Abstract
Multiclass classification of brain tumors from magnetic resonance (MR) images is challenging due to high inter-class similarities. To this end, convolution neural networks (CNN) have been widely adopted in recent studies. However, conventional CNN architectures fail to capture the small lesion patterns of brain tumors. To tackle this issue, in this paper, we propose a global transformer network dubbed GT-Net for multiclass brain tumor classification. The GT-Net mainly comprises a global transformer module (GTM), which is introduced on the top of a backbone network. A generalized self-attention block (GSB) is proposed to capture the feature inter-dependencies not only across spatial dimension but also channel dimension, thereby facilitating the extraction of the detailed tumor lesion information while ignoring less important information. Further, multiple GSB heads are used in GTM to leverage global feature dependencies. We evaluate our GT-Net on a benchmark dataset by adopting several backbone networks, and the results demonstrate the effectiveness of GTM. Further, comparison with state-of-the-art methods validates the superiority of our model.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
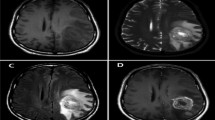
Brain tumor is broadly witnessed as the most dangerous cancer among different cancer types across the globe due to its fast-growing nature. It is generally found in individuals of all ages, including children, and has the lowest survival rate [1]. Therefore, the accurate detection of the brain tumor at an early stage is highly indispensable for timely treatment and better patient care. Based on their shape and location, brain tumors can be categorized into meningioma, glioma, and pituitary. However, detecting such tumors and their types is more challenging due to their similar structures. Magnetic resonance imaging (MRI) is the most commonly used medical imaging technique by physicians and radiologists to diagnose brain and other nervous system disorders [2, 3]. This is primarily due to its non-invasive nature and ability to produce clear images of brain tissues. However, the manual interpretation of brain MR images at huge volumes is tedious, error-prone, and highly dependent on the skill of radiologists. Therefore, it is of utmost necessity to design automated computer-aided diagnosis (CAD) methods to assist doctors for fast and correct detection of brain tumors. The MR images of various types of brain tumors are depicted in Fig. 1.
Considering the above issues, researchers have built several CAD systems for brain tumor detection over the past decade. The CAD systems [1, 4,5,6] proposed earlier were mainly designed based on the traditional multi-stage machine learning structure, in which the typical stages adopted are preprocessing, feature engineering, and classification. With the increase in several sophisticated feature extraction techniques and classifiers, selecting the best techniques for these CAD systems becomes increasingly challenging.
Recent efforts have been directed towards using convolution neural networks (CNNs) for multiclass brain tumor classification due to their ability to learn high-level feature representations and success in many vision tasks, including medical image analysis. Paul et al. [7] evaluated the effectiveness of fully connected networks and CNN for brain tumor classification. Asfhar et al. [2] designed a capsule network (CapsNet) for the classification of brain tumors. Later, they developed a modified CapsNet [8], which fed not only the brain MR images but also tumor coarse boundaries as input for performance improvement. In [9], a block-wise fine-tuning (BFT) approach was adopted with VGG19 to classify brain tumors. In [10], a deep neural network was trained as the discriminator in the generative adversarial network (GAN) model, and the pre-trained discriminator model was later utilized to classify brain tumors. In [11], a boosted capsule network (BoostCaps) was proposed to mitigate the issues of modified CapsNet [8]. Later, Bodapati et al. [12] blended the features captured from Xception and InceptionResNetV2 with the help of average pooling operation and named their architecture a two-channel deep network. In a recent work, a border collie firefly algorithm (BCFA) based GAN [13] was presented to classify tumor severity levels.
