Abstract
Background
Cholangiocarcinoma (CCA), a primary hepatobiliary malignancy, is characterized by a poor prognosis and a lack of effective treatments. Therefore, the need to explore novel therapeutic approaches is urgent. While the role of Peptidylprolyl Cis/Trans Isomerase, NIMA-Interacting 1 (PIN1) has been extensively studied in various tumor types, its involvement in CCA remains poorly understood.
Methods
In this study, we employed tissue microarray (TMA), reverse transcription-polymerase chain reaction (RT-PCR), and The Cancer Genome Atlas (TCGA) database to assess the expression of PIN1. Through in vitro and in vivo functional experiments, we investigated the impact of PIN1 on the adhesion and metastasis of CCA. Additionally, we explored downstream molecular pathways using RNA-seq, western blotting, co-immunoprecipitation, immunofluorescence, and mass spectrometry techniques.
Results
Our findings revealed a negative correlation between PIN1 overexpression and prognosis in CCA tissues. Furthermore, high PIN1 expression promoted CCA cell proliferation and migration. Mechanistically, PIN1 functioned as an oncogene by regulating ANXA2 phosphorylation, thereby promoting CCA adhesion. Notably, the interaction between PIN1 and ANXA2 was facilitated by RACK1. Importantly, pharmacological inhibition of PIN1 using the FDA-approved drug all-trans retinoic acid (ATRA) effectively suppressed the metastatic potential of CCA cells in a nude mouse lung metastasis model.
Conclusion
Overall, our study emphasizes the critical role of the PIN1/RACK1/ANXA2 complex in CCA growth and functionality, highlighting the potential of targeting PIN1 as a promising therapeutic strategy for CCA.
Graphical Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cholangiocarcinoma (CCA) is an aggressive malignancy of the hepatobiliary system, exhibiting a dismal five-year survival rate of less than 20% [1]. CCA is one of the most lethal forms of cancer and is characterized by its aggressive nature. Recurrence and metastasis play crucial roles in determining patient survival outcomes. Even among individuals diagnosed at an early stage, the five-year survival rate remains below 25% [2,3,4,5,6]. Metastasis, the process by which cancer cells spread from the primary site to distant organs or tissues, plays a crucial role in the progression and mortality of CCA. Understanding the mechanisms underlying the metastatic behavior of CCA is essential for the development of targeted therapies and improved patient management [7].
PIN1 belongs to the evolutionarily conserved family of peptidyl-prolyl isomerases (PPIases), which catalyze isomerization by selectively recognizing the phosphorylated Ser/Thr-Pro motif and modulating the conformational state of the phosphorylated protein [8,9,10]. PIN1 is responsible for regulating the functionality and stability of proteins, as evidenced by previous investigations highlighting its critical involvement in modulating the synthesis of various disease-associated proteins. Through manipulation of protein complex formation or facilitation of ubiquitin-mediated proteasomal degradation, PIN1 exerts control over cellular processes. Hence, PIN1 exerts a significant influence on the initiation and/or progression of various types of tumors. Notably, inhibition of PIN1 promotes the degradation of the pathogenic fusion oncogene PML-RARα in acute promyelocytic leukemia (APL), thereby enhancing self-renewal capabilities while impeding promyelocytic differentiation [11]. Furthermore, targeted intervention of PIN1 using all-trans-retinoic acid (ATRA) or arsenic trioxide (ATO) disrupts pro-connective tissue and the immunosuppressive tumor microenvironment (TME), resulting in the upregulation of PD-L1 and ENT1. Consequently, this blockade influences multiple cancer pathways in pancreatic ductal adenocarcinoma (PDAC) [12].
In the realm of intrahepatic cholangiocarcinoma (ICC), prior investigations have unveiled the functional significance of the JNK-PIN1 regulatory axis in governing proliferation and provided substantial evidence supporting the therapeutic potential of inhibiting PIN1 to target JNK activation [13]. Nevertheless, the impact of PIN1 in the treatment of ICC remains uncertain, and further investigations are needed to elucidate its potential therapeutic role. Additionally, the effectiveness of PIN1 inhibitors in eradicating solid malignant tumors is still a subject of ongoing research and necessitates additional exploration. To further investigate the role of PIN1 in CCA, our clinical observations revealed elevated levels of PIN1 expression in CCA tissue compared to the surrounding paraneoplastic tissue. Moreover, patients exhibiting high levels of PIN1 expression faced an unfavorable prognosis. Additionally, through genetic or chemical inhibition of PIN1, we successfully downregulated a multitude of signaling pathways that contribute to cancer progression. This inhibition resulted in the suppression of cell proliferation, colony formation, and adhesion development across various CCA cell lines.
To summarize, these findings strongly suggest that PIN1 is markedly overexpressed in human CCA, thereby highlighting the prospective candidacy of ATRA as a therapeutic agent to impede the metastatic progression of this malignancy.
2 Materials and methods
2.1 Data acquisition
PIN1 expression was extracted from the TCGA, GTEX and GEO databases, and all data were analyzed by the R3.6.3-based bioinformatics online analysis website [14, 15]. GSE26566 and GSE107943 were obtained from the Gene Expression Omnibus (GEO) database.
2.2 CCA patients and tumor microarray (TMA)
The tissues of 227 confirmed CCA and corresponding adjacent non-cancerous tissues treated at the First Affiliated Hospital of Nanjing Medical University from 2010 to 2017 were collected and embedded in paraffin. Relevant clinical information was obtained from hospitalized cases and corresponding follow-up investigations of relevant patients. The study was conducted in accordance with the Declaration of Helsinki, and written informed consent was obtained from all patients. It was also approved by the Ethics Committee of the First Clinical Medical School of Nanjing Medical University. Overall survival (OS) denotes the duration between the completion of the surgical procedure and the demise of the subject resulting from any cause. Disease-free survival (DFS), on the other hand, refers to the period starting from the postoperative phase until the onset of tumor recurrence or metastasis, or the passing of the subject due to any cause.
2.3 Immunohistochemistry (IHC)
Immunohistochemistry was performed using the established streptavidin-peroxidase method, following a standardized protocol as previously described [16]. The anti-PIN1 antibody (Santa Cruz, USA; dilution 1:200; catalog number Sc-46660) was employed for IHC, utilizing the avidin-biotin complex technique. Microwave antigen retrieval was conducted, and the tissue sections were incubated with the primary antibody overnight at 4 °C. Next, the sections were treated with a secondary antibody, and subsequently, a diaminobenzidine solution was used for visualization under a microscope. Counterstaining with hematoxylin was performed. Negative control slides were included in all experimental procedures.
2.4 Cell acquisition and culture
The human normal bile duct cell line HIBEpiC and CCA cell lines QBC939, RBE, HuCCT1, and HCCC9810 used in this study were purchased from the Chinese National Human Genome Center (Shanghai, China). Cholangiocarcinoma cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco, USA) containing 10% fetal bovine serum and antibiotics (1% penicillin/streptomycin; Gibco). All cells were grown at 37 °C in a humidified incubator containing 5% CO2.
2.5 Transfection of small interfering RNA (siRNA) and lentivirus
Small interfering RNA (siRNA) specifically targeting ANXA2, RACK1, and a non-specific control (si-Ctrl) were acquired from GenePharma (Shanghai, China) for transfection. Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA) was employed as the transfection reagent following the manufacturer’s protocol. The cells were cultured until they reached a confluency of 30–40%. The working concentration of siRNA is 50 nM. After a six-hour transfection period, the medium containing the transfection reagents was replaced with fresh medium. Total proteins and RNA were collected 48 h after transfection. The siRNA sequences for ANXA2 knockdown were as follows:
si-ANXA2:
Sense: 5′-GCCGAGUGUACUACUUCAATT-3′
Antisense: 5′-UUGAAGUAGUACACUCGGCTT-3′
si-RACK1:
Sense: 5′-GCUACAUCCAGAAGAUCAATT-3′
Antisense: 5′-UUGAUCUUCUGGAUGUAGCTT-3′
To generate a PIN1 knockdown lentivirus, a lentivirus targeting PIN1 was obtained from Shanghai GeneChem Company. The shRNA target sequences were as follows:
shPIN1 sense: CAGGGAGATTGGATAGAAA
shCtrl sense: TTCTCCGAACGTGTCACGT
CCA cells were infected with the lentivirus for 48 h and subsequently selected with puromycin at a concentration of 2 ng/ml for a duration of 2 weeks.
