Abstract
Autism spectrum disorder (ASD) is characterized by divergent etiological factors, comorbidities, severity levels, genetic influences, and functional connectivity (FC) patterns in the brain. In the literature, ASD classification based on age and severity using fMRI data is extremely limited. This study explores the impact of age, symptom severity, and brain FC patterns on the diagnosis of ASD using deep neural networks (DNNs). The ability to classify ASD by age and severity using fMRI data is extremely limited. This study explores the impact of age, symptom severity, and brain FC patterns on the diagnosis of ASD using deep neural networks (DNNs). The FC measures were extracted using Pearson's correlation coefficient (PCC), fractal connectivity (FrC), and nonfractal connectivity (NFrC) from the ABIDE I and II databases. We studied three age groups (6 to 11, 11 to 18, and 6 to 18 years) and two severity groups (ADOS score ≤ 11 and ADOS score > 11). The FC matrices are constructed from blood oxygen level-dependent (BOLD) time series signals, and the heat maps are used to generate features for the convolutional neural network (CNN), MobileNetV2, and DenseNet201 models. The MobileNetV2 classifier achieved 76.25% accuracy, 77.09% sensitivity, and 79.77% precision in the age group of 6 to 11 years using NFrC feature maps compared to other DNNs. According to ADOS total scores above 11, DenseNet201 demonstrated superior performance with 83.45% accuracy, 87.3% sensitivity, and 79.13% precision. Connectivity measured by NFrC consistently outperformed Frc measures. Various combinations of connectivity measures and classifiers consistently showed promising results for the age group of 6–11 years and the severity group with an ADOS score of more than 11. ASD's inherent heterogeneity can be addressed effectively by developing diagnostic models tailored to age and severity.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
As a neurodevelopmental disorder, autism spectrum disorder is characterized by impairments in social communication and interaction, as well as constrained, repetitive patterns of behavior, interests, or activities [1, 2]. There has been an increase in its prevalence over the last decade [3], with projections ranging from one in 59 to one in 54 children in the USA [4]. Despite its high prevalence, diagnosing ASD remains challenging. There are a variety of diagnostic tools available today, including psychological assessments, medical evaluations, and caregiver interviews, which are highly subjective and susceptible to misdiagnosis or overdiagnosis. In individuals with autism spectrum disorder (ASD), it is usually associated with local underconnectivity or overconnectivity in the cortex. In this sense, the automated identification of significant structural and/or functional biomarkers in the brain is crucial to the diagnosis of autism spectrum disorders.
ASD is characterized by structural and functional abnormalities that are highly heterogeneous, making it difficult to identify distinctive neural signatures [5]. The heterogeneity of demographic samples has always posed a challenge to understanding atypical neural architecture in ASDs, regardless of gender, age, severity of symptoms, or comorbidities [6]. In recent years, there has been an exponential increase in the number of children with autism spectrum disorder (ASD), characterized by a complex behavior problem that affects social skills, communication patterns, and motor skills. Although autism spectrum disorder is linked to atypical brain connectivity across multiple systems, the nature of these differences in young children remains unclear. Many studies have attempted to reduce dataset heterogeneity by identifying neural features associated with age [7, 8], gender [9], severity [10], and site [1]. There has been no study that analyzed functional abnormalities based on both age and severity in the same dataset. Our study aims to determine whether age- and severity-specific datasets can help us diagnose ASDs more accurately.
Neuronal connectivity and its characteristics can be studied using brain imaging methods such as resting-state functional magnetic resonance imaging (rs-fMRI) [11]. Blood-oxygen-level-dependent (BOLD) signals represent changes in deoxyglobin concentrations captured at low frequencies and characteristic of brain activity. By computing the correlation between BOLD time series, functional connectivity (FC) offers insight into the connectivity between brain regions and the interaction between them [12, 13]. Numerous studies have used rs-fMRI to determine if individuals with ASD demonstrate atypical neural development in the amygdala [14], prefrontal cortex [15], cerebellum, inferior occipital gyruses, and posterior inferior temporal gyruses [16, 17]. There is, however, a large disparity in the areas of interest between these studies. Therefore, this study aims to diagnose ASD using brain FC calculated from fMRI data and maximum brain regions parcellated from fMRI data.
