Abstract
Objectives
Ecdysone plays a crucial role in the molting process, is synthesized by neverland (nvd) and regulates the transcription of early response genes, HR3 and E75. However, their roles in the development and reproduction of small crustaceans are still poorly understood at the molecular level. Thus, this study aimed to identify and characterize the nvd, HR3, and E75 genes in the brackish water flea Diaphanosomal celebensis, and to investigate their transcriptional modulation upon exposure to bisphenol (BP) analogs.
Methods
Three genes found in the local D. celebensis transcriptome database were sequenced and identified using BLAST X and conserved domain search. The relative expression patterns of these genes at different ages and under exposure to BP analogs were investigated by real-time reverse transcription polymerase chain reaction.
Results
Sequencing analysis showed that nvd, HR3, and E75 had conserved domains. The mRNA expression patterns of Dc-nvd were highly upregulated at day 5 and day 7 of D. celebensis life cycle. The E75 and HR3 mRNA expression patterns differed according to age. The BP analogue exposure test showed that the three genes were significantly modulated with different patterns.
Conclusions
The nvd, HR3, and E75 had conserved domains, suggesting that they have conserved functions in D. celebensis. Age-dependent expression of these three genes implies their involvement in the molting cycle. Our findings also suggest that BPS, BPF, and BPA may disrupt the edcysteroid signaling pathway in this species by different mechanisms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
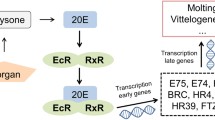
In arthropods, the molting process is an essential step in normal growth, metamorphosis, development, and reproduction. Ecdysone is a key enzyme for modulating molting and synthesized from cholesterol by neverland (encoded by nvd gene). In nvd knockout of Drosophila melanogaster, the larval growth was suppressed, and the larvae finally died, indicating that nvd plays a crucial role in survival and molting [1]. HR3 and E75 are two important transcription regulating factors in ecdysteroid signaling pathway in arthropod, and belong to the early response genes stimulated by ecdysone. They have a ligand-binding domain (LBD) and a DNA-binding domain (DBD) as orphan nuclear receptors (NRs) [2]. HR3 plays a role in activating downstream genes (e.g., vitellogenin and chitin biosynthesis pathway-related genes) in the ecdysteroid signaling pathway, while E75 is involved in suppressing HR3 by binding HR3 or competing with HR3 for DNA binding [3]. Their interaction regulates embryo development and metamorphosis, as well as molting [3]. However, the molecular characteristics and role of these genes in the ecdysteroid signaling pathway are still lacking in small crustaceans.
Bisphenol A (BPA) is a representative endocrine disrupting chemical (EDC) that is widely used in food coatings, cans, and receipt papers as a component of polycarbonate plastics and epoxy resins [4, 5]. Increasing usage of plastic ware has led to increased detection of BPA in aquatic environments [6,7,8]. Because of the negative effects of BPA on development and reproduction, which have been extensively investigated in aquatic organisms, the use and production of BPA have been restricted [9, 10]. Recently, 16 bisphenol (BP) analogs have been developed to replace BPA [5, 8], and bisphenol S (4,40-sulfonyldiphenol; BPS), and bisphenol F (4,40-dihydroxydiphenylmethane; BPF) have been frequently used and broadly detected in South Korea (BPA > BPF > BPS in order of concentration) [7]. Several studies have suggested that BP analogs also have potential toxic effects on the endocrine system (reviewed by Rochester and Bolden) [11]. However, despite their wide distribution in aquatic environments, little information is available on the effects of BP analogs at the molecular level in small crustaceans.
The brackish water flea Diaphanosoma celebensis (Cladocera, Sididae) is mainly found in tropical Asia within a wide range of salinity. They play important roles as primary consumers and transfer energy to higher trophic levels (e.g., fish) in aquatic ecosystems. Because of several advantages, such as short life cycle (4–5 days), easy maintenance under laboratory conditions, single breeding, and small body sizes, D. celebensis is considered a useful non-model species in ecotoxicology [12,13,14]. Our previous studies suggested that some ecdysone-mediated pathway related genes [ecdysone receptors (EcR), ultraspiracle (USP), cyl314a1, vitellogenine, vitellogenine receptor, and estrogen-related receptor] can be transcriptionally modulated by BP analogs in D. celebensis [15, 16].