2.6 RNA extraction and reverse transcription
The adherent cells were harvested and extracted using TRIzol reagent (Ambion, Texas, USA) for mRNA preparation. The total mRNAs were reversely translated using HiScript III RT SuperMix (Vazyme, Nanjing, China), and real-time PCR amplification was performed with AceQ qPCR SYBR Green Master Mix (High ROX Premixed) (Vazyme, Nanjing, China) using the 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific, MA, USA).
Primer sequences: PIN1
F-TCAGGCCGAGTGTACTACTTC
R-TCTTCTGGATGTAGCCGTTGA
β-Actin
F-ATTGCCGACAGGATGCAGAA
R-GCTGATCCACATCTGCTGG AA
qRT-PCR reaction procedure:
Step | Temperature | Time |
|---|---|---|
Pre-denaturation | 95 °C | 5 min |
Denaturation | 95 °C | 10 s |
Annealing/extension | 60 °C | 30 s |
2.7 Library construction and sequencing
A comprehensive analysis of gene expression was conducted by extracting total RNA from SH-CON and SH-PIN1 cells, with three samples included in each group. The purified library products were subjected to rigorous evaluation and subsequently sequenced on the Illumina NovaSeq platform. Total RNA was prepared for RNA-seq analysis by Personalbio (Shanghai, China). P < 0.05 and |log2 FC| > 1 between groups.
2.8 Western blot
Proteins were extracted from tissue or cell lysates [17] as described previously, and protein samples were separated by 10% dodecyl sodium sulfate-polyacrylamide gel electrophoresis (SDS‒PAGE) and transferred to polyvinylidene difluoride membranes (Paarl, NY, USA), incubated with primary and secondary antibodies, and visualized using enhanced chemiluminescence detection reagents (NcmECL High, Suzhou, China). All antibodies are shown in Supplementary Table S2.
2.9 Immunoprecipitation (IP), silver staining, and mass spectrometry
Approximately 1 × 106 Cells were lysed with NP-40 (Beyotime, Shanghai, China) substitute. Protein complexes were captured overnight at 4 °C using PIN1-specific antibodies and IgG, and the next day, antigen-antibody complexes were captured using protein A/G magnetic beads (Beyotime, Shanghai, China). The antigen-antibody complexes were obtained by mixing at room temperature for 2 h. The supernatant was discarded to obtain the antigen-antibody complexes for subsequent experiments. The protein complexes were separated by SDS‒PAGE and detected by a protein blotting assay with the corresponding antibodies. Visualization was performed using silver staining according to the manufacturer’s instructions (Beyotime, Shanghai, China). Candidate targets that differed significantly from the gel were then cut out and sent to the company for mass spectrometry analysis for protein identification.
2.10 CCK-8 assay
Cell Counting Kit-8 was purchased from Biosharp (Shanghai, China). Cells were inoculated in 96-well plates at a density of 1000 cells/well and cultured overnight in complete medium. At 0, 24, 48, 72, 96, and 120 h, 10 ml of CCK-8 solution was added to each well. After 2 h of incubation, absorbance values at 450 nm were measured using a spectrophotometer (Thermo Fisher Scientific, Pittsburgh, PA, USA).
2.11 Cell cycle analysis
Forty-eight hours after transfection, the cells were subjected to serum starvation overnight to synchronize them. Subsequently, they were induced to re-enter the cell cycle by incubating them in medium containing 10% fetal bovine serum for 4 h. The cells were then collected, washed with PBS, and fixed in pre-cooled 70% ethanol at 4 °C overnight. The next day, the cells were washed twice with PBS and resuspended in RNase A, followed by incubation at 37 °C for 30 min. Propidium iodide (PI) was added to the cells in the dark at 4 °C for an additional 30 min. Finally, all samples were analyzed using flow cytometry (BD, FACSCantoTM II).
2.12 5-Ethynyl-20-deoxyuridine (EdU) proliferation assay
The EdU assay was performed by using the EdU Admixture Assay Kit (Beyotime, Shanghai, China) to assess cell growth capacity. Cells (5000/well) were inoculated into 96-well plates and incubated for 12 h, followed by incubation with EdU for 2 h and the remaining steps as described previously [18].
2.13 Colony formation assay
Each experimental group was composed of three technical replicates. A total of 1000 cells were inoculated into 6-well plates containing complete medium. Every 72 h, the six-well plates underwent a medium replacement procedure. The cell culture process was terminated upon the observation of a considerable number of cellular colonies in the control group’s six-well plates. Each well was washed with phosphate-buffered saline (PBS) (×3) and then fixed with formaldehyde for 30 min. This was followed by staining with 1% crystalline violet for 20 min. The quantification of clone cells was performed by counting the number of clones in each well. Finally, the cellular clone count for different experimental groups was calculated and subjected to the appropriate statistical analysis.
2.14 Wound healing assay
Cells at appropriate densities were seeded in 6-well plates, and monolayers were grown in medium without FBS for 24 h. Three replicate wells were established for each experimental group. A vertical cross was gently drawn on the bottom of the six-well plate using a 200-μl pipette tip. The culture medium in the six-well plate was discarded, and the plate was rinsed three times with 1x phosphate-buffered saline (PBS) to remove any detached cells.
After scratching the cells, they were cultured in serum-free high-glucose medium at 37 °C in a CO2 incubator. Using a regular light microscope with a 10× objective, photographs of the cell scratch were taken at 0, 24, and 48 h, respectively, and the width of the cell scratch was measured and recorded.
2.15 Transwell assay
For the migration assay, 30,000 cells were diluted with 300 µL of serum-free medium in the upper chamber, and an additional complete medium containing 20% fetal bovine serum was placed in the lower chamber. After 48 hours in the cell incubator, 4% paraformaldehyde was fixed for 30 minutes, and the cells in the upper chamber were wiped with a cotton swab and stained with 0.1% crystalline substrate for 30 min. Three fields of view were randomly photographed under a bright-field microscope, and the number of cells was counted. For the invasion assay, the procedure was the same as that for the migration assay, except that the upper chamber contained a matrix gel (BD Bioscience Pharmingen).
2.16 Cell adhesion
A 2:1 dilution of DMEM with Matrigel was prepared, and 50 μl of this mixture was incubated in a 96-well plate at 37 °C with 5% CO2 for 4 h. Following the incubation period, the supernatant was discarded. A trypsin-digested cell solution containing 10,000 cells per well was inoculated into the 96-well plates and incubated at 37 °C in a 5% CO2 incubator for either 1 or 3 h. After the designated incubation time, the 96-well plate was gently removed, and the culture medium was carefully discarded. Subsequently, the plate underwent two rounds of PBS washes to eliminate any non-adherent cells. To each well, 100 μl of fresh medium and 10 μl of CCK8 were added, ensuring protection from light. Incubation in darkness continued for 2 h before measuring the optical density (OD) value at 450 nm using an enzyme marker.
To visualize cell adhesion, calcein AM (Beyotime, Shanghai, China) was employed to stain the cells. The subsequent steps followed the aforementioned protocol, culminating in visualization using a fluorescence microscope to capture images. Subsequently, the number of adherent cells was determined through appropriate calculations.
2.17 Immunofluorescence assay (IF)
Approximately 1 × 106 Cells were fixed with 4% paraformaldehyde at 37 °C for 30 min and lysed with 0.3% Triton X-100 for 30 min. Cells were then blocked with 5% donkey serum for 1 h and incubated overnight at 4 °C with a specific primary antibody. We incubated for 2 h at room temperature with 488 and 594-labeled goat anti-rabbit IgG. Cell nuclei were stained with DAPI. Fluorescence images were captured by confocal fluorescence microscopy.