Despite the fact that the CNN-based approaches achieved relatively better classification performance, conventional CNNs may overlook capturing the subtle size variations of brain tumors within the same class and among their types. Further, they face difficulty in capturing global dependency, which makes their practical clinical application more challenging. In recent studies, attention mechanisms have been used as a vital component in CNN architectures to solve several computer vision tasks, allowing the model to focus on vital features while ignoring unnecessary features, thereby improving performance [14,15,16]. However, to our knowledge, only limited studies on these mechanisms have yet been presented for classifying brain tumors [17]. In [18], a model called MSENet was proposed which comprised of squeeze and excitation blocks. In [19], channel-shuffle dual attention block (CSDAB) was coupled with backbone networks to achieve improved classification performance. While in [20], a global attention-based residual multiscale CNN named ARM-Net was introduced to classify brain tumors. Recently, vision transformer (ViT) has gained tremendous interest from the vision community due to its ability to capture wide-range feature interdependencies via self-attention [21]. Inspired by its success, in this paper, a generalized self-attention block (GSB) is proposed to capture the detailed tumor lesion information while ignoring unimportant information. While ViT models are more effective in modeling global information, they lack the ability to capture local information, which CNNs do efficiently [22, 23]. Hence, to address these issues, we propose a transformer-based network using GSB that takes the benefits of both transformer and CNN, thereby modeling both local and global information within the brain MR image.
The primary contributions of this paper are enumerated as follows:
-
We propose a global transformer network called GT-Net for multiclass brain tumor classification, which integrates a novel transformer module with a pre-trained CNN.
-
A global transformer module (GTM) consisting of multiple GSB heads is proposed to adequately exploit the wide-range feature dependencies among the lesion regions. The GSB is introduced to establish global dependencies along both spatial and channel dimensions.
-
We evaluate GT-Net on a publicly available dataset and adopt several pre-trained CNN models as backbones to verify the efficacy of GTM. Additionally, we perform a comparative analysis with contemporary attention mechanisms and state-of-the-art multiclass brain tumor classification approaches.
The remainder of the paper is structured as follows. Section 2 describes the proposed GT-Net model and its components in detail. The dataset description, experimental results, and comparisons are presented in Sect. 3. Eventually, the conclusions are drawn in Sect. 4.
2 Proposed method
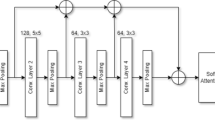
The structure of the proposed GT-Net is shown in Fig. 2, which has three key components: a backbone network, a GTM, and a classifier. The backbone network assists in deriving abstracted feature representations from the input MR images. The GTM includes different GSB heads to learn global dependencies across spatial and channel dimensions, enabling the extraction of more detailed and category-specific features. The classifier comprises a pooling layer followed by a softmax-activated fully connected (FC) layer to classify multiclass brain tumors. The following section provides a detailed description of the GT-Net architecture and its essential parts.
2.1 Backbone
The input to the GT-Net is a brain MR image, which is initially fed to a backbone network, i.e., a CNN architecture pre-trained on ImageNet, facilitating the generation of abstracted feature maps \(F \in \mathbb {R}^{H \times W \times C}\), where, C denotes the number of channels, and H and W indicate spatial dimensions. To acquire hierarchical feature representations, we extract the feature maps from the last convolutional layer of the backbone.
2.2 Proposed global transformer module (GTM)
The GTM consists of multiple GSB heads to explore the long-range global feature relationships from MR images and is designed based on the structure of transformer, but it is structurally different. The traditional ViT works on image patches, and the multi-head attention in this case is employed on the projected input vector representations. On the other hand, the GSB is applied over the feature maps obtained from a backbone without flattening the feature maps, thereby preserving local contextual information. The GTM takes the feature map \(F \in \mathbb {R}^{H \times W \times C}\) and is fed to GSB heads, resulting in different attention feature maps which are fused later to form the output of the block.
2.2.1 Generalized self-attention block (GSB)
The proposed GSB aims at capturing detailed brain lesion information while suppressing the useless information. The structure of GSB as shown in Fig. 3, is inspired by self-attention [15] and GCNet [16]. But different from them, it establishes global dependencies along spatial and channel dimension simultaneously. The spatial-wise attention captures the global feature dependencies along spatial positions, whereas channel-wise attention captures global channel interactions and assesses the importance of each channel.
