Abstract
In Daphnia magna, 20-hydroecdysone (20E) is the main molting hormone and its metabolism is of interest to identify new biomarkers of exposure to contaminants. The present study aimed to (i) assess baseline levels of 20E and transcription levels of four related-genes (shade, neverland, ultraspiracle, and ecdysteroid receptor); and (ii) evaluate effects in D. magna after 21 days of exposure to fenarimol (anti-ecdysteroid) and a mixture of gemfibrozil and clofibric acid (lipid-lowering drugs) at sublethal concentrations. Endpoints included transcription of the target genes and quantification of 20E, mortality, and reproduction of daphnids. Baseline results showed that average responses were relatively similar and did not vary more than 2-fold. However, intra-day variation was generally high and could be explained by sampling individuals with slightly different stages of their development. Exposure tests indicated a significant decrease in daphnid reproduction following chronic exposure to a concentration of 565 μg/L of fenarimol. However, no difference was observed between the control and exposed groups for any of the investigated genes, nor for the levels of 20E after 21 days of exposure. Following exposition to gemfibrozil and clofibric acid at 1 μg/L, no changes were observed for the measured parameters. These results suggest that changes in transcription levels of the target genes and concentrations of 20E may not be sensitive endpoints that can be used as biomarkers of sublethal exposure to the target compounds in D. magna. Measuring multiple time points instead of a single measure as well as additional molecular endpoints obtained from transcriptomic and metabolomic studies could afford more insights on the changes occurring in exposed daphnids to lipid-altering compounds and identify efficient biomarkers of sublethal exposure.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Currently, more than 350,000 chemicals and mixtures have been registered for production and use around the world (Wang et al. 2020). The high production volumes and mobility of compounds lead to their detection in surface waters worldwide at trace concentrations (ng/L to μg/L). Exposure and accumulation in aquatic species can also adversely impact ecosystems (Bradley et al. 2017; Hughes et al. 2013). Some of these compounds, such as pesticides, have been created to specifically affect the endocrine system of arthropods and thus are toxic to nontarget aquatic organisms such as crustaceans (Jansen et al. 2011; Mnif et al. 2011). Pharmaceuticals are also susceptible to causing subtle changes in nontarget species such as feminization and impacting the behavior of different aquatic species (Richmond et al. 2017).
Classic toxicity tests using endpoints such as survival, growth, or reproduction are usually not sensitive enough to detect the effects of these compounds at environmental concentrations (Daughton and Ternes 1999). However, adverse effects such as changes in behavior, metabolic profile, or gene transcription have been observed at sublethal levels (De Lange et al. 2006; Houde et al. 2013; Kovacevic et al. 2016; Wagner et al. 2017). While in-silico techniques based on molecular modeling and docking simulations (Hirano et al. 2020; Li et al. 2023) can be useful to predict interactions of contaminants with key enzymes, in-vivo studies are necessary to identify new biomarkers of sublethal effects in order to assess biological changes in aquatic organisms chronically exposed to low concentrations of contaminants. An interesting model to look for those biomarkers is Daphnia magna.
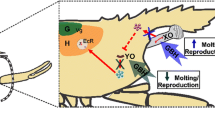
Daphnia is a genus of freshwater crustaceans widely used in ecotoxicology as model species to test the toxicity of chemicals an even wastewaters (Tonkes et al. 1999) because of its easy culture in the laboratory, small size, as well as its parthenogenetic (clonal) reproduction (Dodson and Hanazato 1995). Daphnids occupy a key role in lentic ecosystems as filter feeders and prey of insects and small fishes (Miner et al. 2012). Reproduction and development in daphnids are regulated by ecdysteroids, a group of hormones derived from cholesterol. 20-Hydroxyecdysone (20E) is the main molting hormone in crustaceans and other arthropods and is also involved in the reproduction process and embryonal development of daphnids (LeBlanc 2007). Levels of 20E increase and decrease between successive molts in a pulsative manner, inducing ecdysis through the activation of the ecdysone nuclear receptor (EcR) (Martin-Creuzburg et al. 2007; Song et al. 2017). 20E metabolism is regulated by a group of genes named the Halloween family. These genes encode for cytochrome P450 (CYP450) enzymes that regulate the biosynthesis of ecdysteroids from cholesterol (Fig. 1) (Rewitz and Gilbert 2008).
Given their importance for the survival and reproduction of D. magna, ecdysteroids are potential targets for biomarker studies. Only a few publications have investigated changes in concentrations of ecdysteroids in D. magna after exposure to contaminants. Bodar et al. (1990) reported an increase in ecdysteroid titers of 257% in adult females following exposure to 20 μg/L of cadmium for 8 days and used enzyme immunoassays to quantify ecdysteroids as ecdysone equivalents. They speculated that the observed effect of cadmium on ecdysteroids was the product of cadmium interference with metallo-enzymes involved in the molting process. Mu and LeBlanc (2002) exposed neonates to 497 μg/L of fenarimol, a fungicide principally used on ornamental plants and vegetables, and observed a diminution of around 26% in ecdysteroid levels in exposed neonates and an induced delay for the first and second molting in a dose-dependent manner. These effects were somewhat reverted with the co-administration of 20E, which demonstrated the anti-ecdysteroid activity of fenarimol.
Baseline assessment of both key metabolites and transcription of genes taking part in the synthesis of those metabolites and can help to differentiate natural fluctuations from responses induced by exposure to exogenous compounds. Therefore, in order to understand the effects of contaminants on the metabolism of 20E, it is important to determine first baseline levels of 20E as well as of genes associated with its synthesis and activity.
