Abstract
Background
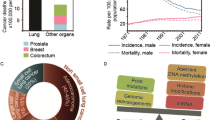
Lung cancer remains the most common cause of cancer-related death, with a 5-year survival rate of only 18%. In recent years, the development of targeted pharmacological agents and immunotherapies has substantially increased the survival of a subset of patients. However, most patients lack such efficacious therapy and are, thus, treated with classical chemotherapy with poor clinical outcomes. Therefore, novel therapeutic strategies are urgently needed. In recent years, the development of epigenetic assays and their application to cancer research have highlighted the relevance of epigenetic regulation in the initiation, development, progression and treatment of lung cancer.
Conclusions
A variety of epigenetic modifications do occur at different steps of lung cancer development, some of which are key to tumor progression. The rise of cutting-edge technologies such as single cell epigenomics is, and will continue to be, crucial for uncovering epigenetic events at a single cell resolution, leading to a better understanding of the biology underlying lung cancer development and to the design of novel therapeutic options. This approach has already led to the development of strategies involving single agents or combined agents targeting epigenetic modifiers, currently in clinical trials. Here, we will discuss the epigenetics of every step of lung cancer development, as well as the translation of these findings into clinical applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is the leading cause of cancer-related death worldwide, with a 5-year survival rate of approximately 18%. In 80% of the patients with lung cancer the disease is inoperable due to late diagnoses. This notion has led to only minor improvements in survival rates. Each year, approximately 220,000 individuals are diagnosed with lung and bronchus cancer in the United States, with tobacco use being the main cause [1].
The two main types of lung cancer are small cell lung cancer (SCLC) and non-SCLC (NSCLC). NSCLC accounts for approximately 85% of all lung cancer cases, including adenocarcinoma (40%), squamous cell carcinoma (40%) and large cell carcinoma (10%). SCLC represents ~15% of all lung cancers [2]. Recent developments in the therapy and understanding of the pathobiology of lung cancer have triggered a refinement of the histologic classification, taking into account the advent of novel targeted therapies and chemotherapy regimens [3]. For instance, several rearrangements in kinases such as EGFR, BRAF, HER2, ALK, ROS1, RET and NTRK have been found to act as oncogenic drivers in adenocarcinomas with therapeutic potential. However, currently, there are few actionable drivers in squamous cell lung carcinomas (i.e., FGFR1, PI3K and DDR2), and their inhibitors have shown low clinical efficacies. In addition, immune checkpoint inhibitors have recently revolutionized lung cancer treatment, leading to durable responses in at least some cases. Despite these advances, the vast heterogeneity of the disease does not allow a generalized clinical approach. Therefore, dedicated molecular analyses are required to (1) elucidate the complexity of clinically relevant phenotypes that determine lung cancer development, (2) define efficient diagnostic methods for early lung cancer detection and (3) develop novel strategies to enhance clinical therapeutic efficacies for lung cancer. Advances in high-resolution and high-throughput molecular technologies have already led to progress in our understanding of the molecular mechanisms underlying lung cancer development, including the complexity of its driving forces. Epigenetic modifications have also been shown to play key roles in lung cancer development, as they are involved in the regulation of gene expression and in maintaining genomic stability. Here, we review the relevance of epigenetic (de)regulation in virtually every step of cancer development, with a focus on lung cancer, and its putative clinical diagnostic and therapeutic applications.
2 Cancer epigenetics
Epigenetic mechanisms can modify gene expression patterns and genome stability without altering the DNA sequence. These modifications include nucleic acid methylation, histone modification, chromatin remodeling and expression alterations of non-coding RNAs, each of which may act in concert with other molecular processes. Epigenetic modifications have been found to be associated with various pathological conditions [4, 5]. Under normal conditions, DNA methylation occurs in a highly regulated manner. It takes place at cytosines, preferentially at CpG nucleotides, usually within gene regulatory elements through the action of DNA methyltransferases (DNMTs) and demethylases [6]. In tumor tissue, however, DNA methylation homeostasis may be disrupted, leading to epigenetic modification of key regulatory regions related to oncogenes or tumor suppressor genes. It has been found that tumors may exhibit a higher variability in DNA methylation changes than normal tissues, involving either reduced (hypo-) or increased (hyper-)methylation [7]. Hypomethylation has been found to be associated with genome instability and aneuploidy, loss of imprinting, reactivation of transposable elements and, importantly, activation of oncogenes [8, 9]. The opposite event, DNA hypermethylation, may play a dual role depending on where it occurs within the human genome. Hypermethylation has been found to act as a silencing mechanism for a vast number of tumor suppressor genes in a variety of tumor types when it occurs at CpG-rich regions (also known as CpG islands, or CGIs) located within gene promoters [10]. DNA hypermethylation may impair the binding of transcription factors directly or indirectly through recruiting protein complexes with a high affinity for methylated DNA (methyl-binding domain complexes, or MBDs). This, in turn, may lead to the recruitment of chromatin remodelers, histone deacetylases and methylases, ultimately leading to the silencing of genes, including tumor suppressor genes, or non-coding RNAs, some of which may play a role in malignant transformation [3, 9,10,11]. Next to gene promoters, gene body regions may exhibit high levels of methylation correlating with a higher expression of the corresponding gene [12, 13]. This suggests that gene body hypermethylation may act as an activator of oncogene transcription [9, 10].
The chromatin state can be dynamically altered by a highly coordinated network of proteins showing different modes of action. The most extensively studied examples are proteins or complexes that recognize, mediate or remove posttranslational modifications of core histone proteins, known as “histone marks” [11, 12]. Histone binding proteins intimately interact with nucleosomes, the fundamental building blocks of eukaryotic chromatin. Histone marks, involving covalent modifications of histones, alter their interaction with DNA and, by doing so, they are able to recruit histone modifiers, ultimately leading to chromatin structure modification, gene silencing and/or activation. These modifications may be diverse in nature, including different variants of histone methylation or lysine acetylation, as well as phosphorylation, glycosylation or ubiquitination of arginine, serine and threonine residues [13, 14]. Within the numerous proteins and molecular complexes involved in chromatin remodeling, some have gained particular interest in cancer research. These include, histone deacetylases (HDACs), enzymes mediating the deacetylation of histones, and polycomb repressive complexes (PRCs), which can interfere with gene expression via histone modification, or directly compacting chromatin in a manner independent of their enzymatic activity [15]. Beyond histone modifications, other examples of chromatin remodeling with a high relevance for cancer include SWI/SNF (SWItch/Sucrose Non-Fermentable) complexes, which can alter gene expression patterns by utilizing ATP energy to reposition nucleosomes and remodel chromatin [16], or histone variants, which can substitute core canonical histones in nucleosomes, thus conferring different structural and functional properties that may affect chromatin compaction [17, 18].
Other cancer-related epigenetic key elements are non-coding RNAs [19, 20]. These RNAs function by themselves and are not translated into proteins. Non-coding RNAs have the ability to regulate gene expression at both the transcriptional and the post-transcriptional level, and have been proposed as key regulators of physiological processes in pathological conditions including cancer, where a number of non-coding RNAs has been catalogued as either oncogenic drivers or as tumor suppressors [21]. Specific subtypes of non-coding RNAs include microRNAs (miRNAs), which are short RNAs targeting mRNAs for degradation, thus hampering target gene expression, and long non-coding RNAs (lncRNAs), which interact with chromatin-modifying enzymes and chromatin remodeling factors, thereby affecting their function [22, 23]. In addition, epigenetic modifications of RNA molecules (derived from protein-coding and non-coding genes) are gaining attention. Several such RNA modifications have been identified, including cytosine or adenine methylation, affecting RNA features such as stability, translation, localization, structure, splicing and/or targeting [24]. Evidence suggests that these modifications may be involved in cancer development by increasing or decreasing tumor characteristics such as proliferation and invasion [25].
3 Epigenetics of lung cancer
Different genome-wide studies of large patient cohorts have highlighted the importance of epigenetic events in several types of cancer, including lung cancer (Fig. 1). Analysis of the prevalence of mutations in genes with epigenetic functions has revealed that lung tumors are among the most frequently affected, with emphasis on HDAC and SWI/SNF component genes, and a mutation frequency of approximately 20% in each case. Some of these mutations may be related to the main risk factor for lung cancer, i.e., smoking. In addition, it has been reported that smoking may have a broad impact on DNA methylation, and even seems to be persistent years beyond cessation [26]. In lung cancer patients, methylation changes in disease-related genes have also been observed in whole blood-derived DNA samples [27,28,29]. Specifically, hypomethylation, as detected in blood, of genes such as those encoding aryl hydrocarbon receptor repressor (AHRR) and F2R like Thrombin or Trypsin Receptor 3 (F2RL3) has been associated with smoking and with an increased lung cancer risk [28]. Concordantly, Bossé et al. found that changes in AHRR expression in non-tumor lung tissues are significantly associated with smoking habits [30]. Others have observed promoter hypermethylation of p16, and to a lesser extent, of death-associated protein (DAP) kinase, in bronchial epithelial cells (BECs) of patients with lung cancer. This aberrant methylation was also found in BECs from current smokers and former smokers without lung cancer, suggesting that it may be tobacco-associated [26]. Also, miRNA expression patterns in BECs have been found to be associated with smoking habits and are, thus, likely due to intrinscic toxins present in cigarrettes [31]. These results should, however, be interpreted with caution since DNA methylation of specific CpG sites may be associated with smoking in a dose- and cell type-specific manner.
Mutation frequency in genes involved in epigenetic alterations in different cancer types. Frequencies of mutations in different groups of genes involved in epigenetic modification shown as percentage of cases per tumor type. Only the 10 tumor types with highest alteration frequencies are shown in each graph. The genes included in each group are: DNA methyltransferases or DNMTs (DNMT1, DNMT3A, DNMT3B), histone deacetylases or HDACs (HDAC1–11, SIRT1–7), SWI/SNF components (BCL7A, BCL7B, BCL7C, BCL11A, SS18, SMARCB1, SMARCA4, SMARCC2, SMARCC1, SMARCD1–3, BRD9, SMARCE1, ARID1A, ARID1B), PCR1 components (CBX2, CBX4, CBX6, CBX7, CBX8, PCGF1–6, HPH1–3, RING1–2) and PCR2 components (JARID2, EZH1–2, SUZ12, BAP1, ASXL1–3). These data are based on studies publicly available at http://www.cbioportal.org/
4 Epigenetics of NSCLC
A large number of studies has addressed the effect of epigenetic alterations on the different stages of NSCLC development. In this section, epigenetic events associated with NSCLC tumor initiation and progression, as well as interactions with the tumor microenvironment (TME) will be discussed (Fig. 2).
Epigenetic alterations with relevance in distinct developmental stages of different lung cancer subtypes. Different epigenetic alterations are indicated by font colors (see color legend at the bottom), and the effects of the epigenetic events on tumor stages are indicated by different arrows (see arrow legend at the bottom). In the “Interaction with TME” box, each arrow color represents a different process. DNMT, DNA methyltransferase; HDAC, Histone deacetylase; CSC, Cancer stem cell; EMT, Epithelial-to-mesenchymal transition; TME, Tumor microenvironment; NK cell, Natural killer cell; TAM, Tumor-associated macrophage
4.1 Epigenetics of NSCLC tumor initiation
Through multi-omics studies, downregulation of cell fate-specifying and other transcription factors has been identified as a potential cause of tumor initiation, with hypermethylation as the key cause of decreased gene expression. Among the potential tumor suppressor genes identified for lung squamous cell carcinoma, several members of the homeobox family were found to be involved, such as HOXA2, HOXA4 and NKX2–1, as well as other transcription factor genes, such as ZNF132 or GATA2. In lung adenocarcinoma, genes such as KCNIP4, ZEB2 and FOXF1 were found to be hypermethylated. In addition, hypermethylation of transcription factor genes such as FOXA2, FOXJ1, HOXA5, TAL1 and HLF has been detected in both lung tumor types [32,33,34]. Interestingly, some of these genes have also been found to be hypermethylated in patients with COPD, which is a predisposing factor for lung cancer [35].
Apart from hypermethylation, several other epigenetic mechanisms have been found to be involved in lung cancer initiation. For example, aberrant expression of certain DNA methyltransferases (DNMTs), such as DNMT1, DNMT3A and DNMT3B, has been found to be associated with lung cancer initiation. These enzymes have been reported to induce aberrant DNA methylation and chromatin remodeling in early stages of NSCLC development and to be associated with an increased lung cancer risk [36, 37]. In lung squamous cell carcinoma (LUSC), DNMT1 has been found to be upregulated during tumor initiation, leading to hypermethylation and silencing of tumor suppressor genes such as RASSF1A, which is involved in RAS signaling, and CDKN2A, a cyclin-dependent kinase inhibitor involved in cell cycle arrest at the G1/S phase [27, 38, 39]. In addition, aberrant promoter methylation has been found to affect the expression of multiple genes in preneoplastic lesions of squamous histology, such as the FHIT gene, whose downregulation is associated with proliferation and invasiveness, or the tumor suppressor miR-47b, whose downregulation is associated with tumor development through the stemness-related Wnt pathway [40, 41]. In addition, chromatin-remodeling complexes have been found to be involved in the early stages of LUSC development. As such, decreased H4K20me3 levels caused by aberrant activity of chromatin remodelers and upregulation of EZH2 expression, involved in H3K27me3 enrichment and DNA methylation-mediated silencing of tumor suppressor miRNAs (i.e., miR-196b, miR-200b, miR-200c or miR-205), have been reported [42,43,44].
Related to lung adenocarcinoma, promoter hypermethylation and subsequent downregulation of different genes, such as MGMT, which is involved in DNA repair, RARb, which is involved in retinoic acid signaling, DAPK, which is related to apoptosis, hTERT, which is involved in immortalization, PTPRN2, which has been implicated in insulin metabolism, as well as CDKN2A, RASSF1A and several antagonists of the Wnt pathway, has been detected in preneoplastic lesions corresponding to this histologic subtype [45, 46].