As shown in the figure, we take the high-level feature map \(F \in \mathbb {R}^{H \times W \times C}\) as input to the GSB, resulting in refined feature maps \(F_{gs} \in \mathbb {R}^{H \times W \times C}\). Similar to transformer, we use three functions \(\textbf{k}\), \(\textbf{q}\), and \(\textbf{v}\) to transform the feature map into key, query, and value, respectively. We implement the function \(\textbf{q}(F)\) using 1\(\times\)1 convolution with \(C' = C/8\) channels and global average pooling (GAP) operations to obtain a vector \(\textbf{q}(F) \in \mathbb {R}^{1 \times C'}\). While functions \(\textbf{k}\) and \(\textbf{v}\) are implemented using 1\(\times\)1 convolution and reshape operations without GAP, resulting in maps \(\textbf{k}(F) \in \mathbb {R}^{HW \times C'}\) and \(\textbf{v}(F) \in \mathbb {R}^{ HW \times C'}\), respectively. Then, we compute the matrix cross product between \(\textbf{q}\) and \(\textbf{k}\), and apply a softmax activation thereafter to generate spatial attention weights as,
where, \(\otimes\) indicates the matrix cross-product, \(\Phi\) is the softmax activation and T in the superscript denotes the matrix transpose operation. Next, we obtain the spatial attention feature map \(F_{sp} \in \mathbb {R}^{H \times W \times C}\) by computing element-wise multiplication between \(F'\) and F as,
where, \(\odot\) represents the element-wise multiplication operation.
Similarly, we perform matrix cross-product between \(F'\) and \(\textbf{v}(F)\) to compute channel attention weights which is further fed to a 1\(\times\)1 convolution and a sigmoid activation to increase the channels from \(C'\) to C (also called linear embedding). This global transform procedure is mathematically expressed as,
The channel-wise attention maps \(F_{ch} \in \mathbb {R}^{H \times W \times C}\) are obtained as,
Finally, we aggregate the output feature maps \(F_{sp}\) and \(F_{ch}\) using their weighted sum to acquire the refined attention feature map \(F_{gs} \in \mathbb {R}^{H \times W \times C}\) and is defined as,
where \(W_1\) and \(W_2\) are the trainable scalar parameters. In a nutshell, GSB enables exploring both channel-wise and spatial-wise feature dependencies simultaneously from MR images, thereby, enhancing the feature representations.
2.2.2 Feature fusion strategy
We combine the feature maps generated from the GSB heads via concatenation, which is followed by a \(1 \times 1\) convolution to obtain the final output of GTM \(F_{tm} \in \mathbb {R}^{H \times W \times C}\). The \(F_{tm}\) is expressed mathematically as,
where h represents the number of GSB heads. It is worth noting that the value of h (i.e., \(h=4\)) has been chosen empirically in this study.
2.3 Classifier
The \(F_{tm}\) is finally fed to a classifier, in which we employ a trainable spatial pooling operation called generalized-mean (GeM) [24] pooling and a softmax-activated FC layer with three nodes to classify tumors into three categories.
3 Experiments and results
In this section, we present the implementation details, experimental results, and a description on the dataset used. We compare the potential of proposed GTM with contemporary attention mechanisms. A comparative analysis with state-of-the-art methods is also provided. Further, we carry out ablation studies to verify the significance of each individual component in the proposed GT-Net model.
3.1 Dataset used and implementation details
We validate our approach on an openly available brain tumor dataset comprises of 3064 T1-weighted contrast-enhanced MR images from three classes [4, 25]. The dataset has 1426, 708, and 930 2D MR images belonging to glioma, meningioma, and pituitary tumor classes, respectively, that are obtained from 233 patients. The images are of resolution \(512 \times 512\) pixels and are available in different views such as axial, coronal, and sagittal view.