Uddin et al. have conducted a study to classify the children with ASD based on the symptoms of severity in 20 children (Male: 16, Female: 4) using fMRI [18]. They achieved a maximum mean classification rate of 83% and 78% using saliency maps of different brain regions and BOLD signal, respectively, using logistic regression classifier. Compared to the saliency map-based features, the BOLD signal gives more meaningful information related to ASD. The major limitations of the study are limited sample size and children with high FIQ are considered for experiment. Children with ASD appear to have functional hyperconnectivity in the brain, which may be a characteristic of the disorder. A major limitation of earlier studies is that only a few female children with ASD were considered compared to male children [19]. A recent study conducted an experiment with an increased number of female and male children with ASD (n = 773) to overcome the problem [19]. Both types of children's fMRI images were fed into a spatiotemporal deep neural network (stDNN) and achieved mean classification rates of 86% in the ABIDE dataset and 83.4% in the CMI-HBN dataset. It was concluded that females with ASD have a different functional organization compared to males with ASD. ASD severity was classified using rest-state functional connectivity patterns from fMRI using regression estimation methods by Liu et.al using ABIDE-I dataset (n = 174) [20]. In classifying ASD with typically developing and normal control (NC), Pearson correlation coefficient (PCC) values of 0.5 were found. Additionally, the results indicate severe differences in functional connectivity indices between NC and ASD in typically developing people. According to a recent study, researchers recorded joint attention behaviors among 45 children with autism spectrum disorder (ASD) ages 2–6 years and fed the data into a deep neural network (DNN) (convolutional neural network–LSTM–attention mechanism) for severity-based ASD classification [21]. With the proposed model, initiation of joint attention was predicted with a maximum AUROC of 99.6%, and symptom severity-based ASD classification was predicted with a maximum AUROC of 93.4%. To the best of our knowledge, numerous studies have utilized either open-source fMRI data or their own fMRI dataset to diagnose ASD using resting-state information based on fMRI. FMRI data have been used very rarely for age- or severity-based diagnosis of ASD in the literature. The age and severity of ASD have not been considered together in an earlier study for a more robust and reliable classification. Thus, the present work aims to classify ASD based on both age- and severity-based fMRI data using deep neural networks (DNNs).
Researchers have found that FC patterns and microscopic synaptic connectivity can serve as biomarkers for autism spectrum disorders. It has been proposed several methods for constructing functional networks in FC modeling, including Pearson correlation coefficient (PCC) [10], Pearson partial correlation coefficient (PPCC) [22, 23], Spearman's rank correlation coefficient (SRCC)[24], mutual information (MI)[25], and Gaussian covariance (GC) [26]. Traditional correlation techniques may not capture the dynamics of spontaneous neuronal activity since non-neuronal physiological operations can affect signals during the resting state [27, 28]. In both the time and frequency domains, rs-fMRI signals exhibit fractal behavior characterized by self-similarity and power-law scaling [29,30,31,32]. Due to fractal behavior, non-fractal connectivity has been proposed as a new method for measuring FC in fMRI. As a result of this approach, the fractal behavior of the signals is removed in order to obtain a more accurate representation of spontaneous neuronal activity's correlation structure. In 2012, You et al. introduced fractal and non-fractal connectivity for a multivariate fractionally integrated noise and proposed several wavelet-based estimators for fractal and non-fractal connectivity [28]. The study demonstrated that FC changes were related to Alzheimer's disease, ASD [33], ASD without language or intellectual disabilities, ASD with language or intellectual disabilities, pervasive developmental disorders, and typically developing (TD) (binary and multiclass classification) [34]. Both binary and multiclass classification outperformed conventional PCC-based connectivity, and these findings indicate that non-oscillatory connectivity approaches have substantial potential. Thus, PCC, fractal connectivity (Frc), and non-fractal connectivity (NFrC) were used as functional connectivity measures in the present work to investigate the impact of age and severity on the diagnosis of autism spectrum disorders.
