurate, and energy-efficient monitoring and personalized management.
References
Hoefler T, Alistarh D, Ben-Nun T, Dryden N, Peste A. Sparsity in deep learning: pruning and growth for efficient inference and training in neural networks. J Mach Learn Res. 2021;22.
Wu H, Judd P, Zhang X, Isaev M, Micikevicius P. Integer quantization for deep learning inference: principles and empirical evaluation. 2020:1–20.
Allen-Zhu Z, Li Y. Towards understanding ensemble, knowledge distillation and self-distillation in deep learning. 2020.
Iandola F, Moskewicz M, Karayev S, Girshick R, Darrell T, Keutzer K, DenseNet. Implement efficient ConvNet descr pyramids. 2014:1–11.
Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and < 0.5 MB model size. 2016:1–13.
Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H. MobileNets: efficient convolutional neural networks for mobile vision applications. 2017.
Davies M, Srinivasa N, Lin TH, Chinya G, Cao Y, Choday SH, Dimou G, Joshi P, Imam N, Jain S, et al. Loihi: a neuromorphic manycore processor with on-chip learning. IEEE Micro. 2018;38:82–99. https://doi.org/10.1109/MM.2018.112130359
Akopyan F, Sawada J, Cassidy A, Alvarez-Icaza R, Arthur J, Merolla P, Imam N, Nakamura Y, Datta P, Nam GJ, et al. TrueNorth: design and tool flow of a 65 MW 1 million neuron programmable neurosynaptic chip. IEEE Trans Comput Des Integr Circuits Syst. 2015;34:1537–57. https://doi.org/10.1109/TCAD.2015.2474396
Painkras E, Plana LA, Garside J, Temple S, Galluppi F, Patterson C, Lester DR, Brown AD, Furber SB, SpiNNaker. A 1-W 18-core system-on-chip for massively-parallel neural network simulation. IEEE J Solid-State Circuits. 2013;48:1943–53. https://doi.org/10.1109/JSSC.2013.2259038
Benjamin BV, Gao P, McQuinn E, Choudhary S, Chandrasekaran AR, Bussat JM, Alvarez-Icaza R, Arthur JV, Merolla PA, Boahen K. Neurogrid: a mixed-analog-digital multichip system for large-scale neural simulations. Proc IEEE. 2014;102:699–716. https://doi.org/10.1109/JPROC.2014.2313565
Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–44. https://doi.org/10.1113/jphysiol.1952.sp004764
Abbott LF. Lapicque’s introduction of the integrate-and-fire model neuron (1907). Brain Res Bull. 1999;50:303–4. https://doi.org/10.1016/S0361-9230(99)00161-6
Izhikevich EM. Which model to use for cortical spiking neurons? IEEE Trans Neural Networks. 2004;15:1063–70. https://doi.org/10.1109/TNN.2004.832719
Eshraghian JK, Ward M, Neftci EO, Wang X, Lenz G, Dwivedi G, Bennamoun M, Jeong DS, Lu WD. Training spiking neural networks using lessons from deep learning. Proc IEEE. 2023;111:1016–54. https://doi.org/10.1109/JPROC.2023.3308088
Zhang M, Gu Z, Zheng N, Ma D, Pan G. Efficient spiking neural networks with logarithmic temporal coding. IEEE Access. 2020;8:98156–67. https://doi.org/10.1109/ACCESS.2020.2994360
Bi G, Poo M. Synaptic modifications in cultured hippocampal neurons: dependence on spike timing, synaptic strength, and postsynaptic cell type. J Neurosci. 1998;18:10464–72. https://doi.org/10.1523/JNEUROSCI.18-24-10464.1998
Song S, Miller KD, Abbott LF. Competitive hebbian learning through spike-timing-dependent synaptic plasticity. Nat Neurosci. 2000;3:919–26. https://doi.org/10.1038/78829
Park KS. Humans and electricity: understanding body electricity and applications. 2023.