For fair comparison, we follow a data split strategy similar to existing studies [2, 4, 7,8,9, 12, 19]. A five-fold cross-validation (CV) setting has been adopted, ensuring the train and test set at the same time do not contain the samples from a single patient. The input images are scaled 224\(\times\)224. Additionally, we employ various transformations such as random rotations, horizontal split, and vertical split for data augmentation. Each model is trained for 50 epochs. We use Adam optimizer and a cross-entropy loss for training purpose. During learning, the batch size and initial learning rate are set to 36 and 0.0002, respectively. The models are implemented on Keras with Tensorflow backend.
3.2 Results
To demonstrate the effectiveness of our GT-Net, several ImageNet pre-trained CNN models such as ResNet-50 [27], SqueezeNet [28], DenseNet-121 [29], MobileNet [30], and EfficientNet B0 [31], are considered as backbones. The size of the abstracted feature maps obtained from these models are \(7 \times 7 \times 2048\), \(13 \times 13 \times 512\), \(7 \times 7 \times 1024\), \(7 \times 7 \times 1024\), and \(7 \times 7 \times 1280\), respectively. These feature maps are then passed to the transformer block. The results of each model with and without transformer block are shown in Table 1. It is clearly evident that the backbone network attached with the proposed GTM significantly improves the classification performance. It is important to note that we fine-tuned all layers of the backbone networks using the target dataset to yield higher performance. To further test the potential of the proposed transformer block, it is compared with contemporary attention mechanisms such as channel split dual attention block (CSDAB) [19], global context (GC) [16], convolutional block attention module (CBAM) [14], self-attention [15], and squeeze and excitation (SE) [26] under similar experimental setup. Table 1 shows that the GTM performs better than other attention mechanisms with all backbone networks. It can be noticed that the CSDAB achieves a comparable performance. Further, the proposed GTM improves the average classification accuracy by 0.64%, 0.79%, 1.04%, 0.6%, and 0.9% with ResNet-50, SqueezeNet, DenseNet-121, MobileNet, and EfficientNet B0, respectively when compared with conventional self-attention mechanism. The DenseNet-121 outperforms other pre-trained models, i.e., it obtains a higher classification accuracy of \(97.11\%\). The DenseNet-121 with GTM is termed as GT-Net in our study. It is worth noting that the reported results are the mean values of the 5-fold CV. The confusion matrices of the proposed GT-Net for each fold are depicted in Fig. 4. As shown, the GT-Net correctly classifies most of the samples from three tumor classes, while considerably fewer samples are misclassified even though the dataset is not balanced. Also, it is observed that the sensitivity of the glioma class is higher than that of other classes.
For better understanding of the interpretability of our GT-Net model, heatmaps are generated using Grad-CAM++ [32] as shown in Fig. 5. It can be clearly seen that the model with GTM precisely locates the lesion regions while the model with the best performing attention block (CSDAB) and the model without attention detects unrelated regions.
The heatmaps generated for a few incorrect predictions of the GT-Net model are shown in Fig. 6. These heatmaps depict the discriminative regions highlighted by the proposed model. It can be observed that our model focuses on other regions instead of the tumor regions in these cases, interpreting the reason behind the misclassification. This is mainly due to the presence of high structural similarity with other regions and high intra-category differences.
3.2.1 Ablation studies
Ablation studies are performed to analyze the effect of each component in our model. We investigate the effect of channel-wise attention (CWA), spatial-wise attention (SWA) and their combination in the proposed GSB. Further, we verify the effect of GAP and GeM pooling in the classification layer of GT-Net. Table 2 demonstrates that the backbone model with only CWA or SWA reduces the classification performance, while the backbone with both CWA and SWA leads to an improved performance. Also, it can be observed that the GeM pooling is performing better than GAP in terms of accuracy and F1-score. Figure 7 depicts the heatmaps obtained from various ablation experiments.