Machine learning algorithms are more popular in ASD diagnosis using fMRI or sMRI or EEG data. Using ABIDE-I data, the researchers have analyzed resting state functional connectivity indices using an attention mechanism-based extra tree algorithm to classify ABD in [35]. The maximum mean accuracy of 72.2% is achieved using CC200 atlas. Support vector machine (SVM) classifiers are used to classify ASDs and NCs using features extracted from multilayer perceptron (MLP) networks [36]. Researchers used four different fMRI datasets and the Auto-Tune Model (ATM) to fine-tune the hyperparameters of an SVM classifier to achieve the highest classification rate. Thus, they achieved a maximum mean accuracy of over 70% across all datasets. According to the authors, they only discussed the classification of ASD based on fMRI and not the severity-based classification. Recently, researchers have used four different machine learning algorithms, namely suport vector machine (SVM), random forest (RF), multilayer perceptron (MLP), and Naïve Bayes (NB), to classify ASDs using fMRI data available in the ABIDE-I dataset [37]. They implemented the SMOTE method to prepare a balanced dataset and achieved an 89.23% classification rate compared with the state-of-the-art methods reported in the literature. Machine learning algorithms are used mostly for limited data, and the performance of these algorithms is not superior when using high-volume data to diagnose ASDs. The state-of-the-art review on the use of unsupervised machine learning algorithms in ASD classification can be found in [38]
Neurological conditions are reflected in high-dimensional FC matrices constructed from different brain regions. Using FC information, convolutional neural networks (CNNs) effectively capture spatial patterns, extracting relevant discriminative features for understanding autism-related brain functioning differences. Studies have used different deep learning models, such as stacked auto-encoders [1], single-layer perceptron [39], recurrent neural networks (RNNs) [40], RNNs with long short-term memory (LSTM) [41], hybrid CNN models [42], multichannel deep attention neural networks [43], configurable CNNs [44], graph neural networks [45], and hierarchical graph convolutional networks [46] to extract patterns from FC matrices. A CNN model, DenseNet201 [47, 48], has been employed to improve the retrieval of information for efficient ASD diagnostic classification using fMRI data, because it is a deeply connected network capable of improving parameter efficiency and gradient flow. Additionally, MobileNetV2, a significantly lighter and less complex architecture capable of striking a great balance between model complexity and performance, has been used with fMRI data for ASD diagnosis [49]. We compare the performance of the more complex DenseNet201 to the computationally inexpensive MobileNetV2, in order to extract age- and severity-specific neural patterns in ASD.
The main motivation of this present study is to investigate the effect of sample heterogeneity, as a function of age and severity on the diagnostic classification of ASD using deep neural networks. Due to the limitations of the availability of extensive publicly accessible datasets, our investigation focused on examining the severity of symptoms based on the autism diagnostic observation schedule [ADOS]. By focusing on FC's diverse nature, we operationalized key factors contributing to its diversity. Data were collected from three distinct cohorts of children and adolescents ranging in age from six to eighteen. These cohorts had varying degrees of heterogeneity incorporated into the severity of ASD symptoms using three FC measures, Pearson’s correlation, fractal connectivity, and non-fractal connectivity. We constructed diagnostic classifiers using rs-fMRI data for each cohort, including MobileNetV2 and DenseNet201. The hypothesis stated that increased homogeneity among cohorts would enhance classification accuracy and that the most influential FC features would be different among cohorts.
The main contributions of this paper are:
-
Deep learning models for the diagnosis of autism spectrum disorders were developed based on age and severity.