2.18 In vivo animal assay
Animal studies were approved by the Nanjing Medical University (NJMU) Institutional Animal Care and Use Committee (Quorum No. IACUC-2203004). BALB/c nude mice (6 weeks old, female) were purchased from the Model Animal Research Centre of Nanjing University. Mice were randomly divided into two groups for tumor xenograft experiments (eight mice in each group). HuCCT1 cells stably expressing SH-PIN1/SH-NC transfected cells were injected subcutaneously into the left inguinal region of the mice. When the tumor became macroscopic, the tumor size was measured every four days for 24 days, and the formula (width2 × length)/2 was used to calculate the tumor volume. A portion of the tumors was preserved in a neutral fixative solution for subsequent tissue immunohistochemistry experiments to assess the expression of PIN1 and Ki-67 within the tumor tissue.
For the tail vein tumor metastasis study, a volume of 0.1 ml consisting of cell suspensions containing 1 × 106 stable cells was intravenously injected into the lateral tail veins of ten mice per experimental group. Subsequently, at the end of a period of 5 weeks, the mice were euthanized, and their lungs were meticulously extracted and fixed using a 4% paraformaldehyde solution. Each lung tissue sample was precisely sectioned at two specified regions, and the presence of lung metastatic nodules was confirmed through microscopic examination of hematoxylin and eosin (HE)-stained sections.
2.19 Statistical analysis
All statistical analyses were performed using SPSS v22.0 (IBM, SPSS, Chicago, Illinois, USA) and GraphPad Prism 9.1 (GraphPad Software, La Jolla, CA, USA). The differences between the two groups were analyzed by Student’s t-test. The X2 test was used to analyze the correlation between PIN1 expression and clinicopathological variables. The Kaplan-Meier method was used to analyze overall survival (OS) and disease-free survival, and the logarithmic rank test was used for comparison. Multivariate analysis using the Cox proportional hazards regression model. The difference is considered statistically significant when *p < 0.05, **p < 0.01, or ***p < 0.001.
3 Results
3.1 PIN1 overexpression predicts unfavorable prognosis in cholangiocarcinoma
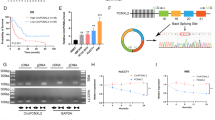
The TCGA database pan-cancer study revealed upregulation of PIN1 expression in various malignancies, including CCA (Fig. 1A). To validate this finding in CCA, we utilized two datasets (GSE26566 and GSE107943) from the GEO database, which confirmed the upregulation of PIN1 in CCA tissues compared to adjacent non-cancerous tissues (Fig. 1B, C) [19, 20]. Moreover, we analyzed both PIN1 mRNA and protein levels in tissue samples, consistently observing a significant increase in CCA tissue samples (Fig. 1D–G). These results provide evidence suggesting the involvement of PIN1 in CCA development.
Elevated PIN1 expression in cholangiocarcinoma correlates with unfavorable prognosis. A PIN1 expression in different cancer types in TCGA and overexpression in cholangiocarcinoma. B, C Increased PIN1 mRNA expression in cholangiocarcinoma datasets derived from the GEO database (GSE26566 and GSE107943). D Comparative analysis of PIN1 mRNA expression between tumor tissues and matched paracancerous tissues using RT-PCR (n = 37). E Western blot results illustrate elevated PIN1 protein expression in cholangiocarcinoma. F, G Immunostaining of tissue microarrays (TMAs) confirming PIN1 overexpression in cholangiocarcinoma (n = 227). H, I High PIN1 expression in CCA patients is associated with poorer overall survival and disease-free survival. All data are presented as the mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001
Continuing our investigation, we examined the correlation between PIN1 expression and clinicopathological variables to assess its clinical significance in CCA tissues. The results revealed a significant association between PIN1 expression and several factors, including age, sex, N-stage, lymph node metastasis, bile duct invasion, and tumor grade (Table 1). Subsequently, by utilizing immunohistochemistry on tissue microarrays and analyzing follow-up data, we found that higher PIN1 expression was associated with worse overall survival (OS, p = 0.0451) and disease-free survival (DFS, p = 0.0044) in CCA patients (Fig. 1H, I).
To further determine the independent prognostic value of PIN1, multivariate Cox regression models were applied, adjusting for N-stage, lymph node metastasis, tumor stage, and PIN1 expression (Supplementary Table S1). Remarkably, PIN1 expression remained an independent prognostic factor for DFS in CCA patients. These findings highlighted that N-stage, lymph node metastasis, tumor stage, and PIN1 expression could serve as prognostic indicators for OS and DFS in CCA patients. In summary, our results demonstrate the overexpression of PIN1 in CCA tissues and suggest that PIN1 may serve as a predictor of prognosis for CCA.
3.2 Suppression of PIN1 attenuates cholangiocarcinoma proliferation in vitro and in vivo
Based on the PIN1 expression profile in CCA cell lines (Fig. 2A), we successfully established two stable knockdown cell lines (SH-PIN1 knockdown group and SH-NC control group), using lentiviral-mediated techniques in HuCCT1 and QBC-939 cells (Fig. 2B, C). To elucidate the functional relevance of PIN1 in CCA cell proliferation, we initially employed the CCK-8 assay. Our results unequivocally demonstrate a significant reduction in cell proliferation upon depletion of PIN1 expression in both HuCCT1 and QBC-939 CCA cells (Supplementary Fig. S1A). Furthermore, consistent with these findings, the EDU (Fig. 2D) and colony formation experiments (Fig. 2E) conducted in HuCCT1 and QBC-939 cells provided additional evidence supporting the inhibitory effect of PIN1 knockdown on cellular proliferation compared to the negative control (SH-NC) group.
Knockdown of PIN1 significantly inhibited the proliferation of CCA in vitro and in vivo. A Analysis of PIN1 expression in CCA cells using qRT-PCR and Western blot. B, C Validation of knockdown efficiency in QBC-939 and HuCCT1 cells through Western blot and qRT-PCR. D Detection of EdU incorporation in PIN1 stable knockdown cells, represented by a scale bar (50 μm). E Evaluation of colony formation ability in PIN1 stable knockdown cells. F Alteration of proliferation-related markers following PIN1 knockdown. G Knockdown of PIN1 increased G0/G1 phase cell population but decreased S and G2 phase cell population, as detected by flow cytometric analysis following Annexin FITC and PI staining. H, I Subcutaneous injection of PIN1 stable knockdown cells into nude mice, followed by measurement of tumor growth. J Immunohistochemistry (IHC) and H&E analysis of PIN1 and Ki-67 in tumors derived from mice, represented by scale bar (50 μm). All data are presented as the mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001
In addition, we investigated alterations in key proliferation-related markers upon PIN1 knockdown. Intriguingly, the results revealed a marked suppression in the expression levels of MMP2, CyclinD1, and PCNA in response to PIN1 knockdown (Fig. 2F), indicating the regulatory role of PIN1 in CCA cell proliferation.
Based on our current findings, we aimed to investigate the underlying mechanism by which PIN1 knockdown inhibited the proliferation of cholangiocarcinoma (CCA) cells. To assess whether the inhibition was due to cell cycle arrest in a specific phase, cell cycle analysis was conducted. Flow cytometric analysis revealed that the downregulation of PIN1 led to a significant increase in the percentage of cells in the G0/G1 phase, while the distribution of cells in the S and G2 phases decreased (Fig. 2G). These results suggest that the downregulation of PIN1 inhibits the G1/S phase transition in the cell cycle, potentially contributing to the suppression of CCA cell proliferation.
To corroborate our in vitro observations and ascertain the proliferative ability of PIN1 in vivo, we subcutaneously implanted HuCCT1 cells into nude mice (Fig. 2H). Remarkably, the knockdown of PIN1 led to a substantial reduction in tumor volume and weight compared to the control group (Fig. 2I). This finding was further supported by histological examination through hematoxylin and eosin (HE) staining and immunohistochemistry (IHC) analysis, which confirmed the favorable effect of PIN1 on CCA proliferation (Fig. 2J). Collectively, our comprehensive findings from both in vitro and in vivo experiments unequivocally indicate that PIN1 plays a pivotal role in promoting CCA cell proliferation.