In the present study, ecdysteroid signaling pathway-related genes (nvd, HR3, and E75) were identified in D. celebensis. Their age-specific expression was examined, and the transcriptional modulation of these genes was further investigated in D. celebensis exposed to different concentrations of BPA, BPS, and BPF. This study is expected to provide a better understanding of the molecular mode of action of BP analogs in cladocerans.
Results and discussion
Identification of nvd, HR3, and E75
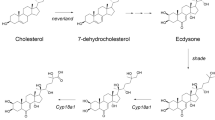
We found two complete sequences of nvd and HR3 and one partial sequence of E75 from a local D. celebensis transcriptome database. The open reading frame (ORF) of D. celebensis nvd (Dc_nvd) cDNA sequence was 1389 bp in length and encoded a polypeptide of 462 amino acids (aa). The putative protein had a theoretical pI of 4.97 and molecular weight of 115.5 kDa. BLASTX search showed a high identity of 61% and 62% with Daphnia magna neverland (KZS09661.1) and cholesterol 7-desaturase-like (XP_032789006.1), respectively. The length of the Dc_nvd was longer than those identified in D. melanogaster (Zhu et al. [17]; 429 aa) and D. magna (Sumiya et al. [18]; 444 aa). D. celebensis nvd had a conserved domain, called the N-terminal Rieske domain (2Fe-2S) (94–203 aa) and non-heme Fe(II) motif (241–253 aa), which is found in the Rieske non-heme iron oxygenase (RO) family. Multiple alignment of a conserved domain and motif showed 44–67% and 69–92% of identity, respectively, with those of other organisms (Fig. 1). The RO family is composed of a large class of aromatic ring- hydroxylating dioxygenases and plays a role in utilizing aromatic compounds for growth in microorganisms [19]. The Rieske domain is similar to the cholesterol 7-desaturase in Caenorhabditis elegans, which catalyzes cholesterol into 7-dehydrocholestoerol in the first step of steroid hormone synthesis [20]. Sequence similarity of nvd suggests that it may be involved in steroid synthesis [21]. The nvd gene has been identified in few crustacean, such as D. magna [18] and the salmon louse (Lepeophtheirus salmonis) [22].
Multiple alignment of the deduced amino acid sequences of conserved domain. A N-terminal Rieske domain and B non-heme Fe(II) motif of D. celebensis neverland with those of other species retrieved from GenBank using Clustal X and GenDoc. Dmag, Daphnia magna (BAQ02388.1, BAQ02389.1); Dmel, Drosophila melanogaster (NP_001097670.1); Bm, Bombyx mori (NP_001037626.1); Pm, Penaeus monodon (APW79685.1); Ci, Ciona Intestinalis (BAK39961.1); Hp, Hemicentrous pulcherrimus (BAK39963.1)
The ORF of D. celebensis HR3 (Dc_HR3) was 1785 bp in length, and encoded a 594 aa polypeptide with a theoretical pI of 6.05 and a molecular weight of 64.9 kDa. The putative protein was highly matched to the HR3 nuclear receptor of D. magna (ACY56690.1) and Daphnia pulex (ACY56691.1) with 73% and 80% identity, respectively, by BLASTX search. Dc_HR3 had two conserved domains, DBD (145–239 residues) and LBD (346–590 residues), which are commonly detected in nuclear receptors. However, because hormone ligand binding to the LBD of HR3 was not identified, it is generally called orphan NR.
D. celebensis E75 (Dc_E75) was obtained partially (2,652 bp ORF encoding an 883 aa polypeptide) and showed 70% and 69% identity with that of D. pulex (ADB79814.1) and D. magna (ABP48738.1), respectively, using BLASTX search. Full-length of E75 ranged from 942 to 954 aa in Daphnia. Two conserved domains, DBD and LBD as orphan NR were found from 15 to 103 aa for DBD, and 171 to 364 aa for LBD. Based on BLASTX and conserved domain searching, sequence information of the three genes was deposited in GenBank (Table 1). HR3 and E75 have been identified in various insects (reviewed by Nakagawa and Henrich [23]), and few crustaceans, such as copepod (Tigriopus japonicus) [24], and cladocerans (Daphnia magna and Daphnia pulex) [3, 25]. While the total sequence similarity was, respectively, low (30% to 74% for HR3; 9–31% for E75) among species, those of DBD (94 to 98% for HR3; 68% to 95% for E75) and LBD (51–86% for HR3; 54 to 90% for E75) were highly conserved (Table 2).