3.2.2 Comparison with ViT models
In this experiment, we compare the performance of GT-Net with traditional ViT [21] and a convolutional transformer model, namely, compact convolutional transformer (CCT) [22]. The comparison results are shown in Table 3. It is worth mentioning here that a variant of ViT, ViT 12/16, has been used for comparison with GT-Net, which includes 12 transformer encoder layers, and uses a \(16 \times 16\) patch size. It is evident from the table that the GT-Net yields higher performance than other ViT models. This can be majorly attributed to the establishment of GTM in the proposed network.
3.2.3 Comparison with state-of-the-art methods
We perform a comparison against the state-of-the-art CNN-based multiclass brain tumor classification approaches on the same dataset and the results are shown in Table 4. It can be observed that our proposed GT-Net model achieves superior performance than existing CNN-based methods. Also, it achieves better classification accuracy than the attention-based methods such as MSENet [18] and CDANet [19]. This is due to the fact that the proposed transformer block facilitates highlighting more detailed and discriminative lesion information in the MR images. It is noteworthy that compared existing approaches have been validated under similar data split strategy and their results are taken directly from the original papers.
4 Conclusion
In this paper, we have introduced a global transformer network called GT-Net for multiclass brain tumor classification, which supports end-to-end learning. Specifically, we present a transformer block on the top of a backbone model which consists of multiple GSB heads to comprehensively capture the global feature dependencies. The GSB helps to exploit more detailed lesion information by learning both channel-wise and spatial-wise attention weights. Experiments and comparisons on a public dataset confirm the effectiveness of GT-Net compared to other state-of-the-art approaches. Further, the GTM achieves improved performance when compared with popular attention mechanisms. In future, the performance of GT-Net can further be tested using a large and diverse datasets. Also, we intend to explore the application of the proposed GTM in other vision tasks.
References
Muhammad K, Khan S. Deep learning for multigrade brain tumor classification in smart healthcare systems: a prospective survey. IEEE Trans Neural Netw Learn Syst. 2020;32(2):507–22.
Afshar P, Mohammadi A, Plataniotis KN. Brain tumor type classification via capsule networks. In: 25th IEEE international conference on image processing (ICIP). 2018, pp. 3129– 3133 . IEEE.
Afshar P, Mohammadi A, Plataniotis KN. Bayescap: a Bayesian approach to brain tumor classification using capsule networks. IEEE Signal Process Lett. 2020;27:2024–8.
Cheng J, Huang W, Cao S. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS One. 2015;10(10):0140381.
Usman K, Rajpoot K. Brain tumor classification from multi-modality MRI using wavelets and machine learning. Pattern Anal Appl. 2017;20(3):871–81.
Sahu O, Anand V, Kanhangad V, Pachori RB. Classification of magnetic resonance brain images using bi-dimensional empirical mode decomposition and autoregressive model. Biomed Eng Lett. 2015;5:311–20.
Paul JS, Plassard AJ. Deep learning for brain tumor classification. In: Medical Imaging 2017: biomedical applications in molecular, structural, and functional imaging, 2017;10137; 253– 268. SPIE.
Afshar P, Plataniotis KN, Mohammadi A. Capsule networks for brain tumor classification based on MRI images and coarse tumor boundaries. In: International conference on acoustics, speech and signal processing (ICASSP). 2019; pp. 1368– 1372 . IEEE.
Swati ZNK, Zhao Q. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput Med Imaging Graph. 2019;75:34–46.
Ghassemi N, Shoeibi A, Rouhani M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control. 2020;57: 101678.
Afshar P, Plataniotis KN, Mohammadi, A. Boostcaps: a boosted capsule network for brain tumor classification. In: 2020 42nd annual international conference of the IEEE engineering in medicine & biology society (EMBC). 2020; pp. 1075– 1079. IEEE.
Bodapati JD, Shaik NS. Joint training of two-channel deep neural network for brain tumor classification. SIViP. 2021;15(4):753–60.