-
Fractal connectivity and non-fractal connectivity-based correlation methods were implemented and compared with Pearson's correlation-based method.
-
Our diagnostic classification of ASD is based on global diagnostic models regardless of the sites or types of data acquisition.
2 Materials and Methods
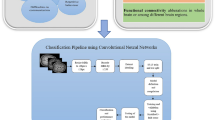
The pipeline used in this study is shown in Fig. 1. Publicly available fMRI data sets, Autism Brain Imaging Data Exchange (ABIDE) I and II, were used in this study [50]. The proposed pipeline uses rs-fMRI data from 317 patients with ASD and 400 controls with TD. The rs-fMRI data are preprocessed, and then the average time series of BOLD signals are extracted as an input dataset for deep learning.
2.1 Dataset
Our study utilized the ABIDE I and ABIDE II databases, which were sourced from worldwide locations and included structural MRIs, diffusion tensor imaging, and rs-fMRIs, all of which had been approved by the local institutional review boards. To ensure a robust data selection process, seven sites were evaluated, each with its own criteria to be included in the rs-fMRI dataset. Moreover, we included rs-fMRI data acquired from subjects with open eyes only to examine the influence of eye state on FC [51].
The study focused on rs-fMRI data from participants aged 6 to 18 years old that retained at least 80% of their original volumes after filtering. Furthermore, we reduced the effects of head movement on BOLD fluctuations by implementing a root mean square deviation of less than 0.2 [52]. A comprehensive demographic profile was constructed by gathering information on the subject's gender, age, intelligence quotient, and severity. Our study included 277 males and 40 females with ASD, as well as 297 males and 103 females with TD. Table 1 presents the demographic data of all participants.
2.2 Preprocessing
In the fMRI data analysis pipeline, preprocessing is extremely important. AFNI (http://afni.nimh.nih.gov) [53] and FSL (http://www.fmrib.ox.ac.uk/fsl) [54] are popular software packages used for preprocessing and analyzing fMRI data. In this present work, we have utilized eight different operations in preprocessing to improve the data quality. The eight operations are: trimming, alignment, normalization, spatial smoothing, temporal filtering, subject-level regression, global signal regression, and nuisance regressor filtering. The detail of each method is given below:
-
Trimming: Trimming is often used to remove initial volumes of fMRI data affected by T1 equilibrium. The fMRI data obtained from the NYU site were trimmed to maintain T1 equilibrium (5 and 3 volumes, respectively, in ABIDE-I and ABIDE-II) [51].
-
Alignment: We used FLIRT and Sinc interpolation to align functional images to anatomical space using six degrees of freedom. The purpose of this step is to align the functional images to the anatomical space and correct any potential misalignments [26].
-
Normalization: To ensure that the images from different sites were aligned in the same space, the aligned images were normalized to MNI152 3 mm, using FNIRT from FSL. Because different scanners have different spatial resolutions and intensities, normalization can help correct these differences.
-
Spatial smoothing: To reduce noise in the images and improve the signal-to-noise ratio (SNR), spatial smoothing was performed. Spatial smoothening helped achieve a global full-width-at-half-maximum of 6 mm in this study. Smoothing extent is an important parameter to optimize, as too much smoothing can reduce spatial resolution, and too little smoothing can result in noisy images.
-
Temporal filtering: The temporal filtering method removes low-frequency drifts and high-frequency noise from fMRI data. Using a second-order band-pass filter with a pass band of 0.008–0.08 Hz, resting-state fMRI data were temporally smoothed [26].
-
Subject-level regression: Denoising was accomplished by performing a subject-level regression on eight nuisance variables and their corresponding first derivatives. A total of six rigid-body motion parameters were estimated by motion correction, along with ventricular cerebrospinal fluid and white-matter signals measured by FSL's FAST method. The RS-fMRI signal is cleaned up using this step to remove noise sources.