Zhang Z, Xiao M, Ji T, Jiang Y, Lin T, Zhou X, Lin Z. Efficient and generalizable cross-patient epileptic seizure detection through a spiking neural network. Front Neurosci. 2023;17. https://doi.org/10.3389/fnins.2023.1303564
Burelo K, Ramantani G, Indiveri G, Sarnthein JA. Neuromorphic spiking neural network detects epileptic high frequency oscillations in the scalp EEG. Sci Rep. 2022;12. https://doi.org/10.1038/s41598-022-05883-8
Li W, Fang C, Zhu Z, Chen C, Song A. Fractal spiking neural network scheme for EEG-based emotion recognition. IEEE J Transl Eng Heal Med. 2024;12:106–18. https://doi.org/10.1109/JTEHM.2023.3320132
Xu FF, Pan D, Zheng H, Ouyang Y, Jia Z, Zeng HEESCN. A novel spiking neural network method for EEG-based emotion recognition. Comput Methods Programs Biomed. 2024;243. https://doi.org/10.1016/j.cmpb.2023.107927
Xu H, Cao K, Chen H, Abudusalamu A, Wu W, Xue Y. Emotional brain network decoded by biological spiking neural network. Front Neurosci. 2023;17. https://doi.org/10.3389/fnins.2023.1200701
Luo Y, Fu Q, Xie J, Qin Y, Wu G, Liu J, Jiang F, Cao Y, Ding X. EEG-based emotion classification using spiking neural networks. IEEE Access. 2020;8:46007–16. https://doi.org/10.1109/ACCESS.2020.2978163
Kasabov NK, NeuCube. A spiking neural network architecture for mapping, learning and understanding of spatio-temporal brain data. Neural Netw. 2014;52:62–76. https://doi.org/10.1016/j.neunet.2014.01.006
Cai S, Li P, Li HA, Bio-Inspired. Spiking attentional neural network for attentional selection in the listening brain. IEEE Trans Neural Networks Learn Syst. 2023. https://doi.org/10.1109/TNNLS.2023.3303308
Faghihi F, Cai S, Moustafa AAA, Neuroscience-Inspired. Spiking neural network for EEG-based auditory spatial attention detection. Neural Netw. 2022;152:555–65. https://doi.org/10.1016/j.neunet.2022.05.003
Liao X, Wu Y, Wang Z, Wang D, Zhang HA. Convolutional spiking neural network with adaptive coding for motor imagery classification. Neurocomputing. 2023;549. https://doi.org/10.1016/j.neucom.2023.126470
Gong P, Wang P, Zhou Y, Zhang DA, Spiking Neural. Network with adaptive graph convolution and LSTM for EEG-based brain-computer interfaces. IEEE Trans Neural Syst Rehabil Eng. 2023;31:1440–50. https://doi.org/10.1109/TNSRE.2023.3246989
Tan C, Šarlija M, Kasabov N, NeuroSense. Short-term emotion recognition and understanding based on spiking neural network modelling of spatio-temporal EEG patterns. Neurocomputing. 2021;434:137–48. https://doi.org/10.1016/j.neucom.2020.12.098
Wu X, Feng Y, Lou S, Zheng H, Hu B, Hong Z, Tan J. Improving NeuCube spiking neural network for EEG-based pattern recognition using transfer learning. Neurocomputing. 2023;529:222–35. https://doi.org/10.1016/j.neucom.2023.01.087
Kennedy J, Eberhart R. Particle swarm optimization. In Proceedings of the Proceedings of ICNN’95 - International Conference on Neural Networks; IEEE; Vol. 4, pp. 1942–1948.
Yang X-S, Deb SC, Search. Recent advances and applications. Neural Comput Appl. 2014;24:169–74. https://doi.org/10.1007/s00521-013-1367-1
Singanamalla SKR, Lin CT. Spiking neural network for augmenting electroencephalographic data for brain computer interfaces. Front Neurosci. 2021;15. https://doi.org/10.3389/fnins.2021.651762
Virgilio G, Sossa CD, Antelis AJH, Falcón JM. Spiking neural networks applied to the classification of motor tasks in EEG signals. Neural Netw. 2020;122:130–43. https://doi.org/10.1016/j.neunet.2019.09.037
Tran LV, Tran HM, Le TM, Huynh TTM, Tran HT, Dao SVT. Application of machine learning in epileptic seizure detection. Diagnostics. 2022;12:2879. https://doi.org/10.3390/diagnostics12112879
Saeedinia SA, Jahed-Motlagh MR, Tafakhori A, Kasabov N. Design of MRI structured spiking neural networks and learning algorithms for personalized modelling, analysis, and prediction of EEG signals. Sci Rep. 2021;11. https://doi.org/10.1038/s41598-021-90029-5
Koelstra S, Muhl C, Soleymani M, Jong-Seok L, Yazdani A, Ebrahimi T, Pun T, Nijholt A, Patras I. DEAP: a database for emotion analysis; using physiological signals. IEEE Trans Affect Comput. 2012;3:18–31. https://doi.org/10.1109/T-AFFC.2011.15
Zheng W-L, Liu W, Lu Y, Lu B-L, Cichocki A, EmotionMeter. A multimodal framework for recognizing human emotions. IEEE Trans Cybern. 2019;49:1110–22. https://doi.org/10.1109/TCYB.2018.2797176
Wei-Long Zheng; Bao-Liang Lu investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans Auton Ment Dev. 2015;7:162–75, https://doi.org/10.1109/TAMD.2015.2431497
Das N, Francart T. and A.B. Auditory attention detection dataset KULeuven.