3.3 PIN1 promoted invasion, migration, and adhesion of CCA cells in vitro and in vivo
To gain insights into the molecular mechanisms underlying PIN1 involvement in CCA, we conducted an RNA-sequencing analysis to compare the differential expression profiles between HuCCT1-SH-NC and HuCCT1-SH-PIN1 cells. This analysis revealed 294 genes that exhibited differential expression, with 202 being up-regulated and 92 down-regulated (Supplementary Fig. S1B). Through KEGG enrichment analysis (Fig. 3A), we observed that the cell adhesion pathway ranked among the top 20 signaling pathways based on P-values. Cell adhesion is a complex process involving numerous molecular interactions that facilitate the attachment and migration of cancer cells to distant sites [21]. We aimed to explore the role of PIN1 in modulating the adhesive properties of CCA cells. Consequently, we performed cell adhesion experiments to assess CCA adhesion following PIN1 knockdown. Our results indicated that the depletion of PIN1 significantly reduced the adhesion capability of CCA cells (Fig. 3B, C).
PIN1 promoted invasion, migration, and adhesion of CCA cells in vitro and in vivo. A Pathway enrichment analysis of differentially expressed genes in HuCCT1-SH-NC and HuCCT1-SH-PIN1 by RNA-sequence. B, C Detection of CCA cell adhesion ability through Cell Counting Kit-8 (CCK-8) assay and fluorescence microscopy (50 μm). D Evaluation of cell migration ability using a wound healing assay, represented by scale bar (50 μm). E Measurement of invasive or migrated cells through a transwell assay with or without matrix, represented by scale bar (100 μm). F Western blot analysis of vimentin, N-cadherin, E-cadherin, SLUG, and SANIL levels in HuCCT1 and QBC939 cells following knockdown of PIN1 expression. G Representative images and HE staining of metastatic nodules in the lungs of mice. The scale bar in one image is 500 μm, providing an overview of the nodules, while the scale bar in another image is 50 μm, allowing for a closer examination of cellular details. All data are presented as the mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001
Furthermore, we investigated the impact of PIN1 on CCA cell migration and invasion using wound-healing assays and transwell assays, respectively. Our observations demonstrated that decreased PIN1 expression impaired both processes (Fig. 3D, E). Subsequently, we examined the expression levels of molecules involved in epithelial-to-mesenchymal transition (EMT), a critical event facilitating tumor invasion and metastasis. Notably, PIN1 knockdown resulted in increased expression of E-cadherin (E-CA) and decreased expression of N-cadherin (N-CA), SNAIL, and SLUG, while vimentin (VIM) expression remained unchanged (Fig. 3F).
To further elucidate the potential role of PIN1 in CCA adhesion and metastasis, we established a lung metastasis model using a CCA cell line with stable PIN1 knockdown. Remarkably, compared to control groups, mice injected with SH-PIN1 cells exhibited minimal metastases in the lungs, as evidenced by histological analysis (Fig. 3G). Overall, our findings provide compelling evidence that PIN1 enhances CCA cell metastasis both in vitro and in vivo.
3.4 PIN1-mediated modulation of ANXA2 phosphorylation enhances cholangiocarcinoma adhesion
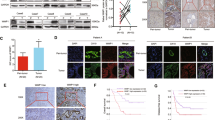
In order to gain insight into the molecular mechanisms by which PIN1 regulates adhesion, we performed silver staining and liquid chromatography‒mass spectrometry/tandem mass spectrometry (LC-MS/MS) to identify proteins that interact with PIN1 in HuCCT1 cells (Supplementary Fig. S1C). Distinct and discernible bands within the molecular weight range of 30–50 kDa were visually detected, subsequently isolated, and subjected to identification. Among the identified bands, we identified Annexin A2 (ANXA2) (Supplementary Table S3). Notably, ANXA2, a member of the membrane-linked protein family, has been reported to be significantly associated with adhesion [22, 23]. Building upon these findings, we postulate that ANXA2 interacts directly with PIN1. To validate this hypothesis, co-immunoprecipitation (co-IP) and immunofluorescence co-localization assays were performed (Fig. 4A, B), confirming the direct interaction between ANXA2 and PIN1.
PIN1 promoted cholangiocarcinoma adhesion by regulating the phosphorylation of ANXA2. A Co-immunoprecipitation (Co-IP) analysis demonstrating the interaction between PIN1 and ANXA2 in HuCCT1 cells. B Confocal immunofluorescence analysis revealing the expression patterns of PIN1 and ANXA2 (5 μm). C, D Verification of the phosphorylation state of P-ANXA2 using SH-PIN1/SH-NC cells and PY-60. PY-60 was dissolved in DMSO at a concentration of 10 μM following the manufacturer’s instructions (MedChemExpress, Shanghai, China). E Evaluation of cell migration ability through a wound healing assay, represented by scale bar (50 μm). F Detection of CCA cell adhesion ability using fluorescence microscopy (50 μm). G Measurement of migrated cells through a transwell assay, represented by scale bar (100 μm). H Representative images and HE staining of metastatic nodules in the lungs of mice. PY-60 was administered via intraperitoneal injection every three days at a concentration of 10 μM dissolved in DMSO. All data are presented as the mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001
To determine whether PIN1 regulates the expression of ANXA2, we investigated ANXA2 levels using Western blot analysis. However, the absence of PIN1 did not have any noticeable effect on ANXA2 expression (Supplementary Fig. S1D). Previous studies have indicated that ANXA2 phosphorylation can influence adhesion capability [24]. Given the role of PIN1, we hypothesized that PIN1 modulates CCA adhesion through the alteration of ANXA2 phosphorylation. Our results showed a significant reduction in ANXA2 phosphorylation upon stable knockdown of PIN1, indicating a relationship between PIN1 and ANXA2 (Fig. 4C).
3.5 PY-60 rescues the PIN1-induced reduction in ANXA2 phosphorylation
To investigate whether PIN1 regulates CCA cell adhesion through ANXA2 phosphorylation, we explored the effects of PY-60, a compound targeting ANXA2, on PIN1-regulated adhesion [25]. Stable PIN1 knockdown cell lines (QBC-939 and HuCCT1) were treated with PY-60 during cultivation. Western blot analysis revealed that the reduction in ANXA2 phosphorylation caused by PIN1 knockdown was significantly reversed by PY-60 treatment (Fig. 4D). Co-IP revealed that the treatment with PY-60 effectively enhanced the phosphorylation levels of ANXA2, while having no impact on the interaction between ANXA2 and PIN1 (Supplementary Fig. S1E). To validate our findings, we employed wound healing assays, transwell assays, and adhesion experiments. The results demonstrated that PY-60 effectively reversed the inhibitory effects of PIN1 knockdown on cell migration, adhesion, and invasion (Fig. 4E–G).
Furthermore, we investigated whether PY-60 could reverse the reduced metastatic capacity induced by PIN1 knockdown in vivo. For this purpose, we utilized a nude mouse lung metastasis model. The results demonstrated a significant augmentation in the extent of CCA cell metastasis, characterized by an increased number and area of metastatic lesions in the lung tissue of mice treated with PY-60, as compared to the PIN1 knockdown group (Fig. 4H).
Overall, our findings suggest that the overexpression of PIN1 can enhance CCA metastasis by activating ANXA2 phosphorylation. These results provide valuable insights into the molecular mechanisms underlying PIN1-mediated regulation of adhesion and metastasis in CCA.
3.6 RACK1 facilitates the interaction between PIN1 and ANXA2
To thoroughly explore the intricate relationship between PIN1 and ANXA2, we integrated our LC-MS/MS findings with relevant literature (Supplementary Table S3). Previous studies have shed light on the pivotal role of RACK1 as a bridging agent in mediating molecular interactions. Building upon this knowledge, we postulated that RACK1, a scaffolding protein involved in protein‒protein interactions and cellular signaling (Supplementary Fig. S1C), might serve as a crucial mediator in facilitating the interaction between PIN1 and ANXA2. To validate this hypothesis, we conducted co-immunoprecipitation (co-IP) experiments to investigate the potential interactions among RACK1, PIN1, and ANXA2. The results undeniably demonstrated robust interactions among these three molecules (Supplementary Fig. S1F–H).
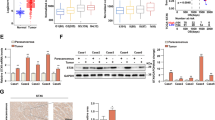
Moreover, immunofluorescence co-localization analysis confirmed the presence of a specific colocalization pattern among PIN1, ANXA2, and RACK1 (Fig. 5A, B). Considering the possible involvement of RACK1 in ANXA2 phosphorylation, we employed si-RACK1 to knock down RACK1 expression. This depletion led to a significant reduction in ANXA2 phosphorylation (Fig. 5C).