4.2 Epigenetic determinants of NSCLC progression and metastasis
NSCLC pro-metastatic phenotypes have been associated with genome-wide DNA methylation alterations [47]. Interestingly, Teschendorff et al. identified a smoking-related DNA methylation signature in buccal cells that can be used to distinguish between progressive and in situ lung tumors, with a high specificity and sensitivity [33]. In lung adenocarcinoma, hypermethylation-dependent downregulation of 164 genes and hypomethylation-dependent upregulation of 57 genes involved in EMT, differentiation and cell cycle progression have been reported [48]. Regarding squamous cell carcinoma, Teixeira et al. recently identified an epigenomic signature predictive of invasive lesions, out of 12064 differentially methylated positions associated with 2695 genes. Within the identified affected genes, several members of the homeobox family, including NKX2–1, were found to exhibit hypermethylation and a concomitant decreased expression in samples with a pro-metastatic phenotype [32]. Others have identified potential key methylation targets related to progression to an invasive phenotype in NSCLC, such as FBP1 [49], MAGE family members [50], CDO1, PTGDR and AJAP1 [51], and revealed methylation pattern alterations in PGC-related coactivator (PRC) family members involved in cell cycle regulation, proliferation and invasion [47, 52].
Epigenetic alterations in miRNA expression have also been found to be involved in the progression of NSCLC. Methylation-dependent silencing of anti-tumorigenic miR-124a, miR-126, miR-34b/c and miR-200c has been reported to be related to pro-tumoral and metastatic potentials [53,54,55,56]. Oncogenic miRNAs (oncomirs), such as miR-135b, have been found to be associated with highly invasive NSCLC through the targeting of components of the Hippo pathway and removing their promoter methylation [57]. In addition, numerous miRNAs have been reported to be involved in the regulation of EMT in NSCLC, such as the miR-200 family, which targets several EMT effectors, including ZEB1, GATA3, miR-132 and miR-149, the latter of which target ZEB2 and FOXM1. By doing so, these miRNAs prevent mesenchymal conversion and inhibit NSCLC invasion, migration and metastasis [56, 58].
Regarding chromatin remodeling, overexpression of certain epigenetic ‘writers’ and ‘erasers’ has been linked to NSCLC invasion and metastasis. Examples of these are KDM2A, a H3K36 demethylase inducing the MAPK pathway, SETDB1, a H3K9 methyltransferase that activates the Wnt pathway and H3F3A, encoding the H3.3 histone variant [17, 18]. In addition, mutations in BAF and PBAF (members of the human SWI/SNF chromatin-remodeling complex) have been observed in NSCLC. One such mutation has been found to occur in the SMARCA4 gene, leading to gene expression changes promoting tumor progression [59].
Currently, hundreds of lncRNAs that are differentially expressed between normal and lung tumor tissues have been identified [60]. Some of them have been found to be involved in lung tumor progression, such as LCAL1, or in NSCLC proliferation, such as LOC146880 and ENST00000439577, which are related to expression of the cancer progression-related genes RCC2 and KPNA2 [61]. Other lncRNAs have been found to be involved in lung cancer metastasis, such as MALAT1 and HOTAIR, which induce a pro-metastatic gene expression signature, or in EMT, such as BANCR and especially SPRY4-ITI, which is downregulated by EZH2 and modulates EMT through the induction of E-cadherin and the repression of vimentin [62,63,64,65]. Some lncRNAs have been found to be involved in both EMT and therapy resistance, including UCA1, BC087858 and GAS5, which are associated with lung cancer metastasis and EGFR-based therapy resistance, potentially through the induction of Akt signaling [66, 67]. Other epigenetic mechanisms have also been found to be involved in NSCLC therapy resistance. A drug-tolerant subpopulation of cells has, for example, been identified in anti-EGFR treated NSCLC cells, which were dependent on a KDM5A histone demethylase-induced altered chromatin state [68].
4.2.1 Epigenetics of interactions between NSCLC cells and the TME
With an increasing interest in the TME and its interplay with tumor cells, different studies have been performed to address the involvement of epigenetics in this interaction. Some epigenetic-mediated tumor-TME interactions have been found to promote lung tumor development. For example, miR-221 and miR-222, which are overexpressed in NSCLC, target TIMP3, a protein involved in the control of the homeostasis of growth factors and cytokines, and whose downregulation promotes tumor progression [69]. Another example is the presence of prostaglandin in the TME. Prostaglandin mediates overexpression of c-Myc in stromal cells, which modulates the expression of the miR-17-92 cluster targeting the tumor suppressor PTEN, thus preventing apoptosis in NSCLC tumor cells [70]. Other tumor-TME interactions have been found to promote angiogenesis, including upregulation of let-7b and miR-126 expression by both tumor and surrounding tissue cells, which leads to an increased micro-vessel density [71]. Tumors can also develop mechanisms to manipulate the TME to create a pro-metastatic environment. NSCLC cells, for example, may induce the secretion of pro-inflammatory cytokines such as IL-6 and TNF-α through the delivery of miR-21 and miR-29a to tumor-associated macrophages (TAMs), which results in increased metastasis [72]. Tumor-TME interactions may also have a tumor suppressive role under certain conditions. Lung tumor exosomal release of miR-92 has, for example, been shown to repress the secretion of pro-angiogenic factors such as IL-8, ICAM or CXCL1 in the TME, leading to a reduced invasion and metastasis [73].
In addition, there are several examples illustrating a role of epigenetics in the modulation of antitumor immune responses in lung cancer. TGFβ has, for example, been shown to induce a release of miR-183 by lung cancer cells, which represses the expression of DAP12, a key signal transduction receptor in natural killer (NK) cells, thus suppressing their antitumor cytotoxic action [74]. Another miRNA, miR-9, which has been found to be overexpressed in lung cancer, acts to downregulate the Major Histocompatibility Complex (MHC) class I gene, thus impairing immune recognition of tumor cells [75, 76].
4.3 Epigenetics of SCLC
Epigenetic alterations also appear to be relevant in SCLC (Fig. 2). Recent analyses have revealed differential DNA methylation patterns in SCLCs compared to other lung cancer types, with a specific methylation signature present in 75% of the cases involving neuroendocrine factors such as NEUROD1 [77, 78]. Furthermore, differential DNA methylation and gene expression clusters have been observed between SCLC samples with highly similar histological and genetic characteristics, thus allowing the identification of SCLC subtypes, which would be impossible by relying exclusively to histological and genetic criteria [79]. Methylation of tumor suppressor genes is a common phenomenon in SCLC. Promoter hypermethylation and, thus, downregulation of RASSF1A has been found in almost all SCLC tumors, suggesting that this event may be involved in tumor initiation. Other examples include the DAPK tumor suppressor gene, methylated in a third of all SCLC cases, and genes involved in signaling pathways involved in death receptor-mediated apoptosis [80, 81].
In addition to aberrations at the methylation level, alterations in chromatin remodeling enzymes and factors have also been identified in SCLC, suggesting that these remodelers may play an important role in the development of these tumors. Supporting this idea, loss of histone H4 methylation has been reported to occur in lung neuroendocrine tumors, including SCLCs, and to be correlated with an increased proliferation [82]. In addition, several mutations have been identified in genes encoding remodeling enzymes. The most frequent are those occurring in the MLL2 gene, encoding a H3K4 histone methyltransferase, which is detected in 8% of all SCLC tumors. These mutations lead to gene inactivation and are associated with an impaired enhancer function [83]. At a lower frequency, inactivating mutations in the histone acetyltransferase CREBBP, EP300 and KAT6B genes, in the histone demethylase UTX gene, and in the chromatin remodeling factor genes PBRM1, ARID1A and ARID1B have also been reported [83,84,85]. Beyond these mutations, altered expression of chromatin remodelers has been noted in these tumors. Examples include upregulation of a PCR2-associated protein, ASXL3, which has been detected in primary SCLC tumors and is associated with an increased growth of SCLC-derived cell lines, and overexpression of the histone methyltransferase EZH2, which promotes E2F-driven tumorigenesis in a subset of SCLC tumors [79, 86, 87].
Epigenetic alterations may also play a role in advanced stages of SCLC development. A recent study has identified a transcription factor, Nfib, that is involved in the increase of chromatin accessibility in a vast number of intergenic regions and in the induction of neuronal gene expression programs driving the metastatic ability of SCLC cells [88]. In addition, MYCL, one of the MYC family members involved in EMT that is frequently overexpressed in SCLC, has been found to be under the control of super enhancers and to depend on histone acetylation for overexpression [89].
The standard of care of advanced-stage SCLC tumors is chemotherapy, but resistance may arise within a short period of time. Recently, upregulation of EZH2 has been found to be associated with chemoresistance through the silencing of SLFN11, a protein involved in DNA damage repair, suggesting that epigenetic alterations may also be involved in resistance to therapy in this setting [90].
5 Translation of epigenetic knowledge to clinical practice
Epigenetic alterations not only represent key events in lung cancer initiation and progression, but they may also play important roles in the design of therapeutic strategies. Several epigenetic biomarkers have, for example, been described as predictors of resistance to chemotherapy, and epigenetic inactivation of some tumor suppressor genes has been proposed to be responsible for the development of chemoresistance. The latter has e.g. been reported for GSTP1 and RARβ2, both of which are downregulated by methylation in lung squamous cell carcinoma and adenocarcinoma cells [91]. In addition, the IGFB3 promoter has been found to be hypermethylated in cisplatin-resistant lung cancer cell lines [92]. Another example involves cancer stem cells (CSCs), which may confer resistance to therapy and reconstitute the tumor when they survive the treatment [93]. The unmethylated status of OCT4 and SLUG, stem-related genes, has been negatively correlated with their expression in NSCLC cells showing resistance to chemotherapy in vitro [91].
Epigenetic therapies can be classified according to functional categories of epigenetic regulators, targeting writers, erasers or readers. For the first category, several inhibitors targeting DNMTs (DNMTis) and histone-modifying enzymes to reactivate silenced tumor suppressor genes or to restore tumor suppressor protein expression have been developed. Both inhibitor types promote tumor cell differentiation, growth inhibition, cell cycle arrest and induction of apoptosis. DNMTis used in research settings include nucleoside analogs and antisense oligonucleotides (ASOs), among others. Within the first group, 5-aza-2′-deoxycytidine (decitabine) and azacitidine (AZA) represent two cytidine nucleoside analogs that induce DNA hypomethylation by irreversibly binding and degrading DNMTs. Both drugs have been approved by the USA Food and Drug Administration (FDA) and the EMA (European Medicines Agency) for the treatment of patients with myelodysplastic syndrome (MDS). AZA was the first drug that conferred a survival benefit to patients with MDS [94]. Decitabine has also been approved for acute myeloid leukemia in the EU. In preclinical lung cancer studies, Zhang et al. found that decitabine decreased epigenetic miR-200/Z1EB axis regulation, thereby reverting TGF-β1-induced epithelial-mesenchymal transition and apoptosis [95]. However, as monotherapy decitabine and AZA have had limited success in NSCLC [96]. Other DNMTis, including zebularine, are promising and they are less toxic than classical drugs. Zebularine is a cytidine deaminase inhibitor that is very stable and has a good affinity for tumor cells, while being harmless to normal cells [97]. To date, studies carried out on adenocarcinoma-derived cell lines have shown that zebularine inhibits their growth through cell cycle arrest and apoptosis, and induces cell death by changing intracellular ROS and glutathione levels [98, 99]. Other analogs, such as CP-4200 and guadecitabine (SGI-110), also appear to exhibit better pharmacokinetic activities. The last compound is currently being evaluated in several clinical trials, including those for the treatment of leukemia (NCT02131597), hepatocarcinoma (NCT01752933), ovarian cancer (NCT01696032) and extensive-stage SCLC (NCT03085849). Additionally, two phase I trials with 4′-thio-2′-deoxycytidine (Tidy) (NCT03366116 and NCT02423057) in solid tumors, and five trials with 5-fluoro-2′-deoxycytidine (FdCyd) in solid tumors, acute myeloid leukemia and MDS (NCT00359606, NCT01479348, NCT01041443, NCT01534598, and NCT00978250) are currently going on.
Despite their specificity and reduced toxicity, these agents still show limited activity when used as monotherapy. For this reason, current efforts are aimed at assessment of the efficacy of combination therapies. It has, for instance, been reported that poly-ADP-ribose polymerase (PARP) inhibitors in combination with DNMTis enhances the binding of PARP1 to chromatin in myeloid leukemia and triple-negative breast cancer [100]. Also, venetoclax, an inhibitor of Bcl-2, in combination with AZA or decitabine has shown a better overall response in patients with relapsed/refractory and previously untreated acute myeloid leukemias [101]. On the other hand, it has been found that azacitidine combined with entinostat does not enhance chemotherapeutic efficacy in most preclinical assays performed. The response to irinotecan appeared, however, promising in lung adenocarcinoma patient-derived primary xenograft models [102]. Recently, a clinical trial has been completed for stage IIIb and IV advanced NSCLC patients treated with decitabine combined with genistein (NCT01628471). The antitumor effect of genistein has, at least partly, been proposed to involve a decrease in DNMT expression levels [103]. The combination of AZA with erlotinib showed interesting activity in a patient with erlotinib-resistant lung adenocarcinoma [104].