At the gene level, baseline information of Halloween genes transcription was evaluated during normal growth in daphnids by Sumiya et al. (2014). The transcription level of the evaluated genes fluctuated between 2- and 3-fold during an 80-h period, equivalent to the duration of molting in adult daphnids. At the metabolite level, two studies have reported ecdysteroid baseline levels in adult D. magna during a single molt cycle (Martin-Creuzburg et al. 2007; Sumiya et al. 2016). In those studies, the authors used immunoassay-based techniques to quantify ecdysteroids, and they both observed an increase in basal levels of ecdysteroids between ≈ 30 to 50 h after ecdysis followed by return to basal levels ≈ 40 h later. However, reported values by these studies diverge. Martin-Creuzburg et al. (2007) used a radioimmunoassay technique and observed that free ecdysteroids increased from ≈ 5 to 10 pg per individual at the end of the first molt to a maximum of ≈ 2 to 50 pg/individual 38–42 h later. Then ecdysteroids declined back to basal levels before the second molt. Sumiya et al. (2016) used enzyme-linked immunosorbent assay to measure ecdysteroids but maximum levels were only about ≈ 2.4 fg/ind. These differences may be due to the distinct techniques and sampling protocols employed. To the author’s knowledge, the only study that has quantified 20E in D. magna is the work of Venne et al. (2016). The authors of that paper used liquid chromatography-triple quadrupole mass spectrometry (LC-QqQMS) to quantify 20E in adult daphnids (19 ± 8 pg/ind). However, only one measure was performed. Therefore, up to now, the baseline of 20E in multiple molt cycles of D. magna is unknown.
The working hypothesis of the present work was that changes in levels of 20E and/or changes in the transcription of 20E-related genes (shade, neverland, ultraspiracle, and ecdysteroid receptor) would be observed in D. magna after exposure to low concentrations of lipid-altering organic contaminants. Fenarimol, a fungicide demethylation inhibitor and known ecdysteroid inhibitor, as well as a mixture of gemfibrozil and clofibric acid, two lipid-lowering molecules commonly found in environmental waters were chosen as target compounds. Thus, the present study aimed to (i) assess the baseline of 20E and transcription of the genes associated with 20E regulation in D. magna over a 21-day period; and (ii) evaluate the effects following a 21-day exposure to the target compounds on the concentration of 20E, expression of target genes, and life history parameters (i.e., fertility and mortality).
Materials and methods
Reagents and chemicals
Standards of 20E (catalog number: SC-202407A, > 98% purity) and makisterone A (SC-202218A, > 95% purity) were obtained from Santa Cruz Biotech (Dallas, TX). Makisterone A is non-endogenous ecdysteroid with 28 carbon atoms that differs from all 27 carbon molting hormones like ecdysone and 20E by having a methyl group at the C-24 position. This compound responds similarly to 20E during extraction and LC-QqQMS analysis; it was therefore used as an internal standard for 20E quantification. Additional purification of makisterone A to remove 20E and ecdysone impurities was done following the method described by Venne et al. (2016). Water, methanol (MeOH), acetonitrile (ACN), methyl tert-butyl ether (MTBE), formic acid (FA), and acetic acid (AA) were LC or LC-MS grade and were purchased from Thermo Fisher Scientific (Waltham, MA). The derivatization reagent hydroxylamine hydrochloride (159417-100G, > 99%), fenarimol (45484-250MG, ≥ 99%), its internal standard nuarimol (31116, ≥ 99%), clofibric acid (90323-100MG, ≥ 99%), and gemfibrozil (91823-100MG, ≥ 98.5%) were purchased from Sigma-Aldrich (St-Louis, MO). Deuterated standards, clofibric acid-d3 (D-6005, 98%), and gemfibrozil-d6 (D-6144, 99%) were purchased from CDN Isotopes (Pointe-Claire, QC, Canada). Stock solutions of 20E were prepared at 0.1 mg/mL in MeOH and working solutions, prepared in 1% FA in MeOH and stored at – 20 °C. The aqueous solution of hydroxylamine hydrochloride solution (100 mg/mL) was prepared before each derivatization. The main properties of the target compounds are found in Table S1 (Supplementary Information).
Daphnia magna culture
D. magna parent stock originated from ephippia acquired from EBPI Canada (Burlington, ON) and maintained in the laboratory in synthetic Moderately Hard Reconstituted Water (Environment Canada 1990). Cultures were kept at 20 ± 1 °C with a 16-h light:8-h dark photoperiod and were renewed every 2 months using neonates from 3rd to 5th broods. Daphnids were fed every second day with 2 mL of green algae Raphidocellis subcapitata (≈ 3.85 × 105 cells/mL). Microalgae were cultured in Bold Modified Basal Freshwater medium from Sigma-Aldrich under the same laboratory conditions described above for D. magna. Algae were regularly harvested while still in the exponential growth phase and inoculated in fresh medium. All experiments were initiated with neonates (> 24 h old), born between the 3rd and 5th broods, derived from a healthy parent stock.
Baseline levels of 20E and transcription of target genes
The first experiment aimed to evaluate the gene transcription of target genes and 20E levels over a 21-day period, the duration of standardized chronic toxicity tests for D. magna. During this experiment, daphnids were maintained under the conditions described above. Every second day, starting from day 9 (organisms at maturity, size needed for LC-QqQMS analysis) until day 21, three replicates of 15 daphnids were sampled for 20E quantification and stored in MeOH at − 80 °C until analysis. Five replicates of one daphnid each were also collected for gene transcription analysis and stored in trizol at − 80 °C until analysis.
Chronic exposure to fenarimol, gemfibrozil, and clofibric acid
Three different exposure tests were performed to evaluate if changes in levels of 20E would be observed in D. magna after exposure to low concentrations of lipid-altering organic contaminants. For test N° 1, gemfibrozil and clofibric acid were used at a concentration of 1 μg/L each which is of the same order of magnitude as the maximum reported environmental concentrations in surface waters reported so far for these two compounds (Ebele et al. 2017). For tests N° 2 and N° 3, two concentrations of fenarimol (113 μg/L and 565 μg/L) corresponding to the no observed adverse effect concentration (NOAEC) and the lowest observed effect concentration (LOEC) for reduced fertility in D. magna, respectively, were employed. Thus, the employed exposure concentrations were selected to reflect environmental levels (test N° 1), as well as reported sublethal concentrations (test N° 2 and N° 3) that are representative of worst-case exposure scenarios such as contaminated discharges due to runoff events near agricultural fields (Lefrancq et al. 2017).