Tangermann M, Müller K-R, Aertsen A, Birbaumer N, Braun C, Brunner C, Leeb R, Mehring C, Miller KJ, Müller-Putz GR, et al. Review of the BCI competition IV. Front Neurosci. 2012;6. https://doi.org/10.3389/fnins.2012.00055
Blankertz B, Muller K-R, Curio G, Vaughan TM, Schalk G, Wolpaw JR, Schlogl A, Neuper C, Pfurtscheller G, Hinterberger T, et al. The BCI competition 2003: progress and perspectives in detection and discrimination of EEG single trials. IEEE Trans Biomed Eng. 2004;51:1044–51. https://doi.org/10.1109/TBME.2004.826692
Soleymani M, Lichtenauer J, Pun T, Pantic MA. Multimodal database for affect recognition and implicit tagging. IEEE Trans Affect Comput. 2012;3:42–55. https://doi.org/10.1109/T-AFFC.2011.25
Schrauwen B, Van Campenhout IBSA. a Fast and Accurate Spike Train Encoding Scheme. In proceedings of the proceedings of the international joint conference on neural networks, 2003.; IEEE; Vol. 4, pp. 2825–2830.
Chu H, Yan Y, Gan L, Jia H, Qian L, Huan Y, Zheng L, Zou Z. A neuromorphic processing system with spike-driven SNN processor for wearable ECG classification. IEEE Trans Biomed Circuits Syst. 2022;16:511–23. https://doi.org/10.1109/TBCAS.2022.3189364
Xing Y, Zhang L, Hou Z, Li X, Shi Y, Yuan Y, Zhang F, Liang S, Li Z, Yan L. Accurate ECG classification based on spiking neural network and attentional mechanism for real-time implementation on personal portable devices. Electron. 2022;11. https://doi.org/10.3390/electronics11121889
Feng Y, Geng S, Chu J, Fu Z, Hong S. Building and training a deep spiking neural network for ECG classification. Biomed Signal Process Control. 2022;77. https://doi.org/10.1016/j.bspc.2022.103749
Jiang J, Tian F, Liang J, Shen Z, Liu Y, Zheng J, Wu H, Zhang Z, Fang C, Zhao Y, et al. MSPAN: a memristive spike-based computing engine with adaptive neuron for edge arrhythmia detection. Front Neurosci. 2021;15. https://doi.org/10.3389/fnins.2021.761127
Yan Z, Zhou J, Wong WF. Energy efficient ECG classification with spiking neural network. Biomed Signal Process Control. 2021;63. https://doi.org/10.1016/j.bspc.2020.102170
Shekhawat D, Chaudhary D, Kumar A, Kalwar A, Mishra N, Sharma D. Binarized spiking neural network optimized with momentum search algorithm for fetal arrhythmia detection and classification from ECG signals. Biomed Signal Process Control. 2024;89. https://doi.org/10.1016/j.bspc.2023.105713
Yin B, Corradi F, Bohté SM. Accurate and efficient time-domain classification with adaptive spiking recurrent neural networks. Nat Mach Intell. 2021;3:905–13. https://doi.org/10.1038/s42256-021-00397-w
Moody GB, Mark RG. The impact of the MIT-BIH arrhythmia database. IEEE Eng Med Biol Mag. 2001;20:45–50. https://doi.org/10.1109/51.932724
Clifford G, Liu C, Moody B, Lehman L, Silva I, Li Q, Johnson A. Mark, R. AF classification from a short single lead ECG recording: the physionet computing in cardiology challenge 2017.; September 14 2017.