Histone deacetylase inhibitors (HDACis) represent a group of drugs with potential anticancer activities in both solid and hematologic malignancies. HDACis are categorized according to their chemical features, such as hydroxamic acids, cyclic peptides, fatty acids, benzamides and electrophilic ketones. Nevertheless, single HDACis have shown limited responses or only transitory effects, and they may exhibit a high toxicity. Romidepsin, a cyclin tetrapeptide, has been found to restore wild-type gene expression in an in vitro lung tumor model. Different mechanisms of action of romidepsin have been proposed, such as cell cycle arrest by overexpression of p21Waf1/Cip1 and hypo-phosphorylated Rb, or repression of the PI3K/Akt pathway [105, 106]. However, despite its preclinical efficacy, romidepsin has been disappointing in the treatment of patients with SCLC [107]. Peifer et al. described histone mutations as rating among the most frequent modifications in SCLC [108]. This notion has led to an increased interest in their use as therapeutic targets. As such, combined treatment with a hydroxamate-based HDACi (vorinostat) and cisplatin showed an enhanced antitumor effect in vitro [109]. Similar results have been reported with valproic acid in NSCLC cells [110]. Currently, panobinostat (a pan-HDACi) has been reported to have a robust anti-NSCLC activity and to sensitize tumors to carboplatin [111]. In addition, panobinostat is known to inhibit TAZ. This inhibition mitigates the EGFR compensatory pathway and enhances sensitivity to gefitinib in NSCLC. Based on this information, a combination of gefitinib with panobinostat has been found to inhibit KRAS-mutant and EGFR-wild-type tumors [112]. Another benzamide-based HDACi, entinostat, has been found to suppress the growth of SALL4 (a stem cell factor)-positive tumors [113]. In a phase II study of combined treatment with entinostat and erlotinib in advanced NSCLC no improved clinical outcome was noted, but a median overall survival improvement was observed in a subset of patients with a high E-cadherin expression compared to the group treated with erlotinib plus placebo (9.4 versus 5.4 months; hazard ratio [HR], 0.35; 95% CI, 0.13 to 0.92; and p = 0.03) [114]. Also, three HDACis (thrichostatin A, valpromide and valproic acid) have been found to repress SCLC cell growth and proliferation in vitro, with thrichostatin A being the most potent agent. It was found that thrichostatin A and valproic acid stimulate Notch signaling. In addition, it was found that a combination of both HDACis with the Notch signaling inhibitor dibenzazepine exhibited a higher efficacy than monotherapy [115]. Thus, there are HDACis that may be suitable for the treatment of this disease. This information has been instrumental for the development of HDACi-based clinical trials in lung cancer, including NCT02151721, NCT00798720, NCT01935947, NCT00738751, NCT00387465, NCT02728492 and NCT01059552. In addition, novel epigenetic drugs are currently emerging, such as those targeting disruptor of telomeric silencing1-like (DOT1L), enhancer of zeste homolog 2 (EZH2), lysin-specific demethylase 1 (LSD1), protein arginine N-methyltransferase and bromodomain and extra-terminal motif (BET). Evanno et al. reported that treating NSCLC cells with a histone deacetylase inhibitor (SAHA) plus a BET inhibitor (PFI-1, a BRD2 and BRD4 inhibitor) could partially reverse epithelial-to-mesenchymal transition [116]. T-3775440, a LSD1 Inhibitor, has been found to decrease the growth and proliferation of SCLC cells in preclinical models [117] and, thus, these drugs may have a potential for treating this disease.
The tumor microenvironment has been proposed to play a key role in the development of tumors [118]. In addition to tumor cells, fibroblasts, vascular endothelial cells, pericytes, adipocytes and immune system cells can be found within the tumor microenvironment. Tumor cells may promote immune checkpoint dysregulation mostly by antitumor cell immunity inhibition. Improved understanding of these mechanisms has resurrected the immunotherapeutic treatment of lung cancer. As a result, immune-based treatment options are expanding. In addition, a role of epigenetics in regulating immune signaling has been reported [119]. As such, epigenetic therapy in combination with immune checkpoint therapy has already been found to reactivate the immune system in several cancer types, including lung cancer [120]. In addition, Topper et al. reported that the combination of AZA with HDACis (givinostat, mocetinostat and entinostat) showed a robust anti-tumor effect in NSCLC, and that givinostat exhibited a high antiproliferative activity. They proposed that AZA induces a decreased expression of MYC and sensitizes tumor cells to HDACis, and modulates T cell phenotypes to memory and effector T cells [121]. These results led to a clinical trial of advanced NSCLC patients with a combination of escalating doses of guadecitabine and mocetinostat, along with an anti-PD1 antibody (pembrolizumab) (NCT03220477). Currently several clinical trials are ongoing exploring the efficacy of combinations of epigenetic drugs and immune checkpoint inhibitors in lung cancer (Table 1). Two phase I/II clinical trials are evaluating decitabine in combination with immune checkpoint inhibitors (nivolumab and pembrolizumab) plus tetrahydrouridine (NCT02664181 and NCT03233724, respectively) in patients with NSCLC. In addition, two trials are evaluating the safety and efficacy of entinostat plus pembrolizumab (NCT02437136 NCT02909452) in advanced NSCLC. All these studies aim to enhance immune activity via epigenetic modulation and to promote a better response to therapy.
6 Conclusions and future perspectives
Epigenetic analyses have provided a deeper understanding of the molecular mechanisms underlying cancer development, with important clinical implications. Epigenetic regulation in lung cancer is highly orchestrated, and plays a role in its different developmental steps, ranging from initiation to resistance to therapy in advanced stages of the disease. A growing number of studies has shown that epigenetic therapies can successfully be used to treat cancer. These therapies were initially developed for hematological tumors and, subsequently, for solid tumors. An in depth understanding of epigenetic regulator proteins has allowed the development novel drugs and therapeutic strategies. In addition, the clinical benefit of a specific therapy can be tailored based on specific characteristics of given patient. In lung cancer patients, epigenetic therapies have shown an improved efficacy when different epigenetic drugs are combined, or after combination with chemotherapy, tyrosine kinase inhibitors and/or immunotherapy. Recently, it has been found that immunotherapy in conjunction with epigenetic drugs may reactivate the immune system and, thus, profoundly affect current immunotherapy approaches. More preclinical and clinical studies in this direction are warranted to develop more specific strategies in the fight against lung cancer.
Since DNA methylation may be cell-type specific [122], cancer cell heterogeneity is an important feature to take into account during sample analysis, acting as a potential confounding factor when performing bulk epigenomics. During recent years, single-cell transcriptomics has demonstrated potential to dissect molecular features of distinct intra-tumoral cell populations (i.e., immune cells, tumor cells and other stromal cell subpopulations). This technology is playing an increasingly important role in precision cancer medicine, provoking a change in the way we understand cancer biology [123]. We predict that the recently developed and optimized single-cell methylation technology will play a key role in the dissection of intra-tumoral heterogeneity, and provide answers to questions related to tumor initiation, progression and therapy response. On the other hand, advances in high-resolution and high-throughput molecular technologies and the use of less invasive methods have shown potential for precision oncology. As such, various body fluids can be used as sources for the study of lung cancer epigenetics, including sputum, saliva, plasma, serum and bronchoalveolar lavage, instead of resection specimens or biopsies [124]. Next to being readily accessible, these sources allow an easy sequential collection [125] . It has been reported that circulating cell-free DNA may be enriched in specific methylation patterns related to the tumor tissue of origin and/or disease stage [126,127,128], which highlights the importance of the use of these samples for such analyses.
In conclusion, the development of cutting-edge technologies and the expansion of the types and numbers of clinical samples that may be used for epigenomic analyses has been, and will continue to be instrumental for our understanding of the biology of lung cancer. This understanding, which is reaching single-cell resolution, is expected to yield novel diagnostic and therapeutic options to combat this recalcitrant tumor type.
References
R.L. Siegel, K.D. Miller and A. Jemal, Secondary R.L. Siegel, K.D. Miller and A. Jemal, Cancer statistics, 2018. CA Cancer J Clin 68, 7–30 (2018)
A.C. MacKinnon, J. Kopatz and T. Sethi, Secondary A.C. MacKinnon, J. Kopatz and T. Sethi, The molecular and cellular biology of lung cancer: identifying novel therapeutic strategies. Br Med Bull 95, 47–61 (2010)
J. Zugazagoitia, S. Molina-Pinelo, F. Lopez-Rios, L. Paz-Ares, Secondary J. Zugazagoitia, S. Molina-Pinelo, F. Lopez-Rios and L. Paz-Ares, biological therapies in non-small cell lung cancer. Eur Respir J 49 (2017)
M.A. Dawson and T. Kouzarides, Secondary M.A. Dawson and T. Kouzarides, Cancer epigenetics: from mechanism to therapy. Cell 150, 12–27 (2012)
M. Esteller, Secondary M. Esteller, Epigenetics in cancer. New Eng J Med 358, 1148–1159 (2008)
P.A. Jones and P.W. Laird, Secondary P.A. Jones and P.W. Laird, Cancer epigenetics comes of age. Nature Genet 21, 163–167 (1999)
R.A. Irizarry, C. Ladd-Acosta, B. Wen, Z. Wu, C. Montano, P. Onyango, H. Cui, K. Gabo, M. Rongione, M. Webster, H. Ji, J. Potash, S. Sabunciyan and A.P. Feinberg, Secondary R.A. Irizarry, C. Ladd-Acosta, B. Wen, Z. Wu, C. Montano, P. Onyango, H. Cui, K. Gabo, M. Rongione, M. Webster, H. Ji, J. Potash, S. Sabunciyan and A.P. Feinberg, The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nature Genet 41, 178–186 (2009)
A.P. Feinberg and B. Vogelstein, Secondary A.P. Feinberg and B. Vogelstein, Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983)