In all assays, neonates (< 24 h) were exposed for 21 days to the contaminant following the OECD guidelines (OECD 2008). Tests were performed thrice using 10 replicate groups (5 control, 5 exposed) of 25 daphnids each. Temperature was kept at 20 ± 1 °C using an incubator, light intensity was 2000 ± 70 lx and a 16-h light:8-h dark photoperiod was maintained. On day 1 of the tests, neonates from the control groups were transferred in 2 L beakers filled with culture medium to which 40 μL of MeOH was added. Neonates from the exposed groups were transferred to 2 L beakers filled with culture medium containing 40 μL of MeOH containing the test compound. Medium was renewed 3 times a week. When performing these renewals, daphnids were sorted by size using a series of sieves, according to a standard protocol used by the Ministry of the Environment of Quebec, Canada (Centre d'expertise analytique environnementale du Québec 2011). Adults were collected on a 900 μm sieve, juveniles on a 560 μm sieve, and neonates on a 300 μm sieve. Offspring (juveniles and neonates) were counted and then eliminated; only adults were transferred to the renewed solutions. At each media renewal, survival was determined by counting and averaging number of non-immobilized adults and reproduction was determined by counting and averaging the number of offspring per adult. Dormant eggs or males were never observed throughout the experiments, which indicated that experimental conditions were adequate to support a healthy population of D. magna. Daphnids were fed with 2 mL of a Raphidocelis subcapitata algae solution at every media renewal and the number of offspring and mortality was recorded. A maximum of 18.9% of mortality was observed in the exposure experiment, thus respecting the sublethality criteria guidelines of the OECD (2008). Detailed mortality curves can be found in Figs. S1, S2 and S3 (Supplementary Information). At the end of the exposure period, D. magna adults were collected in MeOH 1% FA in MeOH or trizol for 20E levels and gene transcription analysis, respectively, and stored at – 80 °C until analysis.
Quantification of 20E using liquid chromatography-triple quadrupole mass spectrometry
20E was extracted and quantified from whole daphnids (15–25 individuals) following the method developed by Venne et al. (2016) with minor modifications. Briefly, D. magna were sorted by size and adult individuals (> 900 μm) were collected on a tissue strainer before being washed with deionized water (18 MΩ). Adult daphnids were homogenized with a mortar and a pestle and sonicated in an ultrasonic bath for 15 min in a volume of 1 mL of 1% FA in MeOH inside a 1.5 mL Eppendorf tube. Next, an aliquot of 800 μL was transferred in a glass tube with 50 μL of internal standard solution (makisterone A) and the solvent was evaporated under a gentle flow of N2(g). 20E and makisterone A were then derivatized to their oxime analogues with 1 mL of a hydroxylamine hydrochloride solution (100 mg/mL). A liquid-liquid extraction with 2 × 1.5 mL of MTBE was carried out with a vortex mixer. The test tubes were then placed at – 20 °C until the water froze, and the organic layer (unfrozen) containing the derivatized analyte and its internal standard was transferred to a test tube and evaporated to dryness. Finally, 250 μL of MeOH were added and the samples were transferred to vials for analysis.
The 20E concentration in daphnid extracts was quantified by LC-QqQMS using an Acquity UPLC system coupled to a Xevo TQ-S micro triple quadrupole mass spectrometer, both from Waters (Billerica, MA). The method was adapted from Venne et al. (2016). Briefly, the compounds were separated on a reversed-phase column Acquity UPLC Cortecs C18+ from Waters of dimensions 50 × 2.1 mm and 1.6 μm particle size. The mobile phase was composed of eluent A (H2O containing 0.1% v/v of AA) and eluent B (mixture of MeOH-ACN 3:2 v/v, containing 0.1% v/v of AA). The elution gradient started with 5% of B, increasing to 55% in 7.9 min, rising immediately to 100% of B and hold for 2 min, then back to initial conditions for column re-equilibration (2.1 min). The sample injection volume was set to 10 μL. Electrospray in the positive mode was used as ionization source and the mass spectrometer was operated in the multiple reaction monitoring (MRM) mode. MRM transitions used were m/z 478.3 → m/z 316.3 for 20E oxime and m/z 492.2 → m/z 316.1 for makisterone A oxime. The concentration of 20E was reported as the average mass per adult individual. The total number of D. magna used and the number of pooled groups varied among the tests due to differences in the mortality of daphnids at the end of the 21-day period.
Quality control (QC) samples were used in order to determine the deviation percentage of the method and thereby verify the accuracy and precision of the measured concentrations. Three concentrations of QC samples were used: low (≈ 100 pg/mL), medium (≈ 300 pg/mL), and high (≈ 500 pg/mL). Matrix effects correction was done by using extracts of frozen daphnids at 50 mg/mL in 1% FA in MeOH. These extracts were used to prepare QC samples. Results were considered acceptable if the QC samples were within ± 20% of the expected value.
Chemical stability
The chemical stability of fenarimol, gemfibrozil, and clofibric acid was evaluated during the tests between two medium renewals. Nuarimol, gemfibrozil-d6, and clofibric acid-d4 were used as internal standards. Aliquots of 50 mL were sampled immediately after and before media renewal at three different moments during the testing. Extraction was performed using Strata-X solid-phase extraction cartridges (polymeric reversed phase with a particle diameter of 33 μm, 200 mg of bed mass, and 6 mL of volume) from Phenomenex (Torrance, CA). Quantification was carried out using the same LC-QqQMS system described previously. The entire protocol is detailed in the Supplementary Information.
RNA extraction
Total RNA was extracted from single D. magna using a Trizol-RNeasy Plus Mini Kit hybrid protocol (Ponton et al. 2011). Briefly, daphnids were homogenized in 500 μL of trizol with a micro pestle before being sonicated for 5 min. A volume of 200 μL of chloroform was added and the tubes were centrifuged at 10,000 g for 18 min at 4 °C. The top layer was then transferred on a RNeasy Plus Mini column from Qiagen Canada (Montreal, QC) and the manufacturer’s instructions were followed from that point (Quiagen 2020). Extractions were performed on 10 independent biological replicates for exposures to 113 μg/L and 15 replicates for the 565 μg/L treatment with fenarimol.
RNA was quantified with a NanoDrop ND-000 spectrophotometer from Thermo Fisher Scientific (Waltham, MA). All samples had a 260 nm/280 nm ratio > 1.8 and a concentration > 54 ng/μL. Chloroform (HPLC, 99%) and ethanol (98%) were purchased from Sigma-Aldrich Canada (Oakville, ON). Random hexamer 5′-NNN NNN-3′ made for L. Gaudreau (IDT, lot 213632751) was used for reverse transcription in addition to dNTP mix (10 mM) from KAPA Biosystems (Cape Town, South Africa), Moloney Murine Leukemia Virus (M-Mulv reverse transcriptase, 200,000 U/mL, lot 12R091118), and 10×M-MulV RT Buffer (lot 081618) from QIAGEN and sterile water (Molecular grade) from Wisent (St-Bruno, QC). Advanced qPCR Mastermix (lot 800431) from Wisent, 96-well plates (Low profile, Clear) from Axygen (Union City, CA), and sealing tapes (optically clear) from Sarstead (Newton, NC) were used for qPCR reaction.