Expression patterns and interactions of RACK1, PIN1, and ANXA2 in CCA cells. A, B Confocal immunofluorescence analysis revealing the expression patterns of RACK1, PIN1, and ANXA2 (5 μm). C Assessment of changes in ANXA2 and P-ANXA2 levels following si-NC/si-RACK1 treatment. D Control or RACK1 knockdown cells were lysed. Immunoprecipitation was performed using antibodies against PIN1 or ANXA2, followed by verification of the interaction between PIN1 and ANXA2 in the two CCA cell lines through protein blot analysis using antibodies against ANXA1, RACK1, or PIN1
Given previous evidence indicating RACK1’s ability to mediate interactions between molecules, we hypothesized that RACK1 may facilitate the interaction between PIN1 and ANXA2. To test this assumption, we examined the impact of RACK1 on the binding of PIN1 and ANXA2. Our results revealed a decrease in the interaction between PIN1 and ANXA2 in the absence of RACK1 (Fig. 5D). Additionally, we explored the effect of PIN1 on the association between RACK1 and ANXA2. Knocking down PIN1 did not affect the expression levels or binding of RACK1 and ANXA2 proteins in the two CCA cell lines (Fig. 6A). Furthermore, we investigated whether ANXA2 was necessary for the interaction between PIN1 and RACK1. The results indicated that the knockout of ANXA2 had no effect on the expression of RACK1 or PIN1, nor did it impact the interaction between RACK1 and PIN1 (Fig. 6B).
RACK1-mediated interaction between PIN1 and ANXA2, as well as the phosphorylation status of P-ANXA2. A Control or PIN1 knockdown cells were lysed. Immunoprecipitation was performed using antibodies against RACK1 or ANXA2, followed by verification of the interaction between RACK1 and ANXA2 in the two CCA cell lines through protein blot analysis using antibodies against ANXA1, RACK1, or PIN1. B Control or ANXA2 knockdown cells were lysed. Immunoprecipitation was performed using antibodies against RACK1 or PIN1, followed by verification of the interaction between RACK1 and PIN1 in the two CCA cell lines through protein blot analysis using antibodies against ANXA1, RACK1, or PIN1. C Evaluation of the phosphorylation levels of P-ANXA2 under treatment with the PIN1 inhibitor ATRA (from Sigma, USA) at different concentrations
In summary, these findings suggest that RACK1 acts as a scaffold molecule, facilitating the interaction between PIN1 and ANXA2. This reveals a potential molecular mechanism through which PIN1 and ANXA2 are interconnected, mediated by RACK1 in CCA cells.
3.7 ATRA, a PIN1 inhibitor, suppresses cholangiocarcinoma cell migration, invasion, and adhesion in vitro through PIN1 downregulation
In our study, we investigated the role of all-trans retinoic acid (ATRA), a chemical targeting inhibitor, in inhibiting the carcinogenic effect of CCA, focusing on the involvement of PIN1. Previous studies have shown that active PIN1 is a key target of ATRA in other cancers [11]. ATRA has been demonstrated to degrade PIN1, thereby blocking the carcinogenic pathway regulated by PIN1 in various malignant tumors, including acute promyelocytic leukemia (APL), cancer, and hepatocellular carcinoma [11, 26, 27].
We treated QBC-939 and HuCCT1 CCA cells with different concentrations of ATRA and observed a significant inhibition of PIN1 expression at various concentrations (Fig. 6C). In the CCK-8 cell proliferation assay, we observed a significant decrease in cell proliferation in both cell lines treated with ATRA compared to the control group (Supplementary Fig. S1I, J). To further investigate the biological function of PIN1 in CCA metastasis, we evaluated the effects of ATRA treatment. Wound healing assays showed that ATRA significantly reduced the migration ability of CCA cells (Fig. 7A). Transwell analysis revealed a significant reduction in the number of migrating and invading cells upon ATRA treatment (Fig. 7B). Moreover, we validated the effect of ATRA on the adhesion ability of CCA cells and found that it inhibited adhesion (Fig. 7C, D). The changes in epithelial-mesenchymal transition (EMT)-related molecules were consistent with previous results, further supporting the inhibitory effect of ATRA on CCA metastasis by suppressing PIN1 expression (Fig. 7E).
ATRA Suppresses Migration, Invasion, and Adhesion of CCA Cells in vitro and in vivo A, B Effect of ATRA on the adhesion ability of cholangiocarcinoma cells (50 μm). C Assessment of cell migration ability through wound healing assay (100 μm). D Transwell assay measuring the invasive or migratory cells treated with ATRA, with or without matrix (50 μm). E Analysis of EMT-related protein levels via Western blot after treatment of CCA cells with ATRA. F Representative images and HE staining of metastatic nodules in the lungs of mice. ATRA-releasing pellets were obtained from Innovative Research of America. All data presented are expressed as the mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001
Furthermore, we sought to ascertain the potential of ATRA in impeding the metastatic capability of CCA in an in vivo setting employing a nude mouse lung metastasis model. Encouragingly, the outcomes revealed a substantial reduction in the extent of CCA cell metastasis within the pulmonary tissue of ATRA-treated mice in comparison to the control group (Fig. 7F).
Overall, our research results suggest that ATRA can inhibit the metastatic potential of CCA by targeting PIN1. These findings provide important insights into the potential therapeutic application of ATRA in suppressing CCA metastasis through PIN1 inhibition.
4 Discussion
Cholangiocarcinoma is a malignancy of epithelial cells that arise from various locations within the biliary tree and exhibit markers of cholangiocyte differentiation [28]. Tumor recurrence is a major obstacle to CCA patient survival [29]. However, the strategies for the diagnosis and treatment of recurrent CCA metastases are limited. Current strategies primarily revolve around chemotherapy and immune checkpoint inhibitors, the efficacy of targeted therapies is limited to patients carrying specific genetic mutations, such as IDH1 and FGFR2 mutations. Even with radical surgical resection for early-stage tumors, the recurrence rate is alarmingly high. Consequently, identifying novel therapeutic targets to aid in the treatment of advanced CCA and prevent postsurgical recurrence is critical. Previous studies have illuminated various roles of PIN1 in different tumors. For instance, targeting PIN1 in hematologic tumors significantly increases treatment efficacy [30]. Moreover, PIN1 is prevalently upregulated in pancreatic cancer and has emerged as an adverse prognostic indicator that maintains redox homeostasis through the c-Myc/NRF2/ARE axis. Encouragingly, combining immunotherapy with PIN1-targeting therapies has shown promising results in reducing pancreatic tumor size in patients with PDAC [12, 31]. KRAS and P53 mutations represent two of the prevailing genetic events in both human CCA and PDAC. PIN1, acting as a pivotal effector within the Kras/ERK axis, synergistically orchestrates a multitude of cellular processes [31]. Furthmore, as an integral part of physiological checkpoint mechanisms, PIN1 can be hijacked during tumorigenesis, assuming a critical role in amplifying the oncogenic functions of mutant p53 [32]. Given the shared biological behaviors and molecular mechanisms observed in pancreatic cancer and cholangiocarcinoma, we aim to evaluate the therapeutic potential associated with targeting PIN1 in this context. Additionally, our research is focused on unraveling the intricate molecular mechanisms governed by PIN1 specifically in cholangiocarcinoma.