M. Vizoso, H.J. Ferreira, P. Lopez-Serra, F.J. Carmona, A. Martinez-Cardus, M.R. Girotti, A. Villanueva, S. Guil, C. Moutinho, J. Liz, A. Portela, H. Heyn, S. Moran, A. Vidal, M. Martinez-Iniesta, J.L. Manzano, M.T. Fernandez-Figueras, E. Elez, E. Munoz-Couselo, R. Botella-Estrada, A. Berrocal, F. Ponten, J. Oord, W.M. Gallagher, D.T. Frederick, K.T. Flaherty, U. McDermott, P. Lorigan, R. Marais, M. Esteller, M. Secondary, H.J. Vizoso, P. Ferreira, F.J. Lopez-Serra, A. Carmona, M.R. Martinez-Cardus, A. Girotti, S. Villanueva, C. Guil, J. Moutinho, A. Liz, H. Portela, S. Heyn, A. Moran, M. Vidal, J.L. Martinez-Iniesta, M.T. Manzano, E. Fernandez-Figueras, E. Elez, R. Munoz-Couselo, A. Botella-Estrada, F. Berrocal, J. Ponten, W.M. Oord, D.T. Gallagher, K.T. Frederick, U. Flaherty, P. McDermott, R. Lorigan, Marais and M. Esteller, Epigenetic activation of a cryptic TBC1D16 transcript enhances melanoma progression by targeting EGFR. Nature Med 21, 741–750 (2015)
S.B. Baylin and P.A. Jones, Secondary S.B. Baylin and P.A. Jones, A decade of exploring the cancer epigenome - biological and translational implications. Nature Rev Cancer 11, 726–734 (2011)
T.K. Barth and A. Imhof, Secondary T.K. Barth and A. Imhof, Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci 35, 618–626 (2010)
A. Tarakhovsky, Secondary A. Tarakhovsky, Tools and landscapes of epigenetics. Nature Immunol 11, 565–568 (2010)
B.M. Turner, Secondary B.M. Turner, Cellular memory and the histone code. Cell 111, 285–291 (2002)
B.D. Strahl and C.D. Allis, Secondary B.D. Strahl and C.D. Allis, The language of covalent histone modifications. Nature 403, 41–45 (2000)
R. Eskeland, M. Leeb, G.R. Grimes, C. Kress, S. Boyle, D. Sproul, N. Gilbert, Y. Fan, A.I. Skoultchi, A. Wutz and W.A. Bickmore, Secondary R. Eskeland, M. Leeb, G.R. Grimes, C. Kress, S. Boyle, D. Sproul, N. Gilbert, Y. Fan, A.I. Skoultchi, A. Wutz and W.A. Bickmore, Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38, 452–464 (2010)
N.J. Francis, A.J. Saurin, Z. Shao and R.E. Kingston, Secondary N.J. Francis, A.J. Saurin, Z. Shao and R.E. Kingston, Reconstitution of a functional core polycomb repressive complex. Mol Cell 8, 545–556 (2001)
K.W. Wagner, H. Alam, S.S. Dhar, U. Giri, N. Li, Y. Wei, D. Giri, T. Cascone, J.H. Kim, Y. Ye, A.S. Multani, C.H. Chan, B. Erez, B. Saigal, J. Chung, H.K. Lin, X. Wu, M.C. Hung, J.V. Heymach and M.G. Lee, Secondary K.W. Wagner, H. Alam, S.S. Dhar, U. Giri, N. Li, Y. Wei, D. Giri, T. Cascone, J.H. Kim, Y. Ye, A.S. Multani, C.H. Chan, B. Erez, B. Saigal, J. Chung, H.K. Lin, X. Wu, M.C. Hung, J.V. Heymach and M.G. Lee, KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest 123, 5231–5246 (2013)
S.M. Park, E.Y. Choi, M. Bae, S. Kim, J.B. Park, H. Yoo, J.K. Choi, Y.J. Kim, S.H. Lee and I.H. Kim, Secondary S.M. Park, E.Y. Choi, M. Bae, S. Kim, J.B. Park, H. Yoo, J.K. Choi, Y.J. Kim, S.H. Lee and I.H. Kim, Histone variant H3F3A promotes lung cancer cell migration through intronic regulation. Nature Commun 7, 12914 (2016)
G.S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C. Polytarchou, K.B. Marcu, A.G. Papavassiliou, R. Sandaltzopoulos and E. Kolettas, Secondary G.S. Markopoulos, E. Roupakia, M. Tokamani, E. Chavdoula, M. Hatziapostolou, C. Polytarchou, K.B. Marcu, A.G. Papavassiliou, R. Sandaltzopoulos and E. Kolettas, A step-by-step microRNA guide to cancer development and metastasis. Cell Oncol 40, 303–339 (2017)
R. Castro-Oropeza, J. Melendez-Zajgla, V. Maldonado, K. Vazquez-Santillan, Secondary R. Castro-Oropeza, J. Melendez-Zajgla, V. Maldonado and K. Vazquez-Santillan, the emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol 41, 585–603 (2018)
E. Anastasiadou, L.S. Jacob and F.J. Slack, Secondary E. Anastasiadou, L.S. Jacob and F.J. Slack, Non-coding RNA networks in cancer. Nat Rev Cancer 18, 5–18 (2018)
P. Han and C.P. Chang, Secondary P. Han and C.P. Chang, Long non-coding RNA and chromatin remodeling. RNA Biol 12, 1094–1098 (2015)
C. Catalanotto, C. Cogoni, G. Zardo, Secondary C. Catalanotto, C. Cogoni and G. Zardo, MicroRNA in control of gene expression: An overview of nuclear functions. Int J Mol Sci 17, E1712 (2016)
M. Esteller and P.P. Pandolfi, Secondary M. Esteller and P.P. Pandolfi, The epitranscriptome of noncoding RNAs in cancer. Cancer Discov 7, 359–368 (2017)
M. Tusup, T. Kundig, S. Pascolo, Secondary M. Tusup, T. Kundig and S. Pascolo, Epitranscriptomics of cancer. World. J Clin Oncol 9, 42–55 (2018)
R. Joehanes, A.C. Just, R.E. Marioni, L.C. Pilling, L.M. Reynolds, P.R. Mandaviya, W. Guan, T. Xu, C.E. Elks, S. Aslibekyan, H. Moreno-Macias, J.A. Smith, J.A. Brody, R. Dhingra, P. Yousefi, J.S. Pankow, S. Kunze, S.H. Shah, A.F. McRae, K. Lohman, J. Sha, D.M. Absher, L. Ferrucci, W. Zhao, E.W. Demerath, J. Bressler, M.L. Grove, T. Huan, C. Liu, M.M. Mendelson, C. Yao, D.P. Kiel, A. Peters, R. Wang-Sattler, P.M. Visscher, N.R. Wray, J.M. Starr, J. Ding, C.J. Rodriguez, N.J. Wareham, M.R. Irvin, D. Zhi, M. Barrdahl, P. Vineis, S. Ambatipudi, A.G. Uitterlinden, A. Hofman, J. Schwartz, E. Colicino, L. Hou, P.S. Vokonas, D.G. Hernandez, A.B. Singleton, S. Bandinelli, S.T. Turner, E.B. Ware, A.K. Smith, T. Klengel, E.B. Binder, B.M. Psaty, K.D. Taylor, S.A. Gharib, B.R. Swenson, L. Liang, D.L. DeMeo, G.T. O'Connor, Z. Herceg, K.J. Ressler, K.N. Conneely, N. Sotoodehnia, S.L. Kardia, D. Melzer, A.A. Baccarelli, J.B. van Meurs, I. Romieu, D.K. Arnett, K.K. Ong, Y. Liu, M. Waldenberger, I.J. Deary, M. Fornage, D. Levy and S.J. London, Secondary R. Joehanes, A.C. Just, R.E. Marioni, L.C. Pilling, L.M. Reynolds, P.R. Mandaviya, W. Guan, T. Xu, C.E. Elks, S. Aslibekyan, H. Moreno-Macias, J.A. Smith, J.A. Brody, R. Dhingra, P. Yousefi, J.S. Pankow, S. Kunze, S.H. Shah, A.F. McRae, K. Lohman, J. Sha, D.M. Absher, L. Ferrucci, W. Zhao, E.W. Demerath, J. Bressler, M.L. Grove, T. Huan, C. Liu, M.M. Mendelson, C. Yao, D.P. Kiel, A. Peters, R. Wang-Sattler, P.M. Visscher, N.R. Wray, J.M. Starr, J. Ding, C.J. Rodriguez, N.J. Wareham, M.R. Irvin, D. Zhi, M. Barrdahl, P. Vineis, S. Ambatipudi, A.G. Uitterlinden, A. Hofman, J. Schwartz, E. Colicino, L. Hou, P.S. Vokonas, D.G. Hernandez, A.B. Singleton, S. Bandinelli, S.T. Turner, E.B. Ware, A.K. Smith, T. Klengel, E.B. Binder, B.M. Psaty, K.D. Taylor, S.A. Gharib, B.R. Swenson, L. Liang, D.L. DeMeo, G.T. O'Connor, Z. Herceg, K.J. Ressler, K.N. Conneely, N. Sotoodehnia, S.L. Kardia, D. Melzer, A.A. Baccarelli, J.B. van Meurs, I. Romieu, D.K. Arnett, K.K. Ong, Y. Liu, M. Waldenberger, I.J. Deary, M. Fornage, D. Levy and S.J. London, Epigenetic signatures of cigarette sking. Circ Cardiovasc Genet 9, 436–447 (2016)
S.A. Belinsky, K.J. Nikula, S.B. Baylin and J.P. Issa, Secondary S.A. Belinsky, K.J. Nikula, S.B. Baylin and J.P. Issa, Increased cytosine DNA-methyltransferase activity is target-cell-specific and an early event in lung cancer. Proc Natl Acad Sci U S A 93, 4045–4050 (1996)
X. Gao, Y. Zhang, L.P. Breitling and H. Brenner, Secondary X. Gao, Y. Zhang, L.P. Breitling and H. Brenner, Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget 7, 59017–59028 (2016)
K.M. Bakulski, J. Dou, N. Lin, S.J. London and J.A. Colacino, Secondary K.M. Bakulski, J. Dou, N. Lin, S.J. London and J.A. Colacino, DNA methylation signature of smoking in lung cancer is enriched for exposure signatures in newborn and adult blood. Sci Rep 9, 4576 (2019)
Y. Bosse, D.S. Postma, D.D. Sin, M. Lamontagne, C. Couture, N. Gaudreault, P. Joubert, V. Wong, M. Elliott, M. van den Berge, C.A. Brandsma, C. Tribouley, V. Malkov, J.A. Tsou, G.J. Opiteck, J.C. Hogg, A.J. Sandford, W. Timens, P.D. Pare and M. Laviolette, Secondary Y. Bosse, D.S. Postma, D.D. Sin, M. Lamontagne, C. Couture, N. Gaudreault, P. Joubert, V. Wong, M. Elliott, M. van den Berge, C.A. Brandsma, C. Tribouley, V. Malkov, J.A. Tsou, G.J. Opiteck, J.C. Hogg, A.J. Sandford, W. Timens, P.D. Pare and M. Laviolette, Molecular signature of smoking in human lung tissues. Cancer Res 72, 3753–3763 (2012)
F. Schembri, S. Sridhar, C. Perdomo, A.M. Gustafson, X. Zhang, A. Ergun, J. Lu, G. Liu, X. Zhang, J. Bowers, C. Vaziri, K. Ott, K. Sensinger, J.J. Collins, J.S. Brody, R. Getts, M.E. Lenburg and A. Spira, Secondary F. Schembri, S. Sridhar, C. Perdomo, A.M. Gustafson, X. Zhang, A. Ergun, J. Lu, G. Liu, X. Zhang, J. Bowers, C. Vaziri, K. Ott, K. Sensinger, J.J. Collins, J.S. Brody, R. Getts, M.E. Lenburg and A. Spira, MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci U S A 106, 2319–2324 (2009)
V.H. Teixeira, C.P. Pipinikas, A. Pennycuick, H. Lee-Six, D. Chandrasekharan, J. Beane, T.J. Morris, A. Karpathakis, A. Feber, C.E. Breeze, P. Ntolios, R.E. Hynds, M. Falzon, A. Capitanio, B. Carroll, P.F. Durrenberger, G. Hardavella, J.M. Brown, A.G. Lynch, H. Farmery, D.S. Paul, R.C. Chambers, N. McGranahan, N. Navani, R.M. Thakrar, C. Swanton, S. Beck, P.J. George, A. Spira, P.J. Campbell, C. Thirlwell and S.M. Janes, Secondary V.H. Teixeira, C.P. Pipinikas, A. Pennycuick, H. Lee-Six, D. Chandrasekharan, J. Beane, T.J. Morris, A. Karpathakis, A. Feber, C.E. Breeze, P. Ntolios, R.E. Hynds, M. Falzon, A. Capitanio, B. Carroll, P.F. Durrenberger, G. Hardavella, J.M. Brown, A.G. Lynch, H. Farmery, D.S. Paul, R.C. Chambers, N. McGranahan, N. Navani, R.M. Thakrar, C. Swanton, S. Beck, P.J. George, A. Spira, P.J. Campbell, C. Thirlwell and S.M. Janes, Deciphering the genomic, epigenomic, and transcriptomic landscapes of pre-invasive lung cancer lesions. Nat Med 25, 517–525 (2019)
A.E. Teschendorff, S.C. Zheng, A. Feber, Z. Yang, S. Beck and M. Widschwendter, Secondary A.E. Teschendorff, S.C. Zheng, A. Feber, Z. Yang, S. Beck and M. Widschwendter, The multi-omic landscape of transcription factor inactivation in cancer. Genome Med 8, 89 (2016)
D.S. Basseres, F. D'Alo, B.Y. Yeap, E.C. Lowenberg, D.A. Gonzalez, H. Yasuda, T. Dayaram, O.N. Kocher, J.J. Godleski, W.G. Richards, M. Meyerson, S. Kobayashi, D.G. Tenen, B. Halmos and D.B. Costa, Secondary D.S. Basseres, F. D'Alo, B.Y. Yeap, E.C. Lowenberg, D.A. Gonzalez, H. Yasuda, T. Dayaram, O.N. Kocher, J.J. Godleski, W.G. Richards, M. Meyerson, S. Kobayashi, D.G. Tenen, B. Halmos and D.B. Costa, Frequent downregulation of the transcription factor Foxa2 in lung cancer through epigenetic silencing. Lung Cancer 77, 31–37 (2012)