Real-time quantitative PCR
Real-time quantitative PCR (RT-qPCR) analyses were conducted on four selected transcripts of the target genes shade (shd), neverland (nvd), ecdysteroid receptor (ecr), and ultraspiracle (usp) and normalized with a combination of the most suitable reference genes cyclophilin (cyc), tubulin α (tuba), ubiquitin (ubi), elongation factor 2 (eef2), and glyceraldehyde 3-phosphate dehydrogenase (gapdh). Genes ubi, eef2, and gapdh were used for the baseline assessment experiment. Genes ubi, tuba, and eef2 were used for test No 2 (exposure to 113 μg/L of fenarimol). Genes ubi, tuba, and gapdh were used for test No 3 (exposure to 565 μg/L of fenarimol). Primer-specific efficiencies and sequences are listed in Table S2 (Supplementary Information) along with sequences.
Total RNA (300 ng) was reverse transcribed using M-Mulv reverse transcriptase according to the manufacturer’s instructions (Qiagen 2023). After dilution of the cDNA samples (1/8 dilution), analyses were then carried out on a CFX96 Connect real-time PCR detection system from Bio-Rad (Hercules, CA) using Advanced qPCR Mastermix with a final concentration of 400 nM for each primer in a total reaction volume of 10 μL. The qPCR conditions were as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 68 °C for 15 s. Each reaction was run in technical triplicate and the mean of all independent biological replicates was calculated. All results were normalized using mRNA level of the reference genes recommended by geNorm depending on the exposition test. Relative expression values were calculated by the qBase relative quantification software (Hellemans et al. 2007). Microcapillary gel electrophoresis (Fig. S4. Supplementary Information) and melt curve analysis were performed on amplicons to verify the specificity of the amplification using a 2100 Bioanalyzer from Agilent (Santa Clara, CA) and CFX96 Connect real-time PCR detection system.
Data analysis
Two-sample t tests (α = 0.05) using Microsoft Excel 365 were employed to evaluate significant differences between exposed and control samples for survival, number of offspring, and 20E levels observed. F test were also performed to compare variances between the two groups. For the baseline analysis of 20E levels and the transcription of target genes, analysis of variance (ANOVA) tests (α = 0.05) were performed between time points using OriginPro version 2023. Before ANOVA tests, data normality and homoscedasticity were verified using Kolmogorov-Smirnov’s test and Levene’s test, respectively. When one of these conditions were not respected, a Kruskal-Wallis ANOVA test was used instead. Tukey’s and Dunn’s post-hoc tests were used to determine which groups were different for ANOVA and Kruskal-Wallis tests, respectively. Graph Pad Prism version 6 was used to plot the results of all assays.
Results
Baseline of target genes and levels of 20E
Transcription of the target genes (shd, nvd, ecr, and usp) as well as 20E levels were evaluated starting from day 9 of the daphnids’ life until day 21 to assess temporal variability during normal development. Most of the transcription of the targeted genes was relatively stable with little to no change over the duration of the observations (Fig. 2). A slight but significant increase at day 19 compared to days 9, 11, and 15 was observed for the transcription levels of ecr (Tukey’s test, p < 0.05) and at day 17 compared to day 15 for usp (Dunn’s test, p = 0.0084). Transcripts of shd were the most stable over time with no significant difference (p > 0.05) at any day and nvd was the most active gene in terms of fluctuation. The relative transcription levels of the latter closely followed the concentrations of 20E (Fig. 3). The gene nvd is responsible for the 7-dehydrogenase, the enzyme catalyzing the first step of ecdysteroid biosynthesis (Song et al. 2017). The transcription analysis results thus suggest that the retroaction in response to 20E levels controlling ecdysteroids production mainly affects this step (7-dehydrogenation of cholesterol).
Relative gene transcription of shd, nvd, ecr, and usp in daphnids for a 12-day period (n = 5). Error bars represent 95% confidence interval limits. Data was normally distributed according to Kolmogorov-Smirnov’s test (for all genes p > 0.1) and the variance was homogeneous according to Levene’s test only for ecr (p = 0.3952)
20E baseline in daphnids over a 12-day period (n = 3 per time point). Error bars represent ± 1 standard error. Means that do not share the same letter were significantly different (p < 0.001) according to Tukey’s post-hoc test. Data was normally distributed according to Kolmogorov-Smirnov’s test (p = 1.0) and the variance was homogeneous according to Levene’s test (p = 0.1477)
Figure 3 shows the 20E levels in D. magna at different time points. Significantly different (p < 0.05) levels of 20E were seen depending on the age of the individuals and the concentration of 20E per daphnid oscillated about every 4 days since day 11 which seems to correspond to the molting cycle duration of D. magna in light and temperature conditions employed (Ebert 2005). However, contrary to previous reports (Martin-Creuzburg et al. 2007; Sumiya et al. 2016), a clear ecdysteroid peak (up to ≈ 50 times the basal level) is absent in Fig. 3.
In the present study, the maximum difference between 20E concentrations measured was only 18 pg/ind and the relative standard deviation was between ± 20 and ± 50%. This observation could be explained by the desynchronization of the molting between sampled daphnids and differences in sampling frequency. Indeed, despite the authors’ efforts to control culture experimental conditions (feeding, temperature, photoperiod, culture medium, etc.), molting cycles of daphnids could not be synchronized and it was hypothesized that small differences in their age (a few hours) could result in significant different levels of 20E at the time of sampling. According to Martin-Creuzburg et al. (2007), 20E concentrations could vary by a factor as high as 50× in less than 36 h. At this point, it cannot be ruled out that other uncontrolled experimental parameters affected the rhythm of the daphnids molting cycles which, like other rhythmic behaviors, could be influenced by numerous environmental cues (Häfker and Tessmar-Raible 2020).