Phylogenetic analysis showed that D. celebensis Nvd was located the same cluster with D. magna nvd1 and nvd2, and E75 and HR3 were also closely clustered with those of Daphnia spp. (Fig. 2). In insects [23] and copepods [24], HR3 and E75 are located in the NR1 family along with EcR in the phylogenetic tree.
Phylogenetic analysis of the deduced amino acid sequences 1) Dc_nvd, B) Dc_E75 and Dc_HR3 with those of other species retrieved from GenBank. The tree was constructed by the neighbor-joining method using MEGA version 6.0 with 1000 bootstrap replicates. GenBank accession no. used in phylogenetic analysis of neverland are as follows: Dmag, Daphnia magna (BAQ02388.1, BAQ02389.1); Dmel, Drosophila melanogaster (NP_001097670.1); Bm, Bombyx mori (NP_001037626.1); Pm, Penaeus monodon (APW79685.1); for outgroup Ci, Ciona Intestinalis (BAK39961.1); Hp, Hemicentrous pulcherrimus (BAK39963.1). Those of E75 and HR3 are indicated in Table 2
Age-dependent gene expression
Ecdysteroid (20-Hydroxyecdysone) is responsible for metamorphosis, development, growth, and reproduction in arthropods [3]. Ecdysteroid synthesis through neverland is the first step in the molting process. RNAi technology revealed that knockout of nvd caused disruption of molting and development in D. magna embryos [26]. Ecdysteroids are transported into the nucleus by binding to EcR and USP [23], which regulate the transcription of ecdysteroid responsive genes, such as the two downstream NRs, HR3, and E75 [27]. E75 acts as a repressor of HR3-mediated gene regulation by forming heterodimers and competitively binding to response elements on DNA [28]. E75 is known to participate in early ecdysone response, oogenesis, and vitellogenesis [2, 23], while HR3 plays an essential role in metamorphosis [23]. More recently, it has been suggested that HR3 is involved in molting by controlling the synthesis and degradation of chitin in Locusta migratoria [29].
In the present study, the mRNA expression patterns of Dc-nvd were highly correlated with the molting period of D. celebensis (Fig. 3A). In et al. [15] showed that molting of adult D. celebensis occurs once every other day since the first molting is observed at day 5, as suggested by Marcial and Hagiwara [12]. The Dc-nvd gene showed a peak at day 5 and 7, while the expression of 20E-hydroxylase (cyp314a1), which is a downstream gene that converts ecdysone into an active form (20-hydroxyecdyson), was highly modulated at day 5 and 8 [15]. This finding suggests a harmony between both genes during the molting process. Similar to Dc_nvd, D. magna nvd1 and nvd2 were highly upregulated during the early inter-molting period (0–10 h after molting) [18]. Martin-Creuzburg et al. [30] suggested that when endogenous ecdysteroid titers are low, molting and vitellogenesis may occur in D. magna. Thus, although little information is available on transcriptional regulation of nvd during the molting period in small crustaceans, fluctuations in the nvd gene seem to play a key role in the regulation of the molting cycle, as suggested by Sumiy et al. [18].
In our results, E75 and HR3 mRNA expression was similarly modulated according to age. The peaks of both genes were observed at day 8 for E75 (Fig. 3B) and at day 6 and 8 for HR3 (Fig. 3C) The fold-change value was higher for HR3 than for E75. In Daphnia, the expression of HR3 mRNA was highly upregulated at 48 h in the molting period, matching with the ecdysteroid levels during the cycle, whereas the level of E75 mRNA level did not change during the molting period [30]. Despite similar pattern on day 8, our results suggest that the expression of both genes was differentially modulated until 7 days, implying the negative regulation of HR3 by E75. However, the interaction of both genes in the molting cycle should be further investigated in small crustaceans.