Abirami S, Venkatesan GP. Deep learning and spark architecture based intelligent brain tumor MRI image severity classification. Biomed Signal Process Control. 2022;76: 103644.
Woo S, Park J, Lee J-Y, Kweon IS. Cbam: convolutional block attention module. In: proceedings of the European conference on computer vision (ECCV). 2018; pp. 3– 19.
Zhang H, Goodfellow I, Metaxas D, Odena A. Self-attention generative adversarial networks. In: International conference on machine learning (ICML); 2019. pp. 7354– 7363.
Cao Y, Xu J, Lin, S. Gcnet: Non-local networks meet squeeze-excitation networks and beyond. In: Proceedings of the IEEE/CVF International Conference on Computer Vision Workshops; 2019.
Sharma P, Nayak DR, Balabantaray BK, Tanveer M, Nayak R. A survey on cancer detection via convolutional neural networks: Current challenges and future directions. Neural Networks; 2023.
Bodapati JD, Shareef SN. Msenet: multi-modal squeeze-and-excitation network for brain tumor severity prediction. Int J Pattern Recognit Artif Intell. 2021;35(07):2157005.
Dutta TK, Nayak DR. CDANet: Channel split dual attention based CNN for brain tumor classification in MR images. In: 2022 IEEE international conference on image processing (ICIP). 2022; pp. 4208– 4212. IEEE.
Dutta TK, Nayak DR, Zhang Y-D. ARM-Net: attention-guided residual multiscale CNN for multiclass brain tumor classification using MR images. Biomed Signal Process Control. 2024;87: 105421.
Dosovitskiy A, Beyer L, Kolesnikov A, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929; 2020.
Hassani A, Walton S, Shah N, Abuduweili A, Li J, Shi H. Escaping the big data paradigm with compact transformers. arXiv preprint arXiv:2104.05704; 2021.
Li K, Wang Y, Zhang J, Gao P, Song G, Liu Y, Li H, Qiao Y. Uniformer: Unifying convolution and self-attention for visual recognition. IEEE Trans Pattern Anal Mach Intell. 2023.
Radenović F, Tolias G, Chum O. Fine-tuning CNN image retrieval with no human annotation. IEEE Trans Pattern Anal Mach Intell. 2018;41(7):1655–68.
Cheng J, Yang W, Huang M, et al. Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. PloS One. 2016;11(6):e0157112.
Hu J, Shen L, Sun G. Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). 2018; pp. 7132– 7141.
He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). 2016; pp. 770– 778.
Iandola FN, Han S, et al. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and \(<\) 0.5 mb model size. arXiv preprint arXiv:1602.07360;2016.
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR). 2017; pp. 4700– 4708.
Howard AG, Zhu M, et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861; 2017.
Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In: International conference on machine learning (ICML). 2019; pp. 6105– 6114.
Chattopadhay A, Sarkar A. Grad-cam++: Generalized gradient-based visual explanations for deep convolutional networks. In: IEEE winter conference on applications of computer vision (WACV). 2018; pp. 839– 847.
Funding
This work was partially supported by the Science and Engineering Research Board, Department of Science and Technology, Govt. of India (Grant No. SRG/2020/001460)
Author information
Authors and Affiliations
Contributions
(1) Tapas Kumar Dutta- Formal Analysis, Methodology, Data Collection and Analysis,, Software, Writing-Original Draft Preparation, (2) Deepak Ranjan Nayak- Formal Analysis, Conceptualization, Methodology, Data Collection and Analysis, Software, Validation, Writing-Reviewing and Editing, Visualization, (3) Ram Bilas Pachori- Formal Analysis, Conceptualization, Investigation, Writing-Reviewing and Editing, Supervision, Validation
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dutta, T.K., Nayak, D.R. & Pachori, R.B. GT-Net: global transformer network for multiclass brain tumor classification using MR images. Biomed. Eng. Lett. 14, 1069–1077 (2024). https://doi.org/10.1007/s13534-024-00393-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13534-024-00393-0