Survival
Daphnids reached adulthood, defined in the present study as a size > 900 μm, between days 7 and 10. Survival was over 80% for all groups in all tests, thus respecting the sublethality criteria of the OECD. According to Figs. S1 to S3 (Supplementary Information), no significant difference (p > 0.05) in the survival was observed between exposed and control groups in test No 1 (gemfibrozil and clofibric acid at 1 μg/L each), but a significant difference in survival was observed for tests No 2 and No 3 (fenarimol at NOAEC and LOEC, respectively) after 21 days. This was surprising for the lowest concentration of fenarimol, but the survival rate was still over 80%.
Reproduction
In test No 1 (gemfibrozil and clofibric acid each at 1 μg/L), only at day 10, a significant difference (p = 0.0218) was observed on the number of offspring (Fig. 4a). Results from daphnids exposed to fenarimol (test No 2: 113 μg/L and No 3: 565 μg/L) indicated that only the highest concentration used in latter test caused a consistent decrease in reproduction (Fig. 4b and c). Indeed, in test No 3, a significant diminution in the number of offspring per adult was observed compared to controls in three consecutive days: 17 (p = 0.0020), 19 (p = 0.0495), and 21 (p = 0.0013).
Offspring production per daphnid exposed for 21 days to a 1 μg/L of gemfibrozil and clofibric acid (n = 10), b 113 μg/L (n = 10), and c 565 μg/L (n = 15) of fenarimol. Error bars represent ± 1 standard error. Asterisks (*) indicate statistically significant differences compared to controls (p < 0.05)
Ecdysteroid levels
For all tests, experiments showed that the level of 20E did not differ significantly (p > 0.05) between controls and organisms exposed (Fig. 5). For the gemfibrozil and clofibric acid exposure (Fig. 5a), control and exposed groups had 20E mean levels of 21 ± 6.1 pg/ind and 16 ± 6.0 pg/ind, respectively. In test No 2 (Fig. 5b), the values observed for control and exposed individuals were 20 ± 17 pg/ind and 9.8 ± 9.1 pg/ind, and for test No 3 (Fig. 5c), 35 ± 24 pg/ind and 24.0 ± 9.7 pg/ind.
Concentration of 20E (pg/individual) in daphnids exposed for 21 days to a 1 μg/L of gemfibrozil and clofibric acid (n = 10), b 113 μg/L (n = 6 for exposed group and n = 9 for control group), and c 565 μg/L (n = 13 for both groups) of fenarimol. Error bars represent ± 1 standard error. Asterisks (*) indicate statistically significant differences compared to controls (p < 0.05)
Transcription of targeted genes
Genes linked to ecdysteroid metabolism such as shd, nvd, usp, and ecr (Goodman and Cusson 2012) were monitored to evaluate if exposure to fenarimol at sublethal levels (test No 2 and No 3) during 21 days could change their transcription levels. The results of these experiments are shown in Fig. 6a and b.
As shown in Fig. 6a and b, expression of any gene at the end of the experiments was not affected at the fenarimol NOAEC (113 μg/L) or LOEC (565 μg/L). These results agree with the 20E data presented in Fig. 5. In Fig. 6a, we can see that nev is the most variable gene in those observed. Since nev regulates the first step of the ecdysteroids biosynthesis from cholesterol, it is possible that this gene is more up and down regulated, initializing and stopping the biosynthesis when needed, thus explaining these variations in transcription levels. However, it not yet clear why such high variability is only observed at the lowest concentration of fenarimol tested.
Discussion
Baseline of target genes and levels of 20E
Results reported by Sumiya et al. (2014) for D. magna were similar to the transcription levels measured in the present study for shd, nvd, usp, and ecr across an intermolt sampling period, with variation ranging between 2 and 3 times the fold change depending on the time point. The relative stability observed here for gene transcription during the normal growth of daphnids could be due to the developmental stage of daphnids sampled and the selection of genes. Many other genes and their isoforms are responsible for the regulation of ecdysteroids. For example, besides shd and nvd other genes of the Halloween family (spook, spookier, disembodied, and phantom) are implicated in the metabolic cascade that biosynthetizes 20E (Song et al. 2017). A time course transcriptomic analysis during the growth period would provide optimal information for the selection of the genes and periods to follow during exposures. When looking at the transcription levels of Halloween genes during normal development in other species, lower differences could be observed between time points. Indeed, for shd and nvd, two genes also measured in the present study, the relative transcription levels in the moth Plutella xylostella oscillated between 0.02 and 0.2 (Peng et al. 2019). The same study also observed differences in gene expression of Halloween genes in P. xylostella according to the developmental stage of individuals and the tissues sampled. These results indicate that even if very slight or non-significant changes in the transcription levels of these genes are observed, developmental changes (growth, reproduction) can still occur.
Finding the right moment for sampling gene transcription levels is crucial, especially for rapidly modulated transcripts. By following several genes of the same family throughout a time-course interval, a better picture of the metabolism regulated by those genes can be drawn. A pulsating pattern was observed during the normal growth of the daphnids, and the basal transcription levels were assessed. However, more genes should be studied, ideally following one another in the metabolic pathway of the studied way. As for the sampling interval, a much shorter time period could give a better picture of the transcription levels across time and will help in correlating these transcription levels with the corresponding metabolites such as ecdysteroids.
The results in Fig. 3 were obtained with pooled individuals (15–25 daphnids). Therefore, daphnids are no longer synchronized by day 9, and as a result, differences in 20E concentration as a function of time are much more subtle and the variability of each measure is higher. Regarding sampling frequency, 7 measurements were done within a 12-day period, while Martin-Creuzburg et al. (2007) performed more than 20 measurements within a 3-day period. Finally, while it is not possible to compare the concentrations observed since previous studies used immunoassay-based techniques rather than mass spectrometry, the reported concentrations herein were about an order of magnitude lower than those reported by Martin-Creuzburg et al. (2007) but about 10,000 times higher than those reported by Sumiya et al. (2016). Besides the techniques employed, those differences can be explained by the sampling of individuals at different stages of their molting cycles.
Reproduction
The results from tests No 1 (gemfibrozil and clofibric acid each at 1 μg/L) (Fig. 4a) are in agreement with a previous study that showed that chronic exposure (30 days) to 1 μg/L of clofibric acid did not affect the reproduction of D. magna (Flaherty and Dodson 2005). Steinkey et al. (2018) also reported that daphnids exposed to low concentrations of gemfibrozil (0.05 μg/L) produced broods at an earlier age and had larger broods than control individuals, but such effect was only observed when food availability was high. The authors also observed that organisms were larger and had higher lipid energy reserves for which mechanisms of action were unexplained by the authors.