In this study, we observed elevated levels of PIN1 mRNA and protein in CCA tissues vs. normal controls. We found that high expression of PIN1 is correlated with a poorer prognosis, which is consistent with the findings of previous research [13, 33,34,35,36]. Furthermore, our experiments demonstrated that PIN1 enhanced the proliferation and metastasis of CCA both in vitro and in vivo. While existing evidence suggests the vital role of PIN1 in CCA, the specific molecular mechanisms involved, particularly the specific targets of direct target of PIN1, remain unclear [33]. Through RNA-seq analysis of CCA cells with PIN1 knockdown, we revealed the potential of PIN1 to promote invasive metastasis by modulating adhesion. In order to further investigate the specific molecular mechanisms underlying PIN1 involvement in cholangiocarcinoma, by employing LC–MS/MS, we identified ANXA2 as a molecule that interacts with PIN1 and functions as a cancer initiator, promoting the formation and progression of CCA. ANXA2, a transmembrane protein, has been implicated in multiple biological processes and clinical associations related to cancer progression [37,38,39,40,41]. Additionally, ANXA2 has been shown to enhance cell migration in ovarian cancer through interactions with HE4 and to control endometrial adhesion via RhoA activation in the endometrium [42, 43]. Intriguingly, our data did not support the initial assumption that PIN1 regulates ANXA2 by altering protein levels, as knocking down PIN1 had no effect on ANXA2 abundance. Previous studies have indicated that ANXA2 phosphorylation can influence adhesion capability [24]. Building upon the findings of previous studies, we propose that PIN1 influences CCA adhesion by modulating ANXA2 phosphorylation. Consistent with this hypothesis, we observed decreased ANXA2 phosphorylation upon PIN1 depletion, resulting in a reduced adhesion capacity of CCA cells. The rescue experiment in our study demonstrated that the addition of PY-60 (a compound targeting ANXA2) significantly attenuated the decrease in adhesion ability caused by PIN1 knockdown. As we all know, PIN1-mediated phosphorylation governs the regulation of signal transduction pathways that rely on phosphorylation, consequently leading to alterations in protein structure, function, and stability. This observation strongly indicates the critical role of ANXA2 phosphorylation in mediating the effects of PIN1 on CCA cell adhesion capacity.
To further investigate the potential mechanisms by which PIN1 regulates the phosphorylation of ANXA2, according to our LC–MS/MS data, RACK1 was identified as a prominent protein among the identified candidates. RACK1, which is well known for its involvement in various malignancies, acts as a critical scaffolding protein that governs the complex processes of migration and invasion [44,45,46,47,48]. As a member of the WD-repeat protein family [45], RACK1 is associated with the epithelial-to-mesenchymal transition (EMT) of cells in the tumor microenvironment [46]. Notably, in multidrug-resistant breast cancer cells, RACK1 acts as a scaffold, promoting invasiveness by increasing the binding affinity between P-glycoproteins and ANXA2 and Src [48, 49]. Moreover, RACK1 exerts a modulatory effect on the tyrosine phosphorylation of ANXA2 through a Src-dependent mechanism [47]. In our investigation, we discerned the capacity of RACK1 to act as a scaffold for PIN1 and ANXA2, facilitating the regulatory effects of PIN1 on ANXA2. Our results revealed a decrease in the interaction between PIN1 and ANXA2 in the absence of RACK1.
This reveals a potential molecular mechanism that establishes an interconnection between PIN1 and ANXA2, mediated by RACK1 in CCA cells. Subsequently, we delved into the feasibility of targeting PIN1 as a therapeutic approach. Research has suggested that ATRA treatment can modulate the expression and activity of PIN1 in certain cancer types. ATRA has yielded promising outcomes in cancer therapy, as evidenced by its profound ability to exert antitumor effects through multiple mechanisms. Primarily, ATRA has been extensively studied for its capacity to induce cellular differentiation and impede proliferation across diverse tumor types. Notably, its differentiation-promoting effect is assumed to be highly important in APL, where ATRA-mediated differentiation therapy has revolutionized treatment outcomes [50]. ATRA, functioning as a PIN1 inhibitor, binds to and inhibits the activity of PIN1. Additionally, ATRA induces the degradation of PIN1, leading to the destabilization of its substrate PML-RARα. This mechanism has shown promising results in treating acute promyelocytic leukemia (APL) in both cellular and animal models, as well as in human patients [11]. Importantly, our experimental findings demonstrated that ATRA effectively inhibits the proliferation and metastasis of CCA cells. In vivo experiments have demonstrated a significant reduction in lung metastasis of cholangiocarcinoma following the administration of ATRA. In light of these compelling findings, the utilization of ATRA as a precision-targeted modality against PIN1 has emerged as a highly auspicious therapeutic approach for inhibiting CCA metastasis. Notably, the ongoing clinical trials investigating this strategy in diverse solid malignancies (ClinicalTrials.gov: NCT04113863) serve as an additional testament to its accelerated translational potential. Further comprehensive investigations are imperative to optimize its clinical implementation and ascertain synergistic combinations with established therapeutic modalities.
While our research represents significant progress, several limitations should be acknowledged. Mechanistically, we have established a connection between PIN1/RACK1/ANXA2 but did not identify the exact binding site or define the region where PIN1 induces ANXA2 phosphorylation. However, our study provides novel evidence supporting the oncogenic role of PIN1 in CCA and highlights its clinical and biological significance.
In conclusion, we propose that PIN1 is a prognostic biomarker for patients with CCA and represents a potential therapeutic target for inhibiting the metastasis of this disease. The potential mechanism underlying the interaction between PIN1 and ANXA2 mediated by RACK1 warrants further investigation.
5 Conclusion
Our study demonstrates that PIN1 regulates the adhesive capacity of CCA by modulating the phosphorylation of ANXA2. Furthermore, the discovery of the RACK1-mediated interaction between PIN1 and ANXA2 provides valuable insights into the complex molecular mechanisms driving CCA progression and identifies potential therapeutic targets. Based on these findings, we propose that targeted inhibition of PIN1 could emerge as a novel anticancer strategy for treating CCA. The potential application of ATRA offers a promising avenue for future investigations and the development of innovative therapeutic approaches in the treatment of CCA.
Data availability
The datasets collected and/or analyzed during the current study are available from the corresponding author upon reasonable request, and the other part of the study data is detailed in the supplementary material.