The mRNA expression of A) Dc_nvd, B) Dc_E75, C) Dc_HR3 during molting of adult Diaphanosoma celebensis (4 to 10 days old). Data are shown as means ± S.D. of 3 replicates. Different lowercase letters indicate significant differences among ages, as determined using a one-way ANOVA followed by Turkey's test
The mRNA expression of Dc_nvd, Dc_E75, and Dc_HR3 in adult Diaphanosoma celebensis exposed to A bisphenol A (0.12, 0.6 and 3.0 mg/L), B bisphenol S (0.92, 4.6 and 23.0 mg/L), and C bisphenol F (0.6, 1.6 and 5.0 mg/L), respectively, for 48 h. Data are shown as means ± S.D. of 3 replicates. Different lowercase letters indicate significant differences among concentrations, as determined using a one-way ANOVA followed by Turkey's test
Regarding the different expression patterns of ecdysteroid signaling pathway-related genes among species, there is a possible explanation. Different target organs can generate different patterns. In insects, ecdysteroids are produced in specific organs, such as Y-organ, whereas it is unknown in crustaceans. Recently, Sumiya et al. [26] suggested that ecdysteroids are synthesized in the gut of D. magna. However, we used the whole body of D. celebensis, which rises the need for further study on the expression of ecdysteroid pathway-related genes in the gut of D. celebensis.
Effects of bisphenol analogs on transcription of three genes
Several studies have reported the modulation of ecdysone pathway genes (e.g., EcR, USP, and cyp314a) by EDCs, including BPA, in crustaceans, such as Chironomus riparius [31,32,33], Gammarus pulex [34], D. celebensis [15], and Tigriopus japonicus [35]. In the present study, transcriptional modulation of the upstream (nvd) and downstream genes (HR3 and E75) of the above-mentioned pathway genes was investigated. As shown in Fig. 4, the three genes were significantly altered upon exposure to BP analogs and showed different patterns. The Dc-nvd mRNA level was highly inhibited after exposure to BPA, whereas its level was significantly upregulated upon BPS and BPF exposures. The expression pattern of Dc_E75 mRNA was sensitively modulated at BPA exposure more than BPS and BPF. In contrast, the Dc_HR mRNA expression was differently changed among BP analogs. Sumiya et al. [26] suggested that nvd may be a target for molting disrupting chemicals in D. magna, as it modulate molting by regulating ecdysteroid synthesis during embryogenesis. In particular, our results support previous studies showing that BPA has anti-ecdysteroidal activity in D. magna [36] and copepods [37].
In addition, in the presence of 20-hydroxyecdysone, HR3 mRNA levels were significantly increased, while E75 mRNA was slightly modulated [30], implying that these genes could also be target genes for ecdysteroid pathway-disrupting chemicals. The different modulation of the BPs was also observed in our previous study using edysteroid pathway-related genes (cyp314a1, EcRA, EcRB, and USP) of D. celebensis [15] and the midge C. riparius [38]. Several studies have suggested that BPS and BPF have similar or low estrogenic activity in aquatic organisms [11, 15, 39]. Although there is as yet little information to compare the impacts of BPs on the expression of nvd, E75, and HR3 in crustaceans, our present findings and previous studies suggest that BPS and BPF may also have endocrine-disrupting properties different from those of BPA in D. celebensis.
Materials and methods
Chemical reagents
All of the chemicals and reagents used in this study were of molecular biology and ultrapure grade. These were purchased from Sigma-Aldrich Co. (Saint Louis, MO, USA) unless otherwise specified.
Experimental organisms
Diaphanosoma celebensis were used in this study the laboratory-cultured strain originally provided by Korea Institute of Ocean Science & Technology (KIOST; Busan, South Korea). The culture medium was 0.2 μm-filtered 15 practical salinity unit (psu) of artificial seawater using an Instant Ocean(Aquarium system, France). D. celebensis were maintained under a 12 h:12 h light/dark photoperiod and at a temperature of 25 ± 1 °C. Chlorella vulgaris cultured in Jaworski’s medium was added as a food source once every two days at a density of 4–4.5 × 108 cells/L.