Concerning tests No 2 (fenarimol 113 μg/L, Fig. 4b) and No 3 (fenarimol 565 μg/L, Fig. 4c), similar observations were also made by Mu and LeBlanc (2002) for D. magna exposed for 21 days to similar concentrations of fenarimol. A reduction in the number of offspring in aquatic invertebrates has been attributed to impairment of energy supply and demand or to endocrine disrupting effects (Barata et al. 2004). Under normal conditions, individuals use energy for their basal metabolism, growth, and reproduction. In the case of chemical exposure, a larger amount of the assimilated energy can be used to cope with the stressor and to maintain or compensate basal metabolism, leaving less energy available for growth and reproduction (Sokolova 2013). Therefore, to survive in test No 3, exposed daphnids had to diminish the production of their offspring.
Ecdysteroid levels
Little information is known on the impact of lipid-lowering molecules on the production of ecdysteroids in D. magna. The present results show that no effect is observed for this parameter in daphnids exposed for 21 days to a mixture of gemfibrozil and clofibric acid at a concentration of 1 μg/L (Fig. 5a) as well as for the two fenarimol exposure assays (113 μg/L and 565 μg/L, Fig. 5b and c, respectively). While the results of the exposure to 113 μg/L of fenarimol could be explained by a concentration too low to have an effect on the metabolism of ecdysteroids, the results for the exposition at 565 μg/L are contrary to those obtained by Mu and Leblanc (2004). In that study, the authors exposed D. magna neonates during the first intermolt period (approximatively during 25 h) to fenarimol at a similar concentration (497 μg/L) than the experiments presented here. This exposure caused a diminution of around 26% in ecdysteroid levels and induced a delay for the first and second molt. However, the discrepancies observed between their study and the present study can be explained by several factors such a difference in methods of quantification of ecdysteroids, the age of the daphnids used as well as the time of exposure. Another hypothesis explaining the similar 20E levels in exposed organisms is the transfer of 20E to embryos. Since ecdysteroids are transferred to neonates during embryogenesis (Subramoniam 2000), and that a lower number of neonates was produced in exposed daphnids, the total 20E present in the colony could have been lower in the exposed groups compared to controls if neonates had been included in the measurements. Unfortunately, the present method used was not sensitive enough to quantify 20E levels in neonates (Venne et al. 2016). When a lower number of neonates are produced, a lower quantity of ecdysteroids and overall resources are needed by the daphnids compared to daphnids producing many neonates. Finally, it could also be possible that cholesterol 7-dehydrogenation was indeed inhibited by exposure to fenarimol, but that the inhibition did not affect the end of the line concentrations of 20E as the organisms were able to cope by other mechanisms. Thus, cholesterol and 7-dehydrocholesterol levels should be assessed and compared between the control and exposed groups to verify this hypothesis.
Transcription of targeted genes
Since fenarimol targets the 7-dehydrogenation step in the biosynthesis of ecdysteroids (LeBlanc 2007), an increase in transcription of nvd would have been expected as this gene encodes for the 7-dehydrogenase enzyme. It is important to note that a single time point (day 21) was measured in control and exposed groups. Therefore, it is possible that differences in transcription levels could have been occurring earlier in time. Soetaert et al. (2007) reported changes in gene expression (cuticula proteins, proteases related genes) in D. magna following exposure to fenarimol for 96 h at a concentration of 1 mg/L using cDNA microarray. When Soetaert et al. (2007) used a concentration similar to the present study (500 μg/L), no changes were observed except for an unknown transcript. These results combined with the ones from the present study suggest that fenarimol may reduce fecundity in D. magna by first delaying molting and development as reported by Hassold and Backhaus (2009); but that shd and nvd genes are not involved in these changes.
Gene expression is a dynamic process where changes can occur within hours (Storey et al. 2005). When changes follow a transient manner, the expression of affected transcripts returns to pre-response levels (Bendjilali et al. 2017). Therefore, finding the right window for the measurements is essential. Only a few transcripts were evaluated in this study; it could be interesting to use techniques with a wider approach such as RNA-sequencing.
Conclusion
The present study aimed to assess the natural baseline of 20E and the transcription of four genes that regulate ecdysteroids in D. magna over a 21-day period, and to evaluate the chronic effects of fenarimol and a mixture of gemfibrozil and clofibric acid on multiple levels of biological responses.
Baseline measurement of 20E and transcription levels of shd, nvd, usp, and ecr indicated concentration of 20E oscillating between 13 and 31 pg/individuals and transcription levels between 1 and − 1 relative to day 9 (first measurement). The hypothesis of the present work that levels of 20E and the transcription of 20E-related genes would be affected by exposure to lipid-altering compounds could not be proved. Although a diminution in number of offspring was observed consistently in the exposition to 565 μg/L of fenarimol, no differences were observed for the molecular assays tested here, i.e., concentration of 20E and transcription levels of shd, nvd, usp, and ecr. Other pathways regulating growth and reproduction, such as the juvenoid hormones (Goodman and Cusson 2012), might be affected and would need to be studied to understand what lies behind this observation. Results suggest that concentrations of 20E and gene transcription quantified in daphnids observed in this work might be too variable when using a single punctual measure to discern effects due to exposure to sublethal concentrations of the target compounds. Therefore, at least for the compounds tested, changes in levels of 20E and/or changes in the transcription of 20E-related genes cannot be used as biomarkers of exposure. Nevertheless, it is not yet clear if the same outcome could be extrapolated to other inorganic or organic contaminants or by modifying the experimental conditions.
Identifying the molting stage of the daphnids is demanding and difficult to synchronize between individuals when exposed for several days. Therefore, another molecule, produced in a more stable way, could be used to normalize the ecdysteroids levels before statistical analysis. Measuring multiple time points instead of a single measure as well as additional molecular endpoints obtained from transcriptomic and metabolomic studies could also afford more insights on the changes occurring in exposed daphnids. Additionally, studying the role of neonates on 20E levels can help understand differences observed with previous studies. Indeed, measuring these parameters sooner in time could offer more insights on the adaptation of the daphnids to lipid-altering compounds on D. magna. In this way, it may be possible to obtain a more comprehensive view of the effects of stressors having common modes of action.