Abbreviations
- APL:
-
Acute promyelocytic leukemia
- ATRA:
-
All-trans-retinoic acid
- ATO:
-
Arsenic trioxide
- CCA:
-
Cholangiocarcinoma
- CCK-8:
-
Cell counting kit-8
- DMSO:
-
Dimethyl sulfoxide
- GEO:
-
Gene expression omnibus
- PIN1:
-
Peptidylprolyl cis/trans isomerase, NIMA-interacting 1
- PDAC:
-
Pancreatic ductal adenocarcinoma
- PBS:
-
Phosphate-buffered saline
- ICC:
-
Intrahepatic cholangiocarcinoma
- IP:
-
Immunoprecipitation
- IF:
-
Immunofluorescence
References
B.J. Dwyer, E.J. Jarman, J. Gogoi-Tiwari, S. Ferreira-Gonzalez, L. Boulter, R.V. Guest, T.J. Kendall, D. Kurian, A.M. Kilpatrick, A.J. Robson, E. O’Duibhir, T.Y. Man, L. Campana, P.J. Starkey Lewis, S.J. Wigmore, J.K. Olynyk, G.A. Ramm, J.E.E. Tirnitz-Parker, S.J. Forbes, TWEAK/Fn14 signalling promotes cholangiocarcinoma niche formation and progression. J. Hepatol. 74, 860–872 (2021)
P. Lindnér, M. Rizell, L. Hafström, The impact of changed strategies for patients with cholangiocarcinoma in this millenium. HPB Surg. 2015, 736049 (2015)
S. Kamsa-Ard, V. Luvira, K. Suwanrungruang, S. Kamsa-Ard, V. Luvira, C. Santong, T. Srisuk, A. Pugkhem, V. Bhudhisawasdi, C. Pairojkul, Cholangiocarcinoma trends, incidence, and relative survival in Khon Kaen, Thailand from 1989 through 2013: a population-based cancer registry study. J. Epidemiol. 29, 197–204 (2019)
E. Alabraba, H. Joshi, N. Bird, R. Griffin, R. Sturgess, N. Stern, C. Sieberhagen, T. Cross, A. Camenzuli, R. Davis, J. Evans, E. O’Grady, D. Palmer, R. Diaz-Nieto, S. Fenwick, G. Poston, H. Malik, Increased multimodality treatment options has improved survival for hepatocellular carcinoma but poor survival for biliary tract cancers remains unchanged. Eur. J. Surg. Oncol. 45, 1660–1667 (2019)
B. Groot Koerkamp, J.K. Wiggers, P.J. Allen, M.G. Besselink, L.H. Blumgart, O.R. Busch, R.J. Coelen, M.I. D’Angelica, R.P. DeMatteo, D.J. Gouma, T.P. Kingham, W.R. Jarnagin, T.M. van Gulik, Recurrence rate and pattern of perihilar cholangiocarcinoma after curative intent resection. J. Am. College Surg. 221, 1041–1049 (2015)
W.A. Cambridge, C. Fairfield, J.J. Powell, E.M. Harrison, K. Søreide, S.J. Wigmore, R.V. Guest, Meta-analysis and meta-regression of survival after liver transplantation for unresectable perihilar cholangiocarcinoma. Ann. Surg. 273, 240–250 (2021)
P.S. Steeg, Targeting metastasis. Nat. Rev. Cancer 16, 201–218 (2016)
E.S. Yeh, A.R. Means, PIN1, the cell cycle and cancer. Nat. Rev. Cancer 7, 381–388 (2007)
A. Galat, Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity–targets–functions. Curr. Top. Med. Chem. 3, 1315–1347 (2003)
K.P. Lu, X.Z. Zhou, The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease, Nature reviews. Mol. Cell Biol. 8, 904–916 (2007)
S. Wei, S. Kozono, L. Kats, M. Nechama, W. Li, J. Guarnerio, M. Luo, M.H. You, Y. Yao, A. Kondo, H. Hu, G. Bozkurt, N.J. Moerke, S. Cao, M. Reschke, C.H. Chen, E.M. Rego, F. Lo-Coco, L.C. Cantley, T.H. Lee, H. Wu, Y. Zhang, P.P. Pandolfi, X.Z. Zhou, K.P. Lu, Active Pin1 is a key target of all-trans retinoic acid in acute promyelocytic leukemia and breast cancer. Nat. Med. 21, 457–466 (2015)
K. Koikawa, S. Kibe, F. Suizu, N. Sekino, N. Kim, T.D. Manz, B.J. Pinch, D. Akshinthala, A. Verma, G. Gaglia, Y. Nezu, S. Ke, C. Qiu, K. Ohuchida, Y. Oda, T.H. Lee, B. Wegiel, J.G. Clohessy, N. London, S. Santagata, G.M. Wulf, M. Hidalgo, S.K. Muthuswamy, M. Nakamura, N.S. Gray, X.Z. Zhou, K.P. Lu, Targeting Pin1 renders pancreatic cancer eradicable by synergizing with immunochemotherapy. Cell 184, 4753–4771.e4727 (2021)
A. Lepore, P.M. Choy, N.C.W. Lee, M.A. Carella, R. Favicchio, M.A. Briones-Orta, S.S. Glaser, G. Alpini, C. D’Santos, R.M. Tooze, M. Lorger, W.K. Syn, A. Papakyriakou, G. Giamas, C. Bubici, S. Papa, Phosphorylation and stabilization of PIN1 by JNK promote intrahepatic cholangiocarcinoma growth. Hepatology 74, 2561–2579 (2021)
D.S. Chandrashekar, S.K. Karthikeyan, P.K. Korla, H. Patel, A.R. Shovon, M. Athar, G.J. Netto, Z.S. Qin, S. Kumar, U. Manne, C.J. Creighton, S. Varambally, UALCAN: an update to the integrated cancer data analysis platform. Neoplasia (New York, N.Y.) 25, 18–27 (2022)
D.S. Chandrashekar, B. Bashel, S.A.H. Balasubramanya, C.J. Creighton, I. Ponce-Rodriguez, B. Chakravarthi, S. Varambally, UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, N.Y.) 19, 649–658 (2017)
Y. Liu, H. Chen, X. Li, F. Zhang, L. Kong, X. Wang, J. Bai, X. Wu, PSMC2 regulates cell cycle progression through the p21/cyclin D1 pathway and predicts a poor prognosis in human hepatocellular carcinoma. Front. Oncol. 11, 607021 (2021)
X. Wu, H. Chen, Q. Gao, J. Bai, X. Wang, J. Zhou, S. Qiu, Y. Xu, Y. Shi, X. Wang, J. Zhou, J. Fan, Downregulation of JWA promotes tumor invasion and predicts poor prognosis in human hepatocellular carcinoma. Mol. Carcinog. 53, 325–336 (2014)
T. Zhou, Y. Zhang, Y. Chen, J. Shan, J. Wang, Y. Wang, J. Chang, W. Jiang, R. Chen, Z. Wang, X. Shi, Y. Yu, C. Li, X. Li, ROBO1 p.E280* loses the inhibitory effects on the proliferation and angiogenesis of wild-type ROBO1 in cholangiocarcinoma by interrupting SLIT2 signal. Front. Oncol. 12, 879963 (2022)
J.B. Andersen, B. Spee, B.R. Blechacz, I. Avital, M. Komuta, A. Barbour, E.A. Conner, M.C. Gillen, T. Roskams, L.R. Roberts, V.M. Factor, S.S. Thorgeirsson, Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 142, 1021–1031.e1015 (2012)
K.S. Ahn, D. O’Brien, Y.N. Kang, T. Mounajjed, Y.H. Kim, T.S. Kim, J.A. Kocher, L.K. Allotey, M.J. Borad, L.R. Roberts, K.J. Kang, Prognostic subclass of intrahepatic cholangiocarcinoma by integrative molecular-clinical analysis and potential targeted approach. Hepatol. Int. 13, 490–500 (2019)
H. Hamidi, J. Ivaska, Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018)
Y. Jung, J. Wang, J. Song, Y. Shiozawa, J. Wang, A. Havens, Z. Wang, Y.X. Sun, S.G. Emerson, P.H. Krebsbach, R.S. Taichman, Annexin II expressed by osteoblasts and endothelial cells regulates stem cell adhesion, homing, and engraftment following transplantation. Blood 110, 82–90 (2007)
C. Zhang, T. Zhou, Z. Chen, M. Yan, B. Li, H. Lv, C. Wang, S. Xiang, L. Shi, Y. Zhu, D. Ai, Coupling of integrin α5 to annexin A2 by flow drives endothelial activation. Circ. Res. 127, 1074–1090 (2020)
Z. Su, T.R. Shelite, Y. Qiu, Q. Chang, M. Wakamiya, J. Bei, X. He, C. Zhou, Y. Liu, E. Nyong, Y. Liang, A. Gaitas, T.B. Saito, B. Gong, Host EPAC1 modulates rickettsial adhesion to vascular endothelial cells via regulation of ANXA2 Y23 phosphorylation. Pathogens (Basel, Switzerland) 10, 1307 (2021)
S.Z. Shalhout, P.Y. Yang, E.M. Grzelak, K. Nutsch, S. Shao, C. Zambaldo, J. Iaconelli, L. Ibrahim, C. Stanton, S.R. Chadwick, E. Chen, M. DeRan, S. Li, M. Hull, X. Wu, A.K. Chatterjee, W. Shen, F.D. Camargo, P.G. Schultz, M.J. Bollong, YAP-dependent proliferation by a small molecule targeting annexin A2. Nat. Chem. Biol. 17, 767–775 (2021)
A.S. Farrell, C. Pelz, X. Wang, C.J. Daniel, Z. Wang, Y. Su, M. Janghorban, X. Zhang, C. Morgan, S. Impey, R.C. Sears, Pin1 regulates the dynamics of c-Myc DNA binding to facilitate target gene regulation and oncogenesis. Mol. Cell. Biol. 33, 2930–2949 (2013)