J. Song, I.H. Heijink, L.E.M. Kistemaker, M. Reinders-Luinge, W. Kooistra, J.A. Noordhoek, R. Gosens, C.A. Brandsma, W. Timens, P.S. Hiemstra, M.G. Rots and M.N. Hylkema, Secondary J. Song, I.H. Heijink, L.E.M. Kistemaker, M. Reinders-Luinge, W. Kooistra, J.A. Noordhoek, R. Gosens, C.A. Brandsma, W. Timens, P.S. Hiemstra, M.G. Rots and M.N. Hylkema, Aberrant DNA methylation and expression of SPDEF and FOXA2 in airway epithelium of patients with COPD. Clin Epigenetics 9, 42 (2017)
I. Rhee, K.E. Bachman, B.H. Park, K.W. Jair, R.W. Yen, K.E. Schuebel, H. Cui, A.P. Feinberg, C. Lengauer, K.W. Kinzler, S.B. Baylin and B. Vogelstein, Secondary I. Rhee, K.E. Bachman, B.H. Park, K.W. Jair, R.W. Yen, K.E. Schuebel, H. Cui, A.P. Feinberg, C. Lengauer, K.W. Kinzler, S.B. Baylin and B. Vogelstein, DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416, 552–556 (2002)
H. Kim, Y.M. Kwon, J.S. Kim, J. Han, Y.M. Shim, J. Park and D.H. Kim, Secondary H. Kim, Y.M. Kwon, J.S. Kim, J. Han, Y.M. Shim, J. Park and D.H. Kim, Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer 107, 1042–1049 (2006)
M. Suzuki, N. Sunaga, D.S. Shames, S. Toyooka, A.F. Gazdar and J.D. Minna, Secondary M. Suzuki, N. Sunaga, D.S. Shames, S. Toyooka, A.F. Gazdar and J.D. Minna, RNA interference-mediated knockdown of DNA methyltransferase 1 leads to promoter demethylation and gene re-expression in human lung and breast cancer cells. Cancer Res 64, 3137–3143 (2004)
J. Lukas, D. Parry, L. Aagaard, D.J. Mann, J. Bartkova, M. Strauss, G. Peters and J. Bartek, Secondary J. Lukas, D. Parry, L. Aagaard, D.J. Mann, J. Bartkova, M. Strauss, G. Peters and J. Bartek, Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375, 503–506 (1995)
G. Sozzi, M.L. Veronese, M. Negrini, R. Baffa, M.G. Cotticelli, H. Inoue, S. Tornielli, S. Pilotti, L. De Gregorio, U. Pastorino, M.A. Pierotti, M. Ohta, K. Huebner and C.M. Croce, Secondary G. Sozzi, M.L. Veronese, M. Negrini, R. Baffa, M.G. Cotticelli, H. Inoue, S. Tornielli, S. Pilotti, L. De Gregorio, U. Pastorino, M.A. Pierotti, M. Ohta, K. Huebner and C.M. Croce, The FHIT gene 3p14.2 is abnormal in lung cancer. Cell 85, 17–26 (1996)
C. Mascaux, B. Martin, J.M. Verdebout, A.P. Meert, V. Ninane and J.P. Sculier, Secondary C. Mascaux, B. Martin, J.M. Verdebout, A.P. Meert, V. Ninane and J.P. Sculier, Fragile histidine triad protein expression in nonsmall cell lung cancer and correlation with Ki-67 and with p53. Eur Resp Journal 21, 753–758 (2003)
A. Van Den Broeck, E. Brambilla, D. Moro-Sibilot, S. Lantuejoul, C. Brambilla, B. Eymin, S. Khochbin, S. Gazzeri, Secondary a. van Den Broeck, E. Brambilla, D. Moro-Sibilot, S. Lantuejoul, C. Brambilla, B. Eymin, S. Khochbin and S. Gazzeri, loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res 14, 7237–7245 (2008)
C.S. Tellez, D.E. Juri, K. Do, M.A. Picchi, T. Wang, G. Liu, A. Spira and S.A. Belinsky, Secondary C.S. Tellez, D.E. Juri, K. Do, M.A. Picchi, T. Wang, G. Liu, A. Spira and S.A. Belinsky, miR-196b is epigenetically silenced during the premalignant stage of lung carcinogenesis. Cancer Res 76, 4741–4751 (2016)
C.S. Tellez, D.E. Juri, K. Do, A.M. Bernauer, C.L. Thomas, L.A. Damiani, M. Tessema, S. Leng and S.A. Belinsky, Secondary C.S. Tellez, D.E. Juri, K. Do, A.M. Bernauer, C.L. Thomas, L.A. Damiani, M. Tessema, S. Leng and S.A. Belinsky, EMT and stem cell-like properties associated with miR-205 and miR-200 epigenetic silencing are early manifestations during carcinogen-induced transformation of human lung epithelial cells. Cancer Res 71, 3087–3097 (2011)
J.D. Licchesi, W.H. Westra, C.M. Hooker, E.O. Machida, S.B. Baylin and J.G. Herman, Secondary J.D. Licchesi, W.H. Westra, C.M. Hooker, E.O. Machida, S.B. Baylin and J.G. Herman, Epigenetic alteration of Wnt pathway antagonists in progressive glandular neoplasia of the lung. Carcinogenesis 29, 895–904 (2008)
J.D. Licchesi, W.H. Westra, C.M. Hooker and J.G. Herman, Secondary J.D. Licchesi, W.H. Westra, C.M. Hooker and J.G. Herman, Promoter hypermethylation of hallmark cancer genes in atypical adenomatous hyperplasia of the lung. Clin Cancer Res 14, 2570–2578 (2008)
A. Hascher, A.K. Haase, K. Hebestreit, C. Rohde, H.U. Klein, M. Rius, D. Jungen, A. Witten, M. Stoll, I. Schulze, S. Ogawa, R. Wiewrodt, L. Tickenbrock, W.E. Berdel, M. Dugas, N.H. Thoennissen and C. Muller-Tidow, Secondary A. Hascher, A.K. Haase, K. Hebestreit, C. Rohde, H.U. Klein, M. Rius, D. Jungen, A. Witten, M. Stoll, I. Schulze, S. Ogawa, R. Wiewrodt, L. Tickenbrock, W.E. Berdel, M. Dugas, N.H. Thoennissen and C. Muller-Tidow, DNA methyltransferase inhibition reverses epigenetically embedded phenotypes in lung cancer preferentially affecting polycomb target genes. Clin Cancer Res 20, 814–826 (2014)
S.A. Selamat, B.S. Chung, L. Girard, W. Zhang, Y. Zhang, M. Campan, K.D. Siegmund, M.N. Koss, J.A. Hagen, W.L. Lam, S. Lam, A.F. Gazdar and I.A. Laird-Offringa, Secondary S.A. Selamat, B.S. Chung, L. Girard, W. Zhang, Y. Zhang, M. Campan, K.D. Siegmund, M.N. Koss, J.A. Hagen, W.L. Lam, S. Lam, A.F. Gazdar and I.A. Laird-Offringa, Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res 22, 1197–1211 (2012)
Y. Dong, S. Huaying, W. Danying, Z. Chihong, J. Ruibin, S. Xiaojiang and F. Jianguo, Secondary Y. Dong, S. Huaying, W. Danying, Z. Chihong, J. Ruibin, S. Xiaojiang and F. Jianguo, Significance of methylation of FBP1 gene in non-small cell lung cancer. Biomed Res Int 2018, 3726091 (2018)
Y. Lian, L. Meng, P. Ding, M. Sang, Secondary Y. Lian, L. Meng, P. Ding and M. Sang, epigenetic regulation of MAGE family in human cancer progression-DNA methylation, histone modification, and non-coding RNAs. Clin. Epigenetics 10(115) (2018)
A. Ooki, Z. Maleki, J.J. Tsay, C. Goparaju, M. Brait, N. Turaga, H.S. Nam, W.N. Rom, H.I. Pass, D. Sidransky, R. Guerrero-Preston and M.O. Hoque, Secondary A. Ooki, Z. Maleki, J.J. Tsay, C. Goparaju, M. Brait, N. Turaga, H.S. Nam, W.N. Rom, H.I. Pass, D. Sidransky, R. Guerrero-Preston and M.O. Hoque, A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin Cancer Res 23, 7141–7152 (2017)
A.I. Robles, E. Arai, E.A. Mathe, H. Okayama, A.J. Schetter, D. Brown, D. Petersen, E.D. Bowman, R. Noro, J.A. Welsh, D.C. Edelman, H.S. Stevenson, Y. Wang, N. Tsuchiya, T. Kohno, V. Skaug, S. Mollerup, A. Haugen, P.S. Meltzer, J. Yokota, Y. Kanai and C.C. Harris, Secondary A.I. Robles, E. Arai, E.A. Mathe, H. Okayama, A.J. Schetter, D. Brown, D. Petersen, E.D. Bowman, R. Noro, J.A. Welsh, D.C. Edelman, H.S. Stevenson, Y. Wang, N. Tsuchiya, T. Kohno, V. Skaug, S. Mollerup, A. Haugen, P.S. Meltzer, J. Yokota, Y. Kanai and C.C. Harris, An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol 10, 1037–1048 (2015)
E. Nadal, G. Chen, M. Gallegos, L. Lin, D. Ferrer-Torres, A. Truini, Z. Wang, J. Lin, R.M. Reddy, R. Llatjos, I. Escobar, J. Moya, A.C. Chang, F. Cardenal, G. Capella and D.G. Beer, Secondary E. Nadal, G. Chen, M. Gallegos, L. Lin, D. Ferrer-Torres, A. Truini, Z. Wang, J. Lin, R.M. Reddy, R. Llatjos, I. Escobar, J. Moya, A.C. Chang, F. Cardenal, G. Capella and D.G. Beer, Epigenetic inactivation of microRNA-34b/c predicts poor disease-free survival in early-stage lung adenocarcinoma. Clin Cancer Res 19, 6842–6852 (2013)
K. Watanabe, N. Emoto, E. Hamano, M. Sunohara, M. Kawakami, H. Kage, K. Kitano, J. Nakajima, A. Goto, M. Fukayama, T. Nagase, Y. Yatomi, N. Ohishi and D. Takai, Secondary K. Watanabe, N. Emoto, E. Hamano, M. Sunohara, M. Kawakami, H. Kage, K. Kitano, J. Nakajima, A. Goto, M. Fukayama, T. Nagase, Y. Yatomi, N. Ohishi and D. Takai, Genome structure-based screening identified epigenetically silenced microRNA associated with invasiveness in non-small-cell lung cancer. Int J Cancer 130, 2580–2590 (2012)
A. Lujambio, S. Ropero, E. Ballestar, M.F. Fraga, C. Cerrato, F. Setien, S. Casado, A. Suarez-Gauthier, M. Sanchez-Cespedes, A. Git, I. Spiteri, P.P. Das, C. Caldas, E. Miska and M. Esteller, Secondary A. Lujambio, S. Ropero, E. Ballestar, M.F. Fraga, C. Cerrato, F. Setien, S. Casado, A. Suarez-Gauthier, M. Sanchez-Cespedes, A. Git, I. Spiteri, P.P. Das, C. Caldas, E. Miska and M. Esteller, Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67, 1424–1429 (2007)
P. Ceppi, G. Mudduluru, R. Kumarswamy, I. Rapa, G.V. Scagliotti, M. Papotti and H. Allgayer, Secondary P. Ceppi, G. Mudduluru, R. Kumarswamy, I. Rapa, G.V. Scagliotti, M. Papotti and H. Allgayer, Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol Cancer Res 8, 1207–1216 (2010)
C.W. Lin, Y.L. Chang, Y.C. Chang, J.C. Lin, C.C. Chen, S.H. Pan, C.T. Wu, H.Y. Chen, S.C. Yang, T.M. Hong and P.C. Yang, Secondary C.W. Lin, Y.L. Chang, Y.C. Chang, J.C. Lin, C.C. Chen, S.H. Pan, C.T. Wu, H.Y. Chen, S.C. Yang, T.M. Hong and P.C. Yang, MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nature Commun 4, 1877 (2013)
D.H. Peng, C. Ungewiss, P. Tong, L.A. Byers, J. Wang, J.R. Canales, P.A. Villalobos, N. Uraoka, B. Mino, C. Behrens, Wistuba, II, R.I. Han, C.A. Wanna, M. Fahrenholtz, K.J. Grande-Allen, C.J. Creighton and D.L. Gibbons, Secondary D.H. Peng, C. Ungewiss, P. Tong, L.A. Byers, J. Wang, J.R. Canales, P.A. Villalobos, N. Uraoka, B. Mino, C. Behrens, Wistuba, II, R.I. Han, C.A. Wanna, M. Fahrenholtz, K.J. Grande-Allen, C.J. Creighton and D.L. Gibbons, ZEB1 induces LOXL2-mediated collagen stabilization and deposition in the extracellular matrix to drive lung cancer invasion and metastasis. Oncogene 36, 1925–1938 (2017)
T. Orvis, A. Hepperla, V. Walter, S. Song, J. Simon, J. Parker, M.D. Wilkerson, N. Desai, M.B. Major, D.N. Hayes, I.J. Davis and B. Weissman, Secondary T. Orvis, A. Hepperla, V. Walter, S. Song, J. Simon, J. Parker, M.D. Wilkerson, N. Desai, M.B. Major, D.N. Hayes, I.J. Davis and B. Weissman, BRG1/SMARCA4 inactivation promotes non-small cell lung cancer aggressiveness by altering chromatin organization. Cancer Res 74, 6486–6498 (2014)
N.M. White, C.R. Cabanski, J.M. Silva-Fisher, H.X. Dang, R. Govindan and C.A. Maher, Secondary N.M. White, C.R. Cabanski, J.M. Silva-Fisher, H.X. Dang, R. Govindan and C.A. Maher, Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 15, 429 (2014)
N. Feng, T. Ching, Y. Wang, B. Liu, H. Lin, O. Shi, X. Zhang, M. Zheng, X. Zheng, M. Gao, Z.J. Zheng, H. Yu, L. Garmire and B. Qian, Secondary N. Feng, T. Ching, Y. Wang, B. Liu, H. Lin, O. Shi, X. Zhang, M. Zheng, X. Zheng, M. Gao, Z.J. Zheng, H. Yu, L. Garmire and B. Qian, Analysis of microarray data on gene expression and methylation to identify long non-coding RNAs in non-small cell lung cancer. Sci Rep 6, 37233 (2016)
Y. Wu, H. Lyu, H. Liu, X. Shi, Y. Song and B. Liu, Secondary Y. Wu, H. Lyu, H. Liu, X. Shi, Y. Song and B. Liu, Downregulation of the long noncoding RNA GAS5-AS1 contributes to tumor metastasis in non-small cell lung cancer. Sci Rep 6, 31093 (2016)