Waterborne exposure tests
Stock solutions of BPA (2,2-Bis(4-hydroxyphenyl)propane; 6 mg/ml), BPF (4,4′-Methylenediphenol; 10 mg/ml) and BPS (4,4′-Sulfonyldiphenol; 46 mg/ml) were made by dissolving these respective chemicals in dimethyl sulfoxide (DMSO). Final concentration of BPA (0, 0.12, 0.6, 3 mg/L), BPF (0, 0.2, 1, 5 mg/L) and BPS (0, 0.92, 4.6, 23 mg/L) were exposed for 48 h in 4 days of D. celebensis (200 individuals/concentration; 200 ml). Exposure concentrations were determined based on acute toxicity values [15]. A final DMSO concentration of less than 0.05% was used, in which no mortality was observed. During the exposure to chemical, food and new media were not supplied. All tests were performed in triplicate.
Sequence analysis
Complete (Nvd and HR3) and partial (E75) cDNA sequences were obtained from local D. celebensis transcriptome database (Sangmyung University, Seoul, South Korea). BLASTX (igBLAST (ver. 1.17)), NCBI conserved domain search and Expasy were used to identity and characterize each gene. Multiple alignment of each gene was analyzed with those of other species retrieved from GenBank using clustalX (1.83) and GeneDoc ver. 2.6. Phylogenetic tree was constructed by the neighbor-joining method using MEGA version 6.0 with 1000 bootstrap replicates.
Total RNA extraction and cDNA synthesis
To quantify temporal gene expression during the molting period, each sample was harvested every 24 h for 7 days of 4-day old D. celebensis. To investigated relative gene expression, D. celebensis of 4-day old was collected after 48-h exposure to BPA and its analogs. Each sample was homogenized in five volumes of TRIzol reagent (Thermo Fisher Scientific Inc., USA). Total RNA was extracted according to the to the manufacturer’s instructions and stored at − 80 °C until it was used for later analyses. Total RNA quality and quantity were confirmed by gel electrophoresis and Nano drop (Maestrogen nano drop, Taiwan). The cDNA was synthesized from 0.5 ug of the total RNA using RevertAid First strand cDNA synthesis kit (ThermoFisher, MA, USA.).
Relative real-time polymerase chain reaction (RT-PCR)
To examine patterns in the transcriptional expression of D. celebensis three genes (Nvd, E75 and HR3) after exposure to BPs, quantitative RT-PCR was performed in a CFX connect opticus module (Bio-Rad, USA). A qRT-PCR reaction including 2 μL of cDNA and 2 μL of a 10 pmol primer set (Table 1). The PCR cycling conditions were as follows: 95 °C for 10 min; 40 cycles of 95 °C for 15 s and then 60 °C for 1 min. To check the amplification of a specific product, melting curves were produced under the following conditions: 95 °C for 15 s and then 60 °C for 1 min with a 0.5 °C increase per second. Agarose gel electrophoresis and sequence analysis were also carried out to check the specific PCR product. In efficiency tests, 90–105% of efficiency was achieved. The PCR conditions were as follows: an initial step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. All of these experiments used SYBR master mix (KAPA Bioassay System, USA), and were performed in triplicate. The threshold cycle (Cq) from each experiment was normalized relative to that of D. celebensis 18 s rRNA (AF144210.1). The fold change was calculated using the 2−△△Ct method [40].
Statistical analyses
Data from all experiments were presented herein as the mean ± standard deviation (S.D.) of three replicates. Relative mRNA expression levels were compared among treatments using one-way analysis of variance (one-way ANOVA) followed by Tukey’s test. The PASW Statistics 18.0 program (SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. A p value below 0.05 was regarded as statistically significant.
Conclusion
In summary, we identified the ecdysteroid signaling pathway- related genes (nvd, HR3, and E75) and investigated the expression of these genes at different ages and upon exposure to three BPs in D. celebensis. Dc-nvd, HR3, and E75 had conserved domains with those of other species, suggesting that they have conserved functions in D. celebensis. Age-dependent expression of Dc-nvd, -E75, and -HR3 implies their involvement in the molting cycle. In addition, real-time RT-PCR results showed the different modulation of these genes upon exposure to BPs, indicating that BPA, BPS, and BPF may disrupt the edcysteroid signaling pathway in this species by different mechanisms.