Additional experiments are required to help explain the lowered fertility observed in D. magna when exposed to an ecdysteroid synthesis inhibitor such as fenarimol. Future work could employ untargeted transcriptomic and lipidomic to evaluate changes in the lipid profile in whole daphnid extracts following exposure to other organic contaminants of interest. Such an approach could offer a better chance of identifying impacted metabolites and lead to a better understanding of mechanisms of toxicity. Moreover, it would be interesting to determine how daphnids adapt to exposure to different organic contaminants in the long term through their whole life cycle and multigenerational experiments since exposure periods of 21 days or less might be too short to clearly observe more gradual effects on survival or reproduction.
Data availability
Available upon reasonable request.
References
Barata C, Porte C, Baird DJ (2004) Experimental designs to assess endocrine disrupting effects in invertebrates a review. Ecotoxicol 13:511–517
Bendjilali N, MacLeon S, Kalra G, Willis SD, Hossian AN, Avery E, Wojtowicz O, Hickman MJ (2017) Time-course analysis of gene expression during the Saccharomyces cerevisiae hypoxic response. G3: Genes Genomes Genet 7:221–231
Bodar C, Voogt P, Zandee D (1990) Ecdysteroids in Daphnia magna: their role in moulting and reproduction and their levels upon exposure to cadmium. Aquat Toxicol 17:339–350
Bradley PM, Journey CA, Romanok KM, Barber LB, Buxton HT, Foreman WT, Furlong ET, Glassmeyer ST, Hladik ML, Iwanowicz LR (2017) Expanded target-chemical analysis reveals extensive mixed-organic-contaminant exposure in US streams. Environ Sci Technol 51:4792–4802
Centre d'expertise analytique environnementale du Québec (2011) Détermination de la toxicité létale CL50 48h Daphnia magna. MA. 500 D.mag 1.1, Rév. 1. Ministère du Développement durable, de l'Environnement et de la Lutlle contre les changements climatiques, Québec, QC, pp 19
Daughton CG, Ternes TA (1999) Pharmaceuticals and personal care products in the environment: agents of subtle change? Environ Health Perspect 107:907–938
De Lange H, Noordoven W, Murk A, Lürling M, Peeters E (2006) Behavioural responses of Gammarus pulex (Crustacea, Amphipoda) to low concentrations of pharmaceuticals. Aquat Toxicol 78:209–216
Dodson SI, Hanazato T (1995) Commentary on effects of anthropogenic and natural organic chemicals on development, swimming behavior, and reproduction of Daphnia, a key member of aquatic ecosystems. Environ Health Perspect 103:7
Ebele AJ, Abdallah MA-E, Harrad S (2017) Pharmaceuticals and personal care products (PPCPs) in the freshwater aquatic environment. Emerg Contam 3:1–16
Ebert D (2005) Ecology, epidemiology, and evolution of parasitism in Daphnia. National Libray of Medicine, National Center for Biotechnology Information, Bethesda, MD, p 98
Environment Canada (1990) Biological test method. Acute Lethality Test Using Daphnia spp. In: Method Development and Applications Section (Hrsg.), Ottawa, ON, p 55. Available at: https://www.canada.ca/en/environment-climatechange/services/wildlife-research-landscape-science/biological-test-method-publications/acute-lethality-testdaphnia-species.html
Flaherty CM, Dodson SI (2005) Effects of pharmaceuticals on Daphnia survival, growth, and reproduction. Chemosphere 61:200–207
Goodman W, Cusson M (2012) The Juvenile Hormones. In: Li G (ed) Insect Endocrinology. Academic Press, San Diego, pp 310–365
Häfker NS, Tessmar-Raible K (2020) Rhythms of behavior: are the times changin’? Curr Opin Neurobiol 60:55–66
Hassold E, Backhaus T (2009) Chronic toxicity of five structurally diverse demethylase-inhibiting fungicides to the crustacean Daphnia magna: a comparative assessment. Environ Toxicol Chem: Int J 28:1218–1226
Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:1–14
Hirano M, Toyota K, Ishibashi H, Tominaga N, Sato T, Tatarazako N, Iguchi T (2020) Molecular insights into structural and ligand binding features of methoprene-tolerant in daphnids. Chem Res Toxicol 33:2785–2792
Houde M, Carter B, Douville M (2013) Sublethal effects of the flame retardant intermediate hexachlorocyclopentadiene (HCCPD) on the gene transcription and protein activity of Daphnia magna. Aquat Toxicol 140:213–219
Hughes SR, Kay P, Brown LE (2013) Global synthesis and critical evaluation of pharmaceutical data sets collected from river systems. Environ Sci Technol 47:661–677
Jansen M, Coors A, Stoks R, De Meester L (2011) Evolutionary ecotoxicology of pesticide resistance: a case study in Daphnia. Ecotoxicol 20:543–551
Kovacevic V, Simpson AJ, Simpson MJ (2016) 1H NMR-based metabolomics of Daphnia magna responses after sub-lethal exposure to triclosan, carbamazepine and ibuprofen. Comp Biochem Physiol Part D: Genom Proteom 19:199–210
LaFont R, Dauphin-Villemant C, Warren JT, Rees H (2012) Ecdysteroid Chemistry and Biochemistry. In: Gilbert LI (ed) Insect Endocrinology. Academic Press, London, UK, pp 106–176
LeBlanc GA (2007) Crustacean endocrine toxicology: a review. Ecotoxicol 16:61–81
Lefrancq M, Jadas-Hécart A, La Jeunesse I, Landry D, Payraudeau S (2017) High frequency monitoring of pesticides in runoff water to improve understanding of their transport and environmental impacts. Sci Total Environ 587:75–86
Li JJ, Dai MJ, Xue JZ (2023) Investigation on toxicity and mechanism to Daphnia magna for 14 disinfection by-products: enzyme activity and molecular docking. Sci Total Environ 905:167059
Martin-Creuzburg D, Westerlund SA, Hoffmann KH (2007) Ecdysteroid levels in Daphnia magna during a molt cycle: determination by radioimmunoassay (RIA) and liquid chromatography-mass spectrometry (LC-MS). Gen Comp Endocrinol 151:66–71
Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG Jr (2012) Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proc R Soc B Biol Sci 279:1873–1882
Mnif W, Hassine AIH, Bouaziz A, Bartegi A, Thomas O, Roig B (2011) Effect of endocrine disruptor pesticides: a review. Int J Environ Res Public Health 8:2265–2303