-
Global signal regression: Preprocessing pipelines incorporate global signal regression to compensate for the effects of using fMRI data from different sites, which also reduces signal-to-noise ratios. In this step, the variability in data is reduced due to differences in acquisition parameters across sites [55].
-
Nuisance regressor filtering: To maintain consistency across the entire dataset, we applied the same second-order Butterworth band-pass filter with a pass band of 0.008–0.08 Hz to all seventeen nuisance regressors. By following the same preprocessing steps for all regressors, it is easier to compare results across different subjects and studies [56, 57].
2.3 BOLD Time Series Extraction
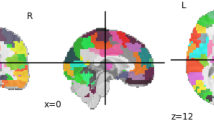
This study constructed a whole-brain mask by identifying voxels in which BOLD signals were detected in at least 95% of participants. We used 333 cortical Regions of Interest (ROI) from Gordon's atlas [58], 14 subcortical ROIs from the Harvard Oxford atlas [59], and 26 cerebellar ROIs from Diedrichsen atlases [60] in combination with some minor cerebellar ROIs. The atlases were selected based on our previous experience categorizing and analyzing the brains of individuals with ASD [61, 62]. A whole-brain mask was then constructed based on the number of voxels contained within each ROI. Our research focused on ROIs that included at least 95% of the voxels in the whole-brain mask [26). As a result, 236 ROIs were obtained, including 213 cortical, 14 subcortical, and 9 cerebellar regions.
2.4 Functional Connectivity Matrix
Three FC metrics were considered in this study: Pearson's correlation coefficient (PCC), fractal connectivity (FrC), and non-fractal connectivity (NFrC). In each of the 236 regions, the average time-series data were calculated for each individual, by correlating every single region with every other region, leading to 236 × 236 sized correlation matrices for each subject. The following subsections explain Pearson's correlation coefficient (PCC), fractal connectivity (FrC), and non-fractal connectivity (NFrC) measures of connectivity.
2.4.1 Pearson Correlation
Pearson correlations are a type of statistical method used to measure FC between different brain regions [63]. In fMRI, it is possible to calculate the linear relationship between two series of brain activity by measuring the BOLD signal. The Pearson correlation coefficient is calculated by dividing the product of the covariances of two time series by the standard deviations of those series. As a result, the values range from -1 to 1, with a value close to 0 indicating no correlation, a value close to 1 indicating a positive correlation (two signals increasing simultaneously), and a value close to 0 indicating a negative correlation.
FC analysis based on Pearson correlation can be used to identify patterns of brain activity that are consistently co-activated across different individuals or conditions. It is often applied to investigate resting-state networks, which are spontaneously synchronized patterns of brain activity that occur in the absence of an external task. For \({a}_{1}\left(t\right)\) and \({a}_{2}\left(t\right)\), which represent stochastic processes where \({\rm E}\left[{a}_{1}\left(t\right)\right]= {\mu }_{1}\) and \({\rm E}\left[ {a}_{2}\left(t\right)\right]= {\mu }_{2}\), the correlation of \({a}_{1}\left(t\right)\) and \({a}_{2}\left(t\right)\) is defined by Eqs. 1 and 2 as:
where, the covariance of \({a}_{1}\) and \({a}_{2}\) is:
2.4.2 Fractal Connectivity
Signals that exhibit long-range dependence and self-similarity are described as fractal behavior [64]. Such behavior can be quantified with the Hurst exponent and the fractal dimension. Basically, it measures the asymptotic wavelet correlation between BOLD signals in different brain regions. The purpose of this type of correlation is to compare two signals over time to see if their values are similar at the same time. The correlation between two signals is high only when their values are similar at the same time. The wavelet transform is applied to the signals to determine the correlation between their wavelet coefficients at different time scales. In this study, a discrete wavelet transform is used, and the level of approximation is determined by the time series length. Depending on the length of the individual time series, the decomposition level will vary. A simple heuristic for automatic level selection is to divide the signal length by a power of two. In this study, the level of decomposition, \(L\), is determined by \(L = \mathrm{floor}({\mathrm{log}}_{2}(N))\), where \(N\) is the signal's length. As a final step, a six-level discrete wavelet transform, focused solely on approximation coefficients, was applied with an 8-length Daubechies least asymmetric filter (\(db8\)). In the following steps, the coefficients of the two signals were compared at each time scale to establish a correlation between them. According to the equations below [28, 29, 34, 65], fractal connectivity is calculated as follows:
where,
where, \({\widehat{\xi }}_{m,n}\) is the non-fractal connectivity, \({B}_{1}\) is a factor describing the shape of the power spectrum, \({d}_{m}\) and \({d}_{n}\) are parameters determining the fractal behavior of the power spectrum.