M. Sun, X.H. Liu, K.M. Wang, F.Q. Nie, R. Kong, J.S. Yang, R. Xia, T.P. Xu, F.Y. Jin, Z.J. Liu, J.F. Chen, E.B. Zhang, W. De and Z.X. Wang, Secondary M. Sun, X.H. Liu, K.M. Wang, F.Q. Nie, R. Kong, J.S. Yang, R. Xia, T.P. Xu, F.Y. Jin, Z.J. Liu, J.F. Chen, E.B. Zhang, W. De and Z.X. Wang, Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer 13, 68 (2014)
L.H. Schmidt, T. Spieker, S. Koschmieder, S. Schaffers, J. Humberg, D. Jungen, E. Bulk, A. Hascher, D. Wittmer, A. Marra, L. Hillejan, K. Wiebe, W.E. Berdel, R. Wiewrodt and C. Muller-Tidow, Secondary L.H. Schmidt, T. Spieker, S. Koschmieder, S. Schaffers, J. Humberg, D. Jungen, E. Bulk, A. Hascher, D. Wittmer, A. Marra, L. Hillejan, K. Wiebe, W.E. Berdel, R. Wiewrodt and C. Muller-Tidow, The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6, 1984–1992 (2011)
R. Wang, Y. Shi, L. Chen, Y. Jiang, C. Mao, B. Yan, S. Liu, B. Shan, Y. Tao and X. Wang, Secondary R. Wang, Y. Shi, L. Chen, Y. Jiang, C. Mao, B. Yan, S. Liu, B. Shan, Y. Tao and X. Wang, The ratio of FoxA1 to FoxA2 in lung adenocarcinoma is regulated by LncRNA HOTAIR and chromatin remodeling factor LSH. Sci Rep 5, 17826 (2015)
H. Pan, T. Jiang, N. Cheng, Q. Wang, S. Ren, X. Li, C. Zhao, L. Zhang, W. Cai and C. Zhou, Secondary H. Pan, T. Jiang, N. Cheng, Q. Wang, S. Ren, X. Li, C. Zhao, L. Zhang, W. Cai and C. Zhou, Long non-coding RNA BC087858 induces non-T790M mutation acquired resistance to EGFR-TKIs by activating PI3K/AKT and MEK/ERK pathways and EMT in non-small-cell lung cancer. Oncotarget 7, 49948–49960 (2016)
N. Cheng, W. Cai, S. Ren, X. Li, Q. Wang, H. Pan, M. Zhao, J. Li, Y. Zhang, C. Zhao, X. Chen, K. Fei, C. Zhou and F.R. Hirsch, Secondary N. Cheng, W. Cai, S. Ren, X. Li, Q. Wang, H. Pan, M. Zhao, J. Li, Y. Zhang, C. Zhao, X. Chen, K. Fei, C. Zhou and F.R. Hirsch, Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 6, 23582–23593 (2015)
S.V. Sharma, D.Y. Lee, B. Li, M.P. Quinlan, F. Takahashi, S. Maheswaran, U. McDermott, N. Azizian, L. Zou, M.A. Fischbach, K.K. Wong, K. Brandstetter, B. Wittner, S. Ramaswamy, M. Classon and J. Settleman, Secondary S.V. Sharma, D.Y. Lee, B. Li, M.P. Quinlan, F. Takahashi, S. Maheswaran, U. McDermott, N. Azizian, L. Zou, M.A. Fischbach, K.K. Wong, K. Brandstetter, B. Wittner, S. Ramaswamy, M. Classon and J. Settleman, A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 (2010)
M. Garofalo, G. Di Leva, G. Romano, G. Nuovo, S.S. Suh, A. Ngankeu, C. Taccioli, F. Pichiorri, H. Alder, P. Secchiero, P. Gasparini, A. Gonelli, S. Costinean, M. Acunzo, G. Condorelli and C.M. Croce, Secondary M. Garofalo, G. Di Leva, G. Romano, G. Nuovo, S.S. Suh, A. Ngankeu, C. Taccioli, F. Pichiorri, H. Alder, P. Secchiero, P. Gasparini, A. Gonelli, S. Costinean, M. Acunzo, G. Condorelli and C.M. Croce, miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509 (2009)
K. Krysan, R. Kusko, T. Grogan, J. O'Hearn, K.L. Reckamp, T.C. Walser, E.B. Garon, M.E. Lenburg, S. Sharma, A.E. Spira, D. Elashoff and S.M. Dubinett, Secondary K. Krysan, R. Kusko, T. Grogan, J. O'Hearn, K.L. Reckamp, T.C. Walser, E.B. Garon, M.E. Lenburg, S. Sharma, A.E. Spira, D. Elashoff and S.M. Dubinett, PGE2-driven expression of c-Myc and oncomiR-17-92 contributes to apoptosis resistance in NSCLC. Mol Cancer Res 12, 765–774 (2014)
E. Jusufovic, M. Rijavec, D. Keser, P. Korosec, E. Sodja, E. Iljazovic, Z. Radojevic and M. Kosnik, Secondary E. Jusufovic, M. Rijavec, D. Keser, P. Korosec, E. Sodja, E. Iljazovic, Z. Radojevic and M. Kosnik, let-7b and miR-126 are down-regulated in tumor tissue and correlate with microvessel density and survival outcomes in non--small--cell lung cancer. PloS one 7, e45577 (2012)
M. Fabbri, A. Paone, F. Calore, R. Galli, E. Gaudio, R. Santhanam, F. Lovat, P. Fadda, C. Mao, G.J. Nuovo, N. Zanesi, M. Crawford, G.H. Ozer, D. Wernicke, H. Alder, M.A. Caligiuri, P. Nana-Sinkam, D. Perrotti and C.M. Croce, Secondary M. Fabbri, A. Paone, F. Calore, R. Galli, E. Gaudio, R. Santhanam, F. Lovat, P. Fadda, C. Mao, G.J. Nuovo, N. Zanesi, M. Crawford, G.H. Ozer, D. Wernicke, H. Alder, M.A. Caligiuri, P. Nana-Sinkam, D. Perrotti and C.M. Croce, MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A 109, E2110–2116 (2012)
K. Valencia, D. Luis-Ravelo, N. Bovy, I. Anton, S. Martinez-Canarias, C. Zandueta, C. Ormazabal, I. Struman, S. Tabruyn, V. Rebmann, J. De Las Rivas, E. Guruceaga, E. Bandres and F. Lecanda, Secondary K. Valencia, D. Luis-Ravelo, N. Bovy, I. Anton, S. Martinez-Canarias, C. Zandueta, C. Ormazabal, I. Struman, S. Tabruyn, V. Rebmann, J. De Las Rivas, E. Guruceaga, E. Bandres and F. Lecanda, miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol 8, 689–703 (2014)
S.S. Donatelli, J.M. Zhou, D.L. Gilvary, E.A. Eksioglu, X. Chen, W.D. Cress, E.B. Haura, M.B. Schabath, D. Coppola, S. Wei and J.Y. Djeu, Secondary S.S. Donatelli, J.M. Zhou, D.L. Gilvary, E.A. Eksioglu, X. Chen, W.D. Cress, E.B. Haura, M.B. Schabath, D. Coppola, S. Wei and J.Y. Djeu, TGF-beta-inducible microRNA-183 silences tumor-associated natural killer cells. Proc Natl Acad Sci U S A 111, 4203–4208 (2014)
T. Xu, X. Liu, L. Han, H. Shen, L. Liu, Y. Shu, Secondary T. Xu, X. Liu, L. Han, H. Shen, L. Liu and Y. Shu, up-regulation of miR-9 expression as a poor prognostic biomarker in patients with non-small cell lung cancer. Clin Transl Oncol 16, 469–475 (2014)
F. Gao, Z.L. Zhao, W.T. Zhao, Q.R. Fan, S.C. Wang, J. Li, Y.Q. Zhang, J.W. Shi, X.L. Lin, S. Yang, R.Y. Xie, W. Liu, T.T. Zhang, Y.L. Sun, K. Xu, K.T. Yao and D. Xiao, Secondary F. Gao, Z.L. Zhao, W.T. Zhao, Q.R. Fan, S.C. Wang, J. Li, Y.Q. Zhang, J.W. Shi, X.L. Lin, S. Yang, R.Y. Xie, W. Liu, T.T. Zhang, Y.L. Sun, K. Xu, K.T. Yao and D. Xiao, miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Comm 431, 610–616 (2013)
N. Reguart, R. Rosell, F. Cardenal, A.F. Cardona, D. Isla, R. Palmero, T. Moran, C. Rolfo, M.C. Pallares, A. Insa, E. Carcereny, M. Majem, J. De Castro, C. Queralt, M.A. Molina and M. Taron, Secondary N. Reguart, R. Rosell, F. Cardenal, A.F. Cardona, D. Isla, R. Palmero, T. Moran, C. Rolfo, M.C. Pallares, A. Insa, E. Carcereny, M. Majem, J. De Castro, C. Queralt, M.A. Molina and M. Taron, Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung Cancer 84, 161–167 (2014)
S. Kalari, M. Jung, K.H. Kernstine, T. Takahashi and G.P. Pfeifer, Secondary S. Kalari, M. Jung, K.H. Kernstine, T. Takahashi and G.P. Pfeifer, The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene 32, 3559–3568 (2013)
J.T. Poirier, E.E. Gardner, N. Connis, A.L. Moreira, E. de Stanchina, C.L. Hann and C.M. Rudin, Secondary J.T. Poirier, E.E. Gardner, N. Connis, A.L. Moreira, E. de Stanchina, C.L. Hann and C.M. Rudin, DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 34, 5869–5878 (2015)
D.G. Burbee, E. Forgacs, S. Zochbauer-Muller, L. Shivakumar, K. Fong, B. Gao, D. Randle, M. Kondo, A. Virmani, S. Bader, Y. Sekido, F. Latif, S. Milchgrub, S. Toyooka, A.F. Gazdar, M.I. Lerman, E. Zabarovsky, M. White and J.D. Minna, Secondary D.G. Burbee, E. Forgacs, S. Zochbauer-Muller, L. Shivakumar, K. Fong, B. Gao, D. Randle, M. Kondo, A. Virmani, S. Bader, Y. Sekido, F. Latif, S. Milchgrub, S. Toyooka, A.F. Gazdar, M.I. Lerman, E. Zabarovsky, M. White and J.D. Minna, Epigenetic inactivation of RASSF1A in lung and breast cancers and malignant phenotype suppression. J Natl Cancer Inst 93, 691–699 (2001)
S. Toyooka, K.O. Toyooka, K. Miyajima, J.L. Reddy, M. Toyota, U.G. Sathyanarayana, A. Padar, M.S. Tockman, S. Lam, N. Shivapurkar and A.F. Gazdar, Secondary S. Toyooka, K.O. Toyooka, K. Miyajima, J.L. Reddy, M. Toyota, U.G. Sathyanarayana, A. Padar, M.S. Tockman, S. Lam, N. Shivapurkar and A.F. Gazdar, Epigenetic down-regulation of death-associated protein kinase in lung cancers. Clin Cancer Res 9, 3034–3041 (2003)
F. Li, B. Ye, L. Hong, H. Xu and M.C. Fishbein, Secondary F. Li, B. Ye, L. Hong, H. Xu and M.C. Fishbein, Epigenetic modifications of histone h4 in lung neuroendocrine tumors. Applied Immunohistochem Mol Morphol 19, 389–394 (2011)
A. Augert, Q. Zhang, B. Bates, M. Cui, X. Wang, G. Wildey, A. Dowlati, D. MacPherson, Secondary a. Augert, Q. Zhang, B. Bates, M. Cui, X. Wang, G. Wildey, A. Dowlati and D. MacPherson, small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol 12, 704–713 (2017)
L. Simo-Riudalbas, M. Perez-Salvia, F. Setien, A. Villanueva, C. Moutinho, A. Martinez-Cardus, S. Moran, M. Berdasco, A. Gomez, E. Vidal, M. Soler, H. Heyn, A. Vaquero, C. de la Torre, S. Barcelo-Batllori, A. Vidal, L. Roz, U. Pastorino, K. Szakszon, G. Borck, C.S. Moura, F. Carneiro, I. Zondervan, S. Savola, R. Iwakawa, T. Kohno, J. Yokota and M. Esteller, Secondary L. Simo-Riudalbas, M. Perez-Salvia, F. Setien, A. Villanueva, C. Moutinho, A. Martinez-Cardus, S. Moran, M. Berdasco, A. Gomez, E. Vidal, M. Soler, H. Heyn, A. Vaquero, C. de la Torre, S. Barcelo-Batllori, A. Vidal, L. Roz, U. Pastorino, K. Szakszon, G. Borck, C.S. Moura, F. Carneiro, I. Zondervan, S. Savola, R. Iwakawa, T. Kohno, J. Yokota and M. Esteller, KAT6B is a tumor suppressor Histone H3 Lysine 23 Acetyltransferase undergoing genomic loss in small cell lung cancer. Cancer Res 75, 3936–3945 (2015)
J. George, J.S. Lim, S.J. Jang, Y. Cun, L. Ozretic, G. Kong, F. Leenders, X. Lu, L. Fernandez-Cuesta, G. Bosco, C. Muller, I. Dahmen, N.S. Jahchan, K.S. Park, D. Yang, A.N. Karnezis, D. Vaka, A. Torres, M.S. Wang, J.O. Korbel, R. Menon, S.M. Chun, D. Kim, M. Wilkerson, N. Hayes, D. Engelmann, B. Putzer, M. Bos, S. Michels, I. Vlasic, D. Seidel, B. Pinther, P. Schaub, C. Becker, J. Altmuller, J. Yokota, T. Kohno, R. Iwakawa, K. Tsuta, M. Noguchi, T. Muley, H. Hoffmann, P.A. Schnabel, I. Petersen, Y. Chen, A. Soltermann, V. Tischler, C.M. Choi, Y.H. Kim, P.P. Massion, Y. Zou, D. Jovanovic, M. Kontic, G.M. Wright, P.A. Russell, B. Solomon, I. Koch, M. Lindner, L.A. Muscarella, A. la Torre, J.K. Field, M. Jakopovic, J. Knezevic, E. Castanos-Velez, L. Roz, U. Pastorino, O.T. Brustugun, M. Lund-Iversen, E. Thunnissen, J. Kohler, M. Schuler, J. Botling, M. Sandelin, M. Sanchez-Cespedes, H.B. Salvesen, V. Achter, U. Lang, M. Bogus, P.M. Schneider, T. Zander, S. Ansen, M. Hallek, J. Wolf, M. Vingron, Y. Yatabe, W.D. Travis, P. Nurnberg, C. Reinhardt, S. Perner, L. Heukamp, R. Buttner, S.A. Haas, E. Brambilla, M. Peifer, J. Sage and R.K. Thomas, Secondary J. George, J.S. Lim, S.J. Jang, Y. Cun, L. Ozretic, G. Kong, F. Leenders, X. Lu, L. Fernandez-Cuesta, G. Bosco, C. Muller, I. Dahmen, N.S. Jahchan, K.S. Park, D. Yang, A.N. Karnezis, D. Vaka, A. Torres, M.S. Wang, J.O. Korbel, R. Menon, S.M. Chun, D. Kim, M. Wilkerson, N. Hayes, D. Engelmann, B. Putzer, M. Bos, S. Michels, I. Vlasic, D. Seidel, B. Pinther, P. Schaub, C. Becker, J. Altmuller, J. Yokota, T. Kohno, R. Iwakawa, K. Tsuta, M. Noguchi, T. Muley, H. Hoffmann, P.A. Schnabel, I. Petersen, Y. Chen, A. Soltermann, V. Tischler, C.M. Choi, Y.H. Kim, P.P. Massion, Y. Zou, D. Jovanovic, M. Kontic, G.M. Wright, P.A. Russell, B. Solomon, I. Koch, M. Lindner, L.A. Muscarella, A. la Torre, J.K. Field, M. Jakopovic, J. Knezevic, E. Castanos-Velez, L. Roz, U. Pastorino, O.T. Brustugun, M. Lund-Iversen, E. Thunnissen, J. Kohler, M. Schuler, J. Botling, M. Sandelin, M. Sanchez-Cespedes, H.B. Salvesen, V. Achter, U. Lang, M. Bogus, P.M. Schneider, T. Zander, S. Ansen, M. Hallek, J. Wolf, M. Vingron, Y. Yatabe, W.D. Travis, P. Nurnberg, C. Reinhardt, S. Perner, L. Heukamp, R. Buttner, S.A. Haas, E. Brambilla, M. Peifer, J. Sage and R.K. Thomas, Comprehensive genomic profiles of small cell lung cancer. Nature 524, 47–53 (2015)