X.H. Liao, A.L. Zhang, M. Zheng, M.Q. Li, C.P. Chen, H. Xu, Q.S. Chu, D. Yang, W. Lu, T.F. Tsai, H. Liu, X.Z. Zhou, K.P. Lu, Chemical or genetic Pin1 inhibition exerts potent anticancer activity against hepatocellular carcinoma by blocking multiple cancer-driving pathways. Sci. Rep. 7, 43639 (2017)
N. Razumilava, G.J. Gores, Cholangiocarcinoma. Lancet (London, England) 383, 2168–2179 (2014)
P.J. Brindley, M. Bachini, S.I. Ilyas, S.A. Khan, A. Loukas, A.E. Sirica, B.T. Teh, S. Wongkham, G.J. Gores, Cholangiocarcinoma. Nat. Rev. Dis. Primers 7, 65 (2021)
X. Lian, Y.M. Lin, S. Kozono, M.K. Herbert, X. Li, X. Yuan, J. Guo, Y. Guo, M. Tang, J. Lin, Y. Huang, B. Wang, C. Qiu, C.Y. Tsai, J. Xie, Z.J. Gao, Y. Wu, H. Liu, X.Z. Zhou, K.P. Lu, Y. Chen, Pin1 inhibition exerts potent activity against acute myeloid leukemia through blocking multiple cancer-driving pathways. J. Hematol. Oncol. 11, 73 (2018)
C. Liang, S. Shi, M. Liu, Y. Qin, Q. Meng, J. Hua, S. Ji, Y. Zhang, J. Yang, J. Xu, Q. Ni, M. Li, X. Yu, PIN1 maintains redox balance via the c-Myc/NRF2 axis to counteract kras-induced mitochondrial respiratory injury in pancreatic cancer cells. Cancer Res. 79, 133–145 (2019)
J.E. Girardini, M. Napoli, S. Piazza, A. Rustighi, C. Marotta, E. Radaelli, V. Capaci, L. Jordan, P. Quinlan, A. Thompson, M. Mano, A. Rosato, T. Crook, E. Scanziani, A.R. Means, G. Lozano, C. Schneider, G. Del Sal, A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell. 20, 79–91 (2011)
U. Jamiyandorj, J.S. Bae, S.J. Noh, S. Jachin, J.E. Choi, K.Y. Jang, M.J. Chung, M.J. Kang, D.G. Lee, W.S. Moon, Expression of peptidyl-prolyl isomerase PIN1 and its role in the pathogenesis of extrahepatic cholangiocarcinoma. Oncol. Lett. 6, 1421–1426 (2013)
L. Ng, V. Kwan, A. Chow, T.C. Yau, R.T. Poon, R. Pang, W.L. Law, Overexpression of Pin1 and rho signaling partners correlates with metastatic behavior and poor recurrence-free survival of hepatocellular carcinoma patients. BMC Cancer 19, 713 (2019)
J.S. Pyo, B.K. Son, I.H. Oh, Cytoplasmic Pin1 expression is correlated with poor prognosis in colorectal cancer. Pathol. Res. Pract. 214, 1848–1853 (2018)
Q. Sun, G. Fan, Q. Zhuo, W. Dai, Z. Ye, S. Ji, W. Xu, W. Liu, Q. Hu, Z. Zhang, M. Liu, X. Yu, X. Xu, Y. Qin, Pin1 promotes pancreatic cancer progression and metastasis by activation of NF-κB-IL-18 feedback loop. Cell Prolif. 53, e12816 (2020)
J.A. Spijkers-Hagelstein, S. Mimoso Pinhanços, P. Schneider, R. Pieters, R.W. Stam, Src kinase-induced phosphorylation of annexin A2 mediates glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia 27, 1063–1071 (2013)
J.S. Menell, G.M. Cesarman, A.T. Jacovina, M.A. McLaughlin, E.A. Lev, K.A. Hajjar, Annexin II and bleeding in acute promyelocytic leukemia. N. Engl. J. Med. 340, 994–1004 (1999)
M.R. Sharma, L. Koltowski, R.T. Ownbey, G.P. Tuszynski, M.C. Sharma, Angiogenesis-associated protein annexin II in breast cancer: selective expression in invasive breast cancer and contribution to tumor invasion and progression. Exp. Mol. Pathol. 81, 146–156 (2006)
K. Emoto, Y. Yamada, H. Sawada, H. Fujimoto, M. Ueno, T. Takayama, K. Kamada, A. Naito, S. Hirao, Y. Nakajima, Annexin II overexpression correlates with stromal tenascin-C overexpression: a prognostic marker in colorectal carcinoma. Cancer 92, 1419–1426 (2001)
B.J. Roseman, A. Bollen, J. Hsu, K. Lamborn, M.A. Israel, Annexin II marks astrocytic brain tumors of high histologic grade. Oncol. Res. 6, 561–567 (1994)
J. Wang, L. Deng, H. Zhuang, J. Liu, D. Liu, X. Li, S. Jin, L. Zhu, H. Wang, B. Lin, Interaction of HE4 and ANXA2 exists in various malignant cells-HE4-ANXA2-MMP2 protein complex promotes cell migration. Cancer Cell Int. 19, 161 (2019)
T. Garrido-Gómez, N. Castillo-Marco, T. Cordero, C. Simón, Decidualization resistance in the origin of preeclampsia. Am. J. Obstet. Gynecol. 226, S886–s894 (2022)
S. Ma, C.C. Lu, L.Y. Yang, J.J. Wang, B.S. Wang, H.Q. Cai, J.J. Hao, X. Xu, Y. Cai, Y. Zhang, M.R. Wang, ANXA2 promotes esophageal cancer progression by activating MYC-HIF1A-VEGF axis. J. Exp. Clin. Cancer Res. 37, 183 (2018)
D.R. Adams, D. Ron, P.A. Kiely, RACK1, a multifaceted scaffolding protein: structure and function. Cell Commun. Signal. 9, 22 (2011)
F. Hu, Z. Tao, M. Wang, G. Li, Y. Zhang, H. Zhong, H. Xiao, X. Xie, M. Ju, RACK1 promoted the growth and migration of the cancer cells in the progression of esophageal squamous cell carcinoma. Tumour. Biol. 34, 3893–3899 (2013)
Y. Fan, W. Si, W. Ji, Z. Wang, Z. Gao, R. Tian, W. Song, H. Zhang, R. Niu, F. Zhang, Rack1 mediates tyrosine phosphorylation of Anxa2 by Src and promotes invasion and metastasis in drug-resistant breast cancer cells. Breast Cancer Res. 21, 66 (2019)
Y. Yang, N. Wu, Z. Wang, F. Zhang, R. Tian, W. Ji, X. Ren, R. Niu, Rack1 mediates the interaction of P-glycoprotein with Anxa2 and regulates migration and invasion of multidrug-resistant breast cancer cells. Int. J. Mol. Sci. 17, 1718 (2016)
Q.L. Lv, Y.T. Huang, G.H. Wang, Y.L. Liu, J. Huang, Q. Qu, B. Sun, L. Hu, L. Cheng, S.H. Chen, H.H. Zhou, Overexpression of RACK1 promotes metastasis by enhancing epithelial-mesenchymal transition and predicts poor prognosis in human glioma. Int. J. Environ. Res. Public Health 13, 1021 (2016)
W. Wu, X. Xue, Y. Chen, N. Zheng, J. Wang, Targeting prolyl isomerase Pin1 as a promising strategy to overcome resistance to cancer therapies. Pharmacol. Res. 184, 106456 (2022)
Acknowledgements
We acknowledge our colleagues for their valuable efforts and comments on this paper.
Funding
We gratefully acknowledge the support of the following grants for this work: National Natural Science Foundation of China (81472306), National Natural Science Foundation of China (82102150), Special Fund for Talents of the Jiangsu Province Hospital (YNRCZN015), and the Natural Science Foundation of Jiangsu Province (BK20210968).
Author information
Authors and Affiliations
Contributions
Conception and design: YMW, GWJ, XFW; Development of methodology: YMW, YWL, WJJ, XX, JJS, JC, TZ; Collection and acquisition of data: YMW, ZGX, HRC, JFW, ALCY; Analysis of data: YMW, ZGX, HRC, SLF, ZFT, KS; Writing, review, and/or revision of the manuscript: YMW, YWL, XCL, XFC, GWJ, XFW. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Research Ethics Committee of the First Affiliated Hospital of Nanjing Medical University, and informed consent was obtained from each participant. Animal studies were approved by the Nanjing Medical University (NJMU) Institutional Animal Care and Use Committee (Quorum No. IACUC-2203004).
Consent for publication
Not applicable.
Conflict of interest
The authors declare that the study was conducted without any commercial or financial relationships that could be interpreted as potential conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Liu, Y., Chen, H. et al. PIN1 promotes the metastasis of cholangiocarcinoma cells by RACK1-mediated phosphorylation of ANXA2. Cell Oncol. 47, 1233–1252 (2024). https://doi.org/10.1007/s13402-024-00924-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-024-00924-y