2.4.3 Non-Fractal Connectivity
Resting-state BOLD signals show a short-term and long-term temporal dependence on short-term and long-term memories, respectively [66]. The short memory describes the relationship between values in a time series that are close in time, whereas the long memory describes the relationship between values that are far apart in time. A Hurst exponent is commonly used to calculate the short and long memories of a time series by dividing the cumulative sum by the standard deviation, raised to a power [64]. By powering the ratio according to the time series scale, the ratio becomes independent of its time series value. The Hurst exponent ranges from 0 to 1. Values closer to 1 indicate a long memory, while values closer to 0 indicate a short memory. In the BOLD time series covariance matrix, non-fractal connectivity between two brain regions is defined using short memory covariance matrices. These matrices can measure the similarity between two signals over a short period of time, in which large covariances are observed when there are similar values for two signals at the same time. A window of fixed width is slid along the two signals during this process. Multivariate fractionally integrated noise, \(x(t)\), is used to model short memory covariance based on a memory parameter, d, and a short memory function, \(u(t)\). Non-fractal connectivity \(\left({\widehat{\xi }}_{m,n}\right)\) between \({u}_{m}(t)\) and \({u}_{n}(t)\) is described by Eq. 5 given below:
where, \({\gamma }_{m,n}\) denotes the covariance of \({u}_{m}(t)\) and \({u}_{n}(t)\) given by \({\gamma }_{m,n} := {\rm E}\left[{u}_{m}(1){ u}_{n}(1)\right]\)
2.5 Data Augmentation and Dataset Segregation
In this study, we created a dataset that included fractal correlations, nonfractal correlations, and Pearson correlations between FC matrices. Therefore, we generated heat maps and converted them into images based on these matrices. A data augmentation procedure was performed on each image, which included rotating by 90 degrees, enhancing edges, blurring with Gaussian noise, zooming, and cropping (four variants: top left, top right, bottom left, bottom right). As a result, we obtained eight different representations of each subject as original heat maps and augmented heat maps.
A group of participants was divided by age to analyze the impact of age on classifier performance. The study examined three age groups: 6 to 11 years, 11 to 18 years, and 6 to 18 years. The demographics of participant datasets are shown in Table 1. A similar analysis was conducted to examine the effects of ADOS severity or score. ADOS is a standardized assessment tool used by healthcare professionals to diagnose ASD in individuals, particularly children. An ADOS score of less than 11 indicates a less severe condition and a score of more than 11 indicates a rather severe case. Two classification sample sets were created by restricting sample heterogeneity in ADOS total scores, comprising ADOS total score ≤ 11, and ADOS total score > 11. There is a very limited number of female participants in the ABIDE dataset, so this analysis was limited to male participants only. Table 2 shows the demographic characteristics of the dataset based on the ADOS score.