V. Shukla, M. Rao, H. Zhang, J. Beers, D. Wangsa, D. Wangsa, F.O. Buishand, Y. Wang, Z. Yu, H.S. Stevenson, E.S. Reardon, K.C. McLoughlin, A.S. Kaufman, E.C. Payabyab, J.A. Hong, M. Zhang, S. Davis, D. Edelman, G. Chen, M.M. Miettinen, N.P. Restifo, T. Ried, P.A. Meltzer and D.S. Schrump, Secondary V. Shukla, M. Rao, H. Zhang, J. Beers, D. Wangsa, D. Wangsa, F.O. Buishand, Y. Wang, Z. Yu, H.S. Stevenson, E.S. Reardon, K.C. McLoughlin, A.S. Kaufman, E.C. Payabyab, J.A. Hong, M. Zhang, S. Davis, D. Edelman, G. Chen, M.M. Miettinen, N.P. Restifo, T. Ried, P.A. Meltzer and D.S. Schrump, ASXL3 is a novel pluripotency factor in human respiratory epithelial cells and a potential therapeutic target in small cell lung cancer. Cancer Res 77, 6267–6281 (2017)
R. Hubaux, K.L. Thu, B.P. Coe, C. MacAulay, S. Lam and W.L. Lam, Secondary R. Hubaux, K.L. Thu, B.P. Coe, C. MacAulay, S. Lam and W.L. Lam, EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 8, 1102–1106 (2013)
S.K. Denny, D. Yang, C.H. Chuang, J.J. Brady, J.S. Lim, B.M. Gruner, S.H. Chiou, A.N. Schep, J. Baral, C. Hamard, M. Antoine, M. Wislez, C.S. Kong, A.J. Connolly, K.S. Park, J. Sage, W.J. Greenleaf and M.M. Winslow, Secondary S.K. Denny, D. Yang, C.H. Chuang, J.J. Brady, J.S. Lim, B.M. Gruner, S.H. Chiou, A.N. Schep, J. Baral, C. Hamard, M. Antoine, M. Wislez, C.S. Kong, A.J. Connolly, K.S. Park, J. Sage, W.J. Greenleaf and M.M. Winslow, Nfib promotes metastasis through a widespread increase in chromatin accessibility. Cell 166, 328–342 (2016)
F. Kato, F.P. Fiorentino, A. Alibes, M. Perucho, M. Sanchez-Cespedes, T. Kohno and J. Yokota, Secondary F. Kato, F.P. Fiorentino, A. Alibes, M. Perucho, M. Sanchez-Cespedes, T. Kohno and J. Yokota, MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget 7, 77378–77388 (2016)
E.E. Gardner, B.H. Lok, V.E. Schneeberger, P. Desmeules, L.A. Miles, P.K. Arnold, A. Ni, I. Khodos, E. de Stanchina, T. Nguyen, J. Sage, J.E. Campbell, S. Ribich, N. Rekhtman, A. Dowlati, P.P. Massion, C.M. Rudin and J.T. Poirier, Secondary E.E. Gardner, B.H. Lok, V.E. Schneeberger, P. Desmeules, L.A. Miles, P.K. Arnold, A. Ni, I. Khodos, E. de Stanchina, T. Nguyen, J. Sage, J.E. Campbell, S. Ribich, N. Rekhtman, A. Dowlati, P.P. Massion, C.M. Rudin and J.T. Poirier, Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell 31, 286–299 (2017)
A. Pasini, G. Paganelli, A. Tesei, W. Zoli, E. Giordano and D. Calistri, Secondary A. Pasini, G. Paganelli, A. Tesei, W. Zoli, E. Giordano and D. Calistri, Specific biomarkers are associated with docetaxeland gemcitabine-resistant NSCLC cell lines. Transl Oncol 5, 461–468 (2012)
I. Ibanez de Caceres, M. Cortes-Sempere, C. Moratilla, R. Machado-Pinilla, V. Rodriguez-Fanjul, C. Manguan-Garcia, P. Cejas, F. Lopez-Rios, L. Paz-Ares, J. de CastroCarpeno, M. Nistal, C. Belda-Iniesta and R. Perona, Secondary I. Ibanez de Caceres, M. Cortes-Sempere, C. Moratilla, R. Machado-Pinilla, V. Rodriguez-Fanjul, C. Manguan-Garcia, P. Cejas, F. Lopez-Rios, L. Paz-Ares, J. de CastroCarpeno, M. Nistal, C. Belda-Iniesta and R. Perona, IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene 29, 1681–1690 (2010)
P.P. Phiboonchaiyanan and P. Chanvorachote, Secondary P.P. Phiboonchaiyanan and P. Chanvorachote, Suppression of a cancer stem-like phenotype mediated by alpha-lipoic acid in human lung cancer cells through down-regulation of beta-catenin and Oct-4. Cell Oncol 40, 497–510 (2017)
S.D. Gore, P. Fenaux, V. Santini, J.M. Bennett, L.R. Silverman, J.F. Seymour, E. Hellstrom-Lindberg, A.S. Swern, C.L. Beach and A.F. List, Secondary S.D. Gore, P. Fenaux, V. Santini, J.M. Bennett, L.R. Silverman, J.F. Seymour, E. Hellstrom-Lindberg, A.S. Swern, C.L. Beach and A.F. List, A multivariate analysis of the relationship between response and survival among patients with higher-risk myelodysplastic syndromes treated within azacitidine or conventional care regimens in the randomized AZA-001 trial. Haematologica 98, 1067–1072 (2013)
N. Zhang, Y. Liu, Y. Wang, M. Zhao, L. Tu and F. Luo, Secondary N. Zhang, Y. Liu, Y. Wang, M. Zhao, L. Tu and F. Luo, Decitabine reverses TGF-beta1-induced epithelial-mesenchymal transition in non-small-cell lung cancer by regulating miR-200/ZEB axis. Drug Des Devel Ther 11, 969–983 (2017)
S.V. Liu, M. Fabbri, B.J. Gitlitz and I.A. Laird-Offringa, Secondary S.V. Liu, M. Fabbri, B.J. Gitlitz and I.A. Laird-Offringa, Epigenetic therapy in lung cancer. Front Oncol 3, 135 (2013)
J.C. Cheng, C.B. Yoo, D.J. Weisenberger, J. Chuang, C. Wozniak, G. Liang, V.E. Marquez, S. Greer, T.F. Orntoft, T. Thykjaer and P.A. Jones, Secondary J.C. Cheng, C.B. Yoo, D.J. Weisenberger, J. Chuang, C. Wozniak, G. Liang, V.E. Marquez, S. Greer, T.F. Orntoft, T. Thykjaer and P.A. Jones, Preferential response of cancer cells to zebularine. Cancer Cell 6, 151–158 (2004)
B.R. You and W.H. Park, Secondary B.R. You and W.H. Park, Zebularine inhibits the growth of A549 lung cancer cells via cell cycle arrest and apoptosis. Mol Carcinog 53, 847–857 (2014)
B.R. You and W.H. Park, Secondary B.R. You and W.H. Park, Zebularine-induced apoptosis in Calu-6 lung cancer cells is influenced by ROS and GSH level changes. Tumour Biol 34, 1145–1153 (2013)
N.E. Muvarak, K. Chowdhury, L. Xia, C. Robert, E.Y. Choi, Y. Cai, M. Bellani, Y. Zou, Z.N. Singh, V.H. Duong, T. Rutherford, P. Nagaria, S.M. Bentzen, M.M. Seidman, M.R. Baer, R.G. Lapidus, S.B. Baylin and F.V. Rassool, Secondary N.E. Muvarak, K. Chowdhury, L. Xia, C. Robert, E.Y. Choi, Y. Cai, M. Bellani, Y. Zou, Z.N. Singh, V.H. Duong, T. Rutherford, P. Nagaria, S.M. Bentzen, M.M. Seidman, M.R. Baer, R.G. Lapidus, S.B. Baylin and F.V. Rassool, Enhancing the cytotoxic effects of PARP inhibitors with DNA demethylating agents - A potential therapy for cancer. Cancer Cell 30, 637–650 (2016)
C.D. DiNardo, K.W. Pratz, A. Letai, B.A. Jonas, A.H. Wei, M. Thirman, M. Arellano, M.G. Frattini, H. Kantarjian, R. Popovic, B. Chyla, T. Xu, M. Dunbar, S.K. Agarwal, R. Humerickhouse, M. Mabry, J. Potluri, M. Konopleva and D.A. Pollyea, Secondary C.D. DiNardo, K.W. Pratz, A. Letai, B.A. Jonas, A.H. Wei, M. Thirman, M. Arellano, M.G. Frattini, H. Kantarjian, R. Popovic, B. Chyla, T. Xu, M. Dunbar, S.K. Agarwal, R. Humerickhouse, M. Mabry, J. Potluri, M. Konopleva and D.A. Pollyea, Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol 19, 216–228 (2018)
F.P. Vendetti, M. Topper, P. Huang, I. Dobromilskaya, H. Easwaran, J. Wrangle, S.B. Baylin, J.T. Poirier and C.M. Rudin, Secondary F.P. Vendetti, M. Topper, P. Huang, I. Dobromilskaya, H. Easwaran, J. Wrangle, S.B. Baylin, J.T. Poirier and C.M. Rudin, Evaluation of azacitidine and entinostat as sensitization agents to cytotoxic chemotherapy in preclinical models of non-small cell lung cancer. Oncotarget 6, 56–70 (2015)
S. Majid, A.A. Dar, V. Shahryari, H. Hirata, A. Ahmad, S. Saini, Y. Tanaka, A.V. Dahiya and R. Dahiya, Secondary S. Majid, A.A. Dar, V. Shahryari, H. Hirata, A. Ahmad, S. Saini, Y. Tanaka, A.V. Dahiya and R. Dahiya, Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer 116, 66–76 (2010)
J. Bauman, C. Verschraegen, S. Belinsky, C. Muller, T. Rutledge, M. Fekrazad, M. Ravindranathan, S.J. Lee and D. Jones, Secondary J. Bauman, C. Verschraegen, S. Belinsky, C. Muller, T. Rutledge, M. Fekrazad, M. Ravindranathan, S.J. Lee and D. Jones, A phase I study of 5-azacytidine and erlotinib in advanced solid tumor malignancies. Cancer Chemother Pharmacol 69, 547–554 (2012)
R. Vinodhkumar, Y.S. Song and T. Devaki, Secondary R. Vinodhkumar, Y.S. Song and T. Devaki, Romidepsin (depsipeptide) induced cell cycle arrest, apoptosis and histone hyperacetylation in lung carcinoma cells (A549) are associated with increase in p21 and hypophosphorylated retinoblastoma proteins expression. Biomed Pharmacother 62, 85–93 (2008)
M. Kodani, T. Igishi, S. Matsumoto, H. Chikumi, Y. Shigeoka, H. Nakanishi, M. Morita, K. Yasuda, Y. Hitsuda, E. Shimizu, Secondary M. Kodani, T. Igishi, S. Matsumoto, H. Chikumi, Y. Shigeoka, H. Nakanishi, M. Morita, K. Yasuda, Y. Hitsuda and E. Shimizu, suppression of phosphatidylinositol 3-kinase/Akt signaling pathway is a determinant of the sensitivity to a novel histone deacetylase inhibitor, FK228, in lung adenocarcinoma cells. Oncol Rep 13, 477–483 (2005)
G.A. Otterson, L. Hodgson, H. Pang, E.E. Vokes, Cancer and B. Leukemia Group, Secondary G.A. Otterson, L. Hodgson, H. Pang, E.E. Vokes, Cancer and B. Leukemia Group, Phase II study of the histone deacetylase inhibitor Romidepsin in relapsed small cell lung cancer (Cancer and Leukemia Group B 30304). J Thorac Oncol 5, 1644–1648 (2010)