Laguna P, Mark RG, Goldberg A, Moody GB. A database for evaluation of algorithms for measurement of QT and other waveform intervals in the ECG. In Proceedings of the Computers in Cardiology 1997; IEEE; pp. 673–676.
Xu M, Chen X, Sun A, Zhang X, Chen XA. Novel event-driven spiking convolutional neural network for electromyography pattern recognition. IEEE Trans Biomed Eng. 2023;70:2604–15. https://doi.org/10.1109/TBME.2023.3258606
Vitale A, Donati E, Germann R, Magno M. Neuromorphic edge computing for biomedical applications: gesture classification using EMG signals. IEEE Sens J. 2022;22:19490–9. https://doi.org/10.1109/JSEN.2022.3194678
Ma Y, Chen B, Ren P, Zheng N, Indiveri G, Donati E. EMG-based gestures classification using a mixed-signal neuromorphic processing system. In Proceedings of the IEEE Journal on Emerging and Selected Topics in Circuits and Systems; Institute of Electrical and Electronics Engineers Inc., December 1 2020; Vol. 10, pp. 578–587.
Ceolini E, Frenkel C, Shrestha SB, Taverni G, Khacef L, Payvand M, Donati E. Hand-gesture recognition based on EMG and event-based camera sensor fusion: a benchmark in neuromorphic computing. Front Neurosci. 2020;14. https://doi.org/10.3389/fnins.2020.00637
Garg N, Balafrej I, Beilliard Y, Drouin D, Alibart F, Rouat J. Signals to spikes for neuromorphic regulated reservoir computing and EMG hand gesture recognition. ACM Int Conf Proceeding Ser. 2021. https://doi.org/10.1145/3477145.3477267
Brodeur S, Rouat J. Regulation toward self-organized criticality in a recurrent spiking neural reservoir. In; 2012; pp. 547–54.
Sun A, Chen X, Xu M, Zhang X, Chen X. Feasibility study on the application of a spiking neural network in myoelectric control systems. Front Neurosci. 2023;17. https://doi.org/10.3389/fnins.2023.1174760
Tanzarella S, Iacono M, Donati E, Farina D, Bartolozzi C. Neuromorphic decoding of spinal motor neuron behaviour during natural hand movements for a new generation of wearable neural interfaces. IEEE Trans Neural Syst Rehabil Eng. 2023;31:3035–46. https://doi.org/10.1109/TNSRE.2023.3295658
Zhang X, Wu L, Yu B, Chen X, Chen X. Adaptive calibration of electrode array shifts enables robust myoelectric control. IEEE Trans Biomed Eng. 2020:1–1. https://doi.org/10.1109/TBME.2019.2952890
Atzori M, Gijsberts A, Heynen S, Hager A-GM, Deriaz O, van der Smagt P, Castellini C, Caputo B, Muller H. Building the Ninapro database: a resource for the biorobotics community. In Proceedings of the 2012 4th IEEE RAS & EMBS International Conference on Biomedical Robotics and Biomechatronics (BioRob); IEEE, June 2012; pp. 1258–1265.
Shupe L, Fetz EA. Integrate-and-fire spiking neural network model simulating artificially induced cortical plasticity. eNeuro. 2021;8:1–22, https://doi.org/10.1523/ENEURO.0333-20.2021
Donati E. EMG from forearm datasets for hand gestures recognition. Zenodo, Inst. Neuroinform.
Atzori M, Gijsberts A, Castellini C, Caputo B, Hager A-GM, Elsig S, Giatsidis G, Bassetto F, Müller H. Electromyography data for non-invasive naturally-controlled robotic hand prostheses. Sci Data. 2014;1. https://doi.org/10.1038/sdata.2014.53
Ceolini E, Taverni G, Payvand M, Donati E. EMG and video dataset for sensor fusion based hand gestures recognition. Eur Comm Bruss. 2020.
Funding
This research was supported by the MSIT(Ministry of Science and ICT), Korea, under the ITRC(Information Technology Research Center) support program(IITP-2024-RS-2022-00156225) supervised by the IITP(Institute for Information & Communications Technology Planning & Evaluation). The present Research has been conducted by the Research Grand of Kwangwoon University in 2024.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The author has no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Choi, S.H. Spiking neural networks for biomedical signal analysis. Biomed. Eng. Lett. 14, 955–966 (2024). https://doi.org/10.1007/s13534-024-00405-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13534-024-00405-z