References
Yoshiyama T, Namiki T, Mita K, Kataoka H, Niwa R (2006) Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133(13):2565–2574
Bialecki M, Shilton A, Fichtenberg C, Segraves WA, Thummel CS (2002) Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev Cell 3(2):209–220
Hannas BR, LeBlanc GA (2010) Expression and ecdysteroid responsiveness of the nuclear receptors HR3 and E75 in the crustacean Daphnia magna. Mol Cell Endocrinol 315:208–218
Vandenberg LN, Colborn T et al (2012) Hormones and endocrine-disrupting chemicals: low-dose effects and nonmonotonic dose responses. Endocr Rev 33:378–455
Chen D, Kannan K et al (2016) Bisphenol analogues other than BPA: Environmental occurrence, human exposure, and toxicity—a review. Environ Sci Technol 50:5438–5453
Suzuki T, Nakagawa Y, Takano I, Yaguchi K, Yasuda K (2004) Environmental fate of bisphenol A and its biological metabolites in river water and their xenoestrogenic activity. Environ Sci Technol 38:2389–2396
Liao C, Liu F, Moon H-B, Yamashita N, Yun S, Kannan K (2012) Bisphenol analogues in sediments from industrialized areas in the United States, Japan, and Korea: spatial and temporal distributions. Environ Sci Technol 46:11558–11565
Canesi L, Fabbri E (2015) Environmental effects of BPA: Focus on aquatic species. Dose Response 13:1–14
Segner H, Caroll K et al (2003) Identification of endocrine-disrupting effects in aquatic vertebrates and invertebrates: report from the European IDEA project. Ecotoxicol Environ Saf 54:302–314
Gibert Y, Sassi-Messai S et al (2011) Bisphenol A induces otolith malformations during vertebrate embryogenesis. BMC Dev Biol 11:4
Rochester JR, Bolden AL (2015) Bispehnol S and F: a systematic review and comparison of the hormonal activity of bisphenol A substitutes. Environ Health Perspect 123:643–650
Marcial HS, Hagiwara A (2007) Multigenerational effects of 17b-estradiol and nonylphenol on euryhaline cladoceran Diaphanosoma celebensis. Fish Sci 73:324–330
Kim BM, Kang S, Kim RO, Jung JH, Lee KW, Rhee JS, Lee YM (2018) De novo transcriptome assembly of brackish water flea Diaphanosoma celebensis based on short-term cadmium and benzo[a]pyrene exposure experiments. Hereditas 155:36
Bae C, Kim RO, Kim JS, Lee YM (2018) Acute toxicity and modulation of an antioxidant defence system in the brackish water flea Diaphanosoma celebensis exposed to cadmium and copper. Toxicol Environ Health Sci 10:186–193
In S, Yoon HW et al (2019) Acute toxicity of bisphenol A and its structural analogues and transcriptional modulation of the ecdysone-mediated pathway in the brackish water flea Diaphanosoma celebensis. Ecotoxicol Environ Saf 179:310–317
In S, Cho H, Lee KW, Won EJ, Lee YM (2020) Cloning and molecular characterization of estrogen-related receptor (ERR) and vitellogenin genes in the brackish water flea Diaphanosoma celebensis exposed to bisphenol A and its structural analogues. Mar Pollut Bull 154:111063
Zhu Z, Li C, Cheng X, Chen Y, Zhu M, Liu X, Mao S, Qin HM, Lu F (2019) Soluble expression, purification and biochemical characterization of a C-7 cholesterol dehydrogenase from Drosophila melanogaster. Steroids 152:108495
Sumiya E, Ogino Y et al (2014) Roles of ecdysteroids for progression of reproductive cycle in the fresh water crustacean Daphnia magna. Front Zool 11(1):60
Schneider D, Schmidt CL (2005) Multiple Rieske proteins in prokaryotes: where and why? Biochim Biophys Acta 1710(1):1–12
Wollam J, Magomedova L et al (2011) The Rieske oxygenase DAF-36 functions as a cholesterol 7-desaturase in steroidogenic pathways governing longevity. Aging Cell 10:879–884
Yoshiyama-Yanagawa T, Enya S et al (2011) The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J Biol Chem 286(29):25756–25762
Sandlund L, Kongshaug H et al (2018) Identification andcharacterisation of the ecdysone biosynthetic genes neverland, disembodied and shade in the salmon louse Lepeophtheirus salmonis (Copepoda, Caligidae). PLoS ONE 13(2):e0191995