M. Peifer, L. Fernandez-Cuesta, M.L. Sos, J. George, D. Seidel, L.H. Kasper, D. Plenker, F. Leenders, R. Sun, T. Zander, R. Menon, M. Koker, I. Dahmen, C. Muller, V. Di Cerbo, H.U. Schildhaus, J. Altmuller, I. Baessmann, C. Becker, B. de Wilde, J. Vandesompele, D. Bohm, S. Ansen, F. Gabler, I. Wilkening, S. Heynck, J.M. Heuckmann, X. Lu, S.L. Carter, K. Cibulskis, S. Banerji, G. Getz, K.S. Park, D. Rauh, C. Grutter, M. Fischer, L. Pasqualucci, G. Wright, Z. Wainer, P. Russell, I. Petersen, Y. Chen, E. Stoelben, C. Ludwig, P. Schnabel, H. Hoffmann, T. Muley, M. Brockmann, W. Engel-Riedel, L.A. Muscarella, V.M. Fazio, H. Groen, W. Timens, H. Sietsma, E. Thunnissen, E. Smit, D.A. Heideman, P.J. Snijders, F. Cappuzzo, C. Ligorio, S. Damiani, J. Field, S. Solberg, O.T. Brustugun, M. Lund-Iversen, J. Sanger, J.H. Clement, A. Soltermann, H. Moch, W. Weder, B. Solomon, J.C. Soria, P. Validire, B. Besse, E. Brambilla, C. Brambilla, S. Lantuejoul, P. Lorimier, P.M. Schneider, M. Hallek, W. Pao, M. Meyerson, J. Sage, J. Shendure, R. Schneider, R. Buttner, J. Wolf, P. Nurnberg, S. Perner, L.C. Heukamp, P.K. Brindle, S. Haas and R.K. Thomas, Secondary M. Peifer, L. Fernandez-Cuesta, M.L. Sos, J. George, D. Seidel, L.H. Kasper, D. Plenker, F. Leenders, R. Sun, T. Zander, R. Menon, M. Koker, I. Dahmen, C. Muller, V. Di Cerbo, H.U. Schildhaus, J. Altmuller, I. Baessmann, C. Becker, B. de Wilde, J. Vandesompele, D. Bohm, S. Ansen, F. Gabler, I. Wilkening, S. Heynck, J.M. Heuckmann, X. Lu, S.L. Carter, K. Cibulskis, S. Banerji, G. Getz, K.S. Park, D. Rauh, C. Grutter, M. Fischer, L. Pasqualucci, G. Wright, Z. Wainer, P. Russell, I. Petersen, Y. Chen, E. Stoelben, C. Ludwig, P. Schnabel, H. Hoffmann, T. Muley, M. Brockmann, W. Engel-Riedel, L.A. Muscarella, V.M. Fazio, H. Groen, W. Timens, H. Sietsma, E. Thunnissen, E. Smit, D.A. Heideman, P.J. Snijders, F. Cappuzzo, C. Ligorio, S. Damiani, J. Field, S. Solberg, O.T. Brustugun, M. Lund-Iversen, J. Sanger, J.H. Clement, A. Soltermann, H. Moch, W. Weder, B. Solomon, J.C. Soria, P. Validire, B. Besse, E. Brambilla, C. Brambilla, S. Lantuejoul, P. Lorimier, P.M. Schneider, M. Hallek, W. Pao, M. Meyerson, J. Sage, J. Shendure, R. Schneider, R. Buttner, J. Wolf, P. Nurnberg, S. Perner, L.C. Heukamp, P.K. Brindle, S. Haas and R.K. Thomas, Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nature Genet 44, 1104–1110 (2012)
C.H. Pan, Y.F. Chang, M.S. Lee, B.C. Wen, J.C. Ko, S.K. Liang and M.C. Liang, Secondary C.H. Pan, Y.F. Chang, M.S. Lee, B.C. Wen, J.C. Ko, S.K. Liang and M.C. Liang, Vorinostat enhances the cisplatin-mediated anticancer effects in small cell lung cancer cells. BMC Cancer 16, 857 (2016)
J.H. Chen, Y.L. Zheng, C.Q. Xu, L.Z. Gu, Z.L. Ding, L. Qin, Y. Wang, R. Fu, Y.F. Wan and C.P. Hu, Secondary J.H. Chen, Y.L. Zheng, C.Q. Xu, L.Z. Gu, Z.L. Ding, L. Qin, Y. Wang, R. Fu, Y.F. Wan and C.P. Hu, Valproic acid (VPA) enhances cisplatin sensitivity of non-small cell lung cancer cells via HDAC2 mediated down regulation of ABCA1. Biol Chem 398, 785–792 (2017)
L. Wang, N.L. Syn, V.V. Subhash, Y. Any, W.L. Thuya, E.S.H. Cheow, L. Kong, F. Yu, P.C. Peethala, A.L. Wong, H.J. Laljibhai, A. Chinnathambi, P.S. Ong, P.C. Ho, G. Sethi, W.P. Yong and B.C. Goh, Secondary L. Wang, N.L. Syn, V.V. Subhash, Y. Any, W.L. Thuya, E.S.H. Cheow, L. Kong, F. Yu, P.C. Peethala, A.L. Wong, H.J. Laljibhai, A. Chinnathambi, P.S. Ong, P.C. Ho, G. Sethi, W.P. Yong and B.C. Goh, Pan-HDAC inhibition by panobinostat mediates chemosensitization to carboplatin in non-small cell lung cancer via attenuation of EGFR signaling. Cancer Lett 417, 152–160 (2018)
W.Y. Lee, P.C. Chen, W.S. Wu, H.C. Wu, C.H. Lan, Y.H. Huang, C.H. Cheng, K.C. Chen and C.W. Lin, Secondary W.Y. Lee, P.C. Chen, W.S. Wu, H.C. Wu, C.H. Lan, Y.H. Huang, C.H. Cheng, K.C. Chen and C.W. Lin, Panobinostat sensitizes KRAS-mutant non-small-cell lung cancer to gefitinib by targeting TAZ. Int J Cancer 141, 1921–1931 (2017)
K.J. Yong, A. Li, W.B. Ou, C.K. Hong, W. Zhao, F. Wang, H. Tatetsu, B. Yan, L. Qi, J.A. Fletcher, H. Yang, R. Soo, D.G. Tenen and L. Chai, Secondary K.J. Yong, A. Li, W.B. Ou, C.K. Hong, W. Zhao, F. Wang, H. Tatetsu, B. Yan, L. Qi, J.A. Fletcher, H. Yang, R. Soo, D.G. Tenen and L. Chai, Targeting SALL4 by entinostat in lung cancer. Oncotarget 7, 75425–75440 (2016)
S.E. Witta, R.M. Jotte, K. Konduri, M.A. Neubauer, A.I. Spira, R.L. Ruxer, M. Varella-Garcia, P.A. Bunn, Jr. and F.R. Hirsch, Secondary S.E. Witta, R.M. Jotte, K. Konduri, M.A. Neubauer, A.I. Spira, R.L. Ruxer, M. Varella-Garcia, P.A. Bunn, Jr. and F.R. Hirsch, Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J Clin Oncol 30, 2248–2255 (2012)
L. Sun, Q. He, C. Tsai, J. Lei, J. Chen, L. Vienna Makcey and D.H. Coy, Secondary L. Sun, Q. He, C. Tsai, J. Lei, J. Chen, L. Vienna Makcey and D.H. Coy, HDAC inhibitors suppressed small cell lung cancer cell growth and enhanced the suppressive effects of receptor-targeting cytotoxins via upregulating somatostatin receptor II. Am J Transl Res 10, 545–553 (2018)
E. Evanno, J. Godet, N. Piccirilli, J. Guilhot, S. Milin, J.M. Gombert, B. Fouchaq and J. Roche, Secondary E. Evanno, J. Godet, N. Piccirilli, J. Guilhot, S. Milin, J.M. Gombert, B. Fouchaq and J. Roche, Tri-methylation of H3K79 is decreased in TGF-beta1-induced epithelial-to-mesenchymal transition in lung cancer. Clin Epigenetics 9, 80 (2017)
S. Takagi, Y. Ishikawa, A. Mizutani, S. Iwasaki, S. Matsumoto, Y. Kamada, T. Nomura, K. Nakamura, Secondary S. Takagi, Y. Ishikawa, A. Mizutani, S. Iwasaki, S. Matsumoto, Y. Kamada, T. Nomura and K. Nakamura, LSD1 inhibitor T-3775440 inhibits SCLC cell proliferation by disrupting LSD1 interactions with SNAG domain proteins INSM1 and GFI1B. Cancer Res 77, 4652–4662 (2017)
P. Nilendu, S.C. Sarode, D. Jahagirdar, I. Tandon, S. Patil, G.S. Sarode, J.K. Pal and N.K. Sharma, Secondary P. Nilendu, S.C. Sarode, D. Jahagirdar, I. Tandon, S. Patil, G.S. Sarode, J.K. Pal and N.K. Sharma, Mutual concessions and compromises between stromal cells and cancer cells: driving tumor development and drug resistance. Cell Oncol 41, 353–367 (2018)
E. Heninger, T.E. Krueger and J.M. Lang, Secondary E. Heninger, T.E. Krueger and J.M. Lang, Augmenting antitumor immune responses with epigenetic modifying agents. Front Immunol 6, 29 (2015)
J. Wrangle, W. Wang, A. Koch, H. Easwaran, H.P. Mohammad, F. Vendetti, W. Vancriekinge, T. Demeyer, Z. Du, P. Parsana, K. Rodgers, R.W. Yen, C.A. Zahnow, J.M. Taube, J.R. Brahmer, S.S. Tykodi, K. Easton, R.D. Carvajal, P.A. Jones, P.W. Laird, D.J. Weisenberger, S. Tsai, R.A. Juergens, S.L. Topalian, C.M. Rudin, M.V. Brock, D. Pardoll and S.B. Baylin, Secondary J. Wrangle, W. Wang, A. Koch, H. Easwaran, H.P. Mohammad, F. Vendetti, W. Vancriekinge, T. Demeyer, Z. Du, P. Parsana, K. Rodgers, R.W. Yen, C.A. Zahnow, J.M. Taube, J.R. Brahmer, S.S. Tykodi, K. Easton, R.D. Carvajal, P.A. Jones, P.W. Laird, D.J. Weisenberger, S. Tsai, R.A. Juergens, S.L. Topalian, C.M. Rudin, M.V. Brock, D. Pardoll and S.B. Baylin, Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget 4, 2067–2079 (2013)
M.J. Topper, M. Vaz, K.B. Chiappinelli, C.E. DeStefano Shields, N. Niknafs, R.C. Yen, A. Wenzel, J. Hicks, M. Ballew, M. Stone, P.T. Tran, C.A. Zahnow, M.D. Hellmann, V. Anagnostou, P.L. Strissel, R. Strick, V.E. Velculescu and S.B. Baylin, Secondary M.J. Topper, M. Vaz, K.B. Chiappinelli, C.E. DeStefano Shields, N. Niknafs, R.C. Yen, A. Wenzel, J. Hicks, M. Ballew, M. Stone, P.T. Tran, C.A. Zahnow, M.D. Hellmann, V. Anagnostou, P.L. Strissel, R. Strick, V.E. Velculescu and S.B. Baylin, Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell 171, 1284–1300 e1221 (2017)
A.E. Teschendorff and C.L. Relton, Secondary A.E. Teschendorff and C.L. Relton, Statistical and integrative system-level analysis of DNA methylation data. Nature Rev Genet 19, 129–147 (2018)
D. Lambrechts, E. Wauters, B. Boeckx, S. Aibar, D. Nittner, O. Burton, A. Bassez, H. Decaluwe, A. Pircher, K. Van den Eynde, B. Weynand, E. Verbeken, P. De Leyn, A. Liston, J. Vansteenkiste, P. Carmeliet, S. Aerts and B. Thienpont, Secondary D. Lambrechts, E. Wauters, B. Boeckx, S. Aibar, D. Nittner, O. Burton, A. Bassez, H. Decaluwe, A. Pircher, K. Van den Eynde, B. Weynand, E. Verbeken, P. De Leyn, A. Liston, J. Vansteenkiste, P. Carmeliet, S. Aerts and B. Thienpont, Phenotype molding of stromal cells in the lung tumor microenvironment. Nature Med 24, 1277–1289 (2018)
H. Fettke, E.M. Kwan and A.A. Azad, Secondary H. Fettke, E.M. Kwan and A.A. Azad, Cell-free DNA in cancer: current insights. Cell Oncol 42, 13–28 (2019)
M. Sromek, M. Glogowski, M. Chechlinska, M. Kulinczak, L. Szafron, K. Zakrzewska, J. Owczarek, P. Wisniewski, R. Wlodarczyk, L. Talarek, M. Turski and J.K. Siwicki, Secondary M. Sromek, M. Glogowski, M. Chechlinska, M. Kulinczak, L. Szafron, K. Zakrzewska, J. Owczarek, P. Wisniewski, R. Wlodarczyk, L. Talarek, M. Turski and J.K. Siwicki, Changes in plasma miR-9, miR-16, miR-205 and miR-486 levels after non-small cell lung cancer resection. Cell Oncol 40, 529–536 (2017)
S.Y. Shen, R. Singhania, G. Fehringer, A. Chakravarthy, M.H.A. Roehrl, D. Chadwick, P.C. Zuzarte, A. Borgida, T.T. Wang, T. Li, O. Kis, Z. Zhao, A. Spreafico, T.D.S. Medina, Y. Wang, D. Roulois, I. Ettayebi, Z. Chen, S. Chow, T. Murphy, A. Arruda, G.M. O'Kane, J. Liu, M. Mansour, J.D. McPherson, C. O'Brien, N. Leighl, P.L. Bedard, N. Fleshner, G. Liu, M.D. Minden, S. Gallinger, A. Goldenberg, T.J. Pugh, M.M. Hoffman, S.V. Bratman, R.J. Hung and D.D. De Carvalho, Secondary S.Y. Shen, R. Singhania, G. Fehringer, A. Chakravarthy, M.H.A. Roehrl, D. Chadwick, P.C. Zuzarte, A. Borgida, T.T. Wang, T. Li, O. Kis, Z. Zhao, A. Spreafico, T.D.S. Medina, Y. Wang, D. Roulois, I. Ettayebi, Z. Chen, S. Chow, T. Murphy, A. Arruda, G.M. O'Kane, J. Liu, M. Mansour, J.D. McPherson, C. O'Brien, N. Leighl, P.L. Bedard, N. Fleshner, G. Liu, M.D. Minden, S. Gallinger, A. Goldenberg, T.J. Pugh, M.M. Hoffman, S.V. Bratman, R.J. Hung and D.D. De Carvalho, Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018)
S.W. Um, H.K. Kim, Y. Kim, B.B. Lee, D. Kim, J. Han, H. Kim, Y.M. Shim and D.H. Kim, Secondary S.W. Um, H.K. Kim, Y. Kim, B.B. Lee, D. Kim, J. Han, H. Kim, Y.M. Shim and D.H. Kim, Bronchial biopsy specimen as a surrogate for DNA methylation analysis in inoperable lung cancer. Clin Epigenetics 9, 131 (2017)
S. Leng, G. Wu, D.M. Klinge, C.L. Thomas, E. Casas, M.A. Picchi, C.A. Stidley, S.J. Lee, S. Aisner, J.M. Siegfried, S. Ramalingam, F.R. Khuri, D.D. Karp and S.A. Belinsky, Secondary S. Leng, G. Wu, D.M. Klinge, C.L. Thomas, E. Casas, M.A. Picchi, C.A. Stidley, S.J. Lee, S. Aisner, J.M. Siegfried, S. Ramalingam, F.R. Khuri, D.D. Karp and S.A. Belinsky, Gene methylation biomarkers in sputum as a classifier for lung cancer risk. Oncotarget 8, 63978–63985 (2017)
Funding
SMP was supported by the Consejería de Salud y Bienestar Social of Junta de Andalucía through the “‘Nicolás Monardes’ program [C-0040-2016] and ISCIII (PI17/00033) and cofounder by FEDER from Regional Development European Funds (European Union).”
Author information
Authors and Affiliations
Contributions
AQV and SMP performed the review and wrote and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Quintanal-Villalonga, Á., Molina-Pinelo, S. Epigenetics of lung cancer: a translational perspective. Cell Oncol. 42, 739–756 (2019). https://doi.org/10.1007/s13402-019-00465-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-019-00465-9