2.6 Classification
We resized the high-dimensional heat map images to 224 × 224 × 3 and divided them into training samples, which made up 75% of the total samples; validation samples, which made up 12.5% of the total samples; and testing samples, which made up 12.5% of the total samples. Finally, the training data are fed into two pre-trained CNN classifiers, MobileNetV2 and DenseNet201. A transfer learning algorithm is used to initialize these classifiers with ImageNet weights. As a result of fine-tuning for model training, pre-trained models can be refined to suit new or similar tasks. A fine-tuning process can improve the performance of a model on new data by initializing it with pre-existing knowledge. Models were implemented in Python (3.9.16) and TensorFlow (2.10.1) and trained on a workstation running Windows 11 Pro on a Dell Inc Precision 3660 x-64 based PC. The system employs a 12th Gen Intel(R) Core (TM) i7-12700K, with a processing speed of 3600 MHz with 12 Core(s), 20 logical processors, and an NVIDIA RTX A200 graphics processing unit with 12 GB of memory. We used the available non-fractal toolbox in Mathworks, MATLAB (Version R2017b) for FC matrix calculations.
2.6.1 MobileNetV2
A MobileNetV2 classifier is trained for 500 epochs at a learning rate of 0.0001. Once a normal run is completed, the model is fine-tuned, which means that a certain number of layers are unfrozen and allowed to adapt to the current dataset. Among the 154 layers in MobileNetV2, 54 layers were fine-tuned and then used to evaluate the test dataset for another 500 epochs using the model.
2.6.2 DenseNet201
The procedure used for DenseNet201 was similar to that used for MobileNetV2. It is important to emphasize that the model was trained normally, without fine-tuning, for 500 epochs and with a learning rate of 0.0001. The model was then fine-tuned on 57 of the 707 layers of the dataset and trained for another 500 epochs before it was used on the test dataset in order to evaluate its performance.
3 Numerical Results
This study analyzed FC matrices obtained from resting-state fMRIs for persons with TD and autism spectrum disorders with three different approaches: Pearson correlations, fractal correlations, and nonfractal correlations. Using the BOLD time-series signals of 236 different brain regions, each subject was mapped out into a matrix of 236 by 236. Figure 2a–c shows six representative FC heat maps based on Pearson correlation, fractal connectivity, and non-fractal connectivity for a TD subject. Figure 3a–c shows similar representations of an ASD subject. All matrices showed a strong correlation between their diagonal values, indicating that each region is highly self-correlated. A significant difference was seen in the patterns of the correlation matrices between the three methods. Compared to fractal and nonfractal methods, the Pearson method showed a completely different pattern, which can be attributed to the different calculations involved in each method.
There was an interesting finding that the difference between the correlation matrices between fractals and non-fractals was related to the memory parameter in the correlation matrices. It has been shown that when the difference between fractal and non-fractal connectivity parameters is close to zero, then the fractal connectivity is almost identical to the non-fractal connectivity. It has been shown that non-fractal connectivity can remove non-physiological characteristics from fMRI data, and it can therefore be inferred that fractal connectivity retains these characteristics when non-fractal connectivity is used. This means that fractal and non-fractal connectivity can provide complementary information to each other.
According to the correlation matrices, the FC matrices of the ASD subjects and those of the TD subjects exhibited different patterns as seen in the FC matrices. Several regions in the TD subjects showed strong connections, as shown by the white color in the middle of these regions, while no such patterns could be seen in the ASD subjects. As a result of all three methods used in this study, there was a difference in FC matrices, which may suggest that ASD is characterized by this characteristic feature. It is important to keep in mind that not all samples will exhibit the same patterns at the same time, and thus visual distinction may not be possible.
To train the deep-learning models, the FC heat maps for TD and ASD participants derived from these analyses were fed into the models as inputs. As part of the evaluation, we tested the performance of the model on all the datasets under similar conditions, that is, using the same weights (ImageNet) and architectures (MobileNetV2 and DenseNet201) with similar augmentation methods. To calculate the performance metrics given for each dataset, we calculated the accuracy, sensitivity, precision, and F1 score for each dataset [67]. It is imperative to emphasize that these performance measures calculations were repeated twice for each dataset: once without fine-tuning, and then once with fine-tuning.