Nakagawa Y, Henrich VC (2009) Arthropod nuclear receptors and their role in molting. FEBS J 276:6128–6157
Hwang DS, Lee BY et al (2014) Genome-wide identification of nuclear receptor (NR) superfamily genes in the copepod Tigriopus japonicus. BMC Genom 15:993
Thomson SA, Baldwin WS, Wang YH, Kwon G, LeBlanc GA (2009) Annotation, phylogenetics, and expression of the nuclear receptors in Daphnia pulex. BMC Genom 10:500
Sumiya E, Ogino Y et al (2016) Neverland regulates embryonic moltings through the regulation of ecdysteroid synthesis in the water flea Daphnia magna, and may thus act as a target for chemical disruption of molting. J Appl Toxicol 36:1476–1485
Thummel CS (1996) Flies on steroids–Drosophila metamorphosis and the mechanisms of seroid hormone action. Trends Genet 12:306–310
Swevers L, Ito K, Iatrou K (2002) The BmE75 nuclear receptors function as dominant repressors of the nuclear receptor BmHR3A. J Biol Chem 277:41637–41644
Zhao X, Qin Z et al (2018) Nuclear receptor HR3 controls locust molt by regulating chitin synthesis and degradation genes of Locusta migratoria. Insect Biochem Mol Biol 92:1–11
Martin-Creuzburg D, Westerlund SA, Hoffmann KH (2007) Ecdysteroid levels in Daphnia magna during a molt cycle: determination by radioimmunoassay (RIA) and liquid chromatography–mass spectrometry (LC–MS). Gen Comp Endocrinol 151:66–71
Planelló R, Martínez-Guitarte JL, Morcillo G (2008) The endocrine disruptor bisphenol A increases the expression of HSP70 and ecdysone receptor genes in the aquatic larvae of Chironomus riparius. Chemosphere 71:1870–1876
Nair PMG, Choi J (2012) Modulation in the mRNA expression of ecdysone receptor gene in aquatic midge, Chironomus riparius upon exposure to nonylphenol and silver nanoparticles. Environ Toxicol Pharmacol 33:98–106
Morales M, Martínez-Paz P et al (2014) Transcriptional changes induced by in vivo exposure to pentachlorophenol (PCP) in Chironomus riparius (Diptera) aquatic larvae. Aquat Toxicol 157:1–9
Gismondi E (2018) Identification of molt-inhibiting hormone and ecdysteroid receptor cDNA sequences in Gammarus pulex, and variations after endocrine disruptor exposures. Ecotoxicol Environ Saf 158:9–17
Hwang DS, Han J et al (2016) BDE-47 causes developmental retardation with down-regulated expression profiles of ecdysteroid signaling pathway-involved nuclear receptor (NR) genes in the copepod Tigriopus japonicus. Aquat Toxicol 177:285–294
Mu X, Rider CV, Hwang GP, Hoy H, LeBlanc GA (2005) Covert signal disruption: anti-ecdysteroidal activity of bisphenol A involves cross-talk between signaling pathways. Environ Toxicol Chem 24:146–152
Andersen HR, Wollenberger L, Halling-Sørensen B, Kusk KO (2001) Development of copepod nauplii to copepodites—a parameter for chronic toxicity including endocrine disruption. Environ Toxicol Chem 20:2821–2829
Herrero Ó, Planelló R, Morcillo G (2015) The plasticizer benzyl butyl phthalate (BBP) alters the ecdysone hormone pathway, the cellular response to stress, the energy metabolism, and several detoxication mechanisms in Chironomus riparius larvae. Chemosphere 128:266–277
Rosenmai AK, Dybdahl M et al (2014) Are structural analogues to bisphenol A safe alternatives? Toxicol Sci 139:35–47
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Acknowledgements
This work was supported by a 2020 research grant from Sangmyung University funded to Young-Mi Lee.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Soyeon In, Hayoung Cho and Young‑Mi Lee declare that we have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
In, S., Cho, H. & Lee, YM. Identification of ecdysteroid pathway-related genes and their transcriptional modulation in the brackish water flea Diaphanosoma celebensis exposed to bisphenol analogs. Toxicol. Environ. Health Sci. 13, 261–268 (2021). https://doi.org/10.1007/s13530-021-00103-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13530-021-00103-8