Mu X, LeBlanc GA (2002) Environmental antiecdysteroids alter embryo development in the crustacean Daphnia magna. J Exp Zool 292:287–292
Mu X, Leblanc GA (2004) Synergistic interaction of endocrine-disrupting chemicals: model development using an ecdysone receptor antagonist and a hormone synthesis inhibitor. Environ Toxicol Chem 23:1085–1091
OECD (2008) Test No. 211: Daphnia magna reproduction test. In: OECD Guidelines for the Testing of Chemicals, Section 2: Effects on Biotic Systems. OCDE Publishing
Peng L, Wang L, Zou M-M, Vasseur L, Chu L-N, Qin Y-D, Zhai Y-L, You M-S (2019) Identification of halloween genes and RNA interference-mediated functional characterization of a Halloween gene Shadow in Plutella xylostella. Front Physiol 10:1120
Ponton F, Chapuis M-P, Pernice M, Sword GA, Simpson SJ (2011) Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol 57:840–850
Qiagen (2023) M-MuLV Reverse Transcriptase. Qiagen, Ipswich, MA
Quiagen (2020) RNeasy Plus Mini Handbook (HB-0405-005). Quiagen, Hilden, Germany, p 47
Rewitz KF, Gilbert LI (2008) Daphnia Halloween genes that encode cytochrome P450s mediating the synthesis of the arthropod molting hormone: evolutionary implications. BMC Evol Biol 8:1–8
Richmond EK, Grace MR, Kelly JJ, Reisinger AJ, Rosi EJ, Walters DM (2017): Pharmaceuticals and personal care products (PPCPs) are ecological disrupting compounds (EcoDC). Elem Sci Anth 5:1–8
Soetaert A, van der Ven K, Moens LN, Vandenbrouck T, van Remortel P, De Coen WM (2007) Daphnia magna and ecotoxicogenomics: gene expression profiles of the anti-ecdysteroidal fungicide fenarimol using energy-, molting-and life stage-related cDNA libraries. Chemosphere 67:60–71
Sokolova IM (2013) Energy-limited tolerance to stress as a conceptual framework to integrate the effects of multiple stressors. Integr Comp Biol 53:597–608
Song Y, Villeneuve DL, Toyota K, Iguchi T, Tollefsen KE (2017) Ecdysone receptor agonism leading to lethal molting disruption in arthropods: review and adverse outcome pathway development. Environ Sci Technol 51:4142–4157
Steinkey D, Lari E, Woodman SG, Luong KH, Wong CS, Pyle GG (2018) Effects of gemfibrozil on the growth, reproduction, and energy stores of Daphnia magna in the presence of varying food concentrations. Chemosphere 192:75–80
Storey JD, Xiao W, Leek JT, Tompkins RG, Davis RW (2005) Significance analysis of time course microarray experiments. P Natl Acad Sci USA 102:12837–12842
Subramoniam T (2000) Crustacean ecdysteriods in reproduction and embryogenesis. Comp Biochem Physiol C: Pharmacol Toxicol Endocrinol 125:135–156
Sumiya E, Ogino Y, Miyakawa H, Hiruta C, Toyota K, Miyagawa S, Iguchi T (2014) Roles of ecdysteroids for progression of reproductive cycle in the fresh water crustacean Daphnia magna. Front Zool 11:60
Sumiya E, Ogino Y, Toyota K, Miyakawa H, Miyagawa S, Iguchi T (2016) Neverland regulates embryonic moltings through the regulation of ecdysteroid synthesis in the water flea Daphnia magna, and may thus act as a target for chemical disruption of molting. J Appl Toxicol 36:1476–1485
Tonkes M, De Graaf PJ, Graansma J (1999) Assessment of complex industrial effluents in the Netherlands using a whole effluent toxicity (or WET) approach. Water Sci Technol 39:55–61
Venne P, Yargeau V, Segura PA (2016) Quantification of ecdysteroids and retinoic acids in whole daphnids by liquid chromatography-triple quadrupole mass spectrometry. J Chromatogr A 1438:57–64
Wagner ND, Simpson AJ, Simpson MJ (2017) Metabolomic responses to sublethal contaminant exposure in neonate and adult Daphnia magna. Environ Toxicol Chem 36:938–946
Wang Z, Walker GW, Muir DC, Nagatani-Yoshida K (2020) Toward a global understanding of chemical pollution: a first comprehensive analysis of national and regional chemical inventories. Environ Sci Technol 54:2575–2584
Funding
This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) through a discovery grant awarded to P.A. Segura and by the Faculty of Sciences of the Université de Sherbrooke. We would like to thank Hydro-Québec, the Environmental Decontamination Technologies and Integrated Water and Wastewater Management (TEDGIEER) NSERC CREATE program, and the Quebec Ecotoxicology Research Centre (EcotoQ) for scholarships granted to H. Alarie. Funding sources did not have any involvement in the design, experiments, data interpretation, writing, revision, or submission of this study.
Author information
Authors and Affiliations
Contributions
Hugo Alarie: conceptualization, formal analysis, investigation, methodology, software, validation, visualization, writing—original draft, writing—review and editing. Nadia Côté: supervision. Magali Houde: supervision, resources, writing—review and editing. Luc R. Gaudreau: writing—review and editing, resources. Pedro A. Segura: conceptualization, funding acquisition, methodology, resources, supervision, writing—review and editing, project administration.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Ludek Blaha
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
Table S1. Properties of target compounds. Table S2. Name, symbol and oligonucleotide primers used for RT-PCR analyses. Figure S1. Survival curve (%) of Daphnia magna exposed to 0.113 mg/L of fenarimol for 21 days. Figure S2. Survival curve (%) of Daphnia magna exposed to 0.565 mg/L of fenarimol for 21 days. Figure S3. Survival curve (%) of Daphnia magna exposed to 1 μg/L of gemfibrozil and clofibric acid for 21 days. Figure S4. Capillary electrophoresis of Shade amplicon on bioanalyzer 2100. Section S1. Chemical stability assays: Ferarimol, Gemfibrozil and clofibric acid. (DOCX 136 kb).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alarie, H., Côté, N., Gaudreau, L. et al. Are 20-hydroxyecdysone and related genes potential biomarkers of sublethal exposure to lipid-altering contaminants?. Environ Sci Pollut Res 30, 126104–126115 (2023). https://doi.org/10.1007/s11356-023-31087-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-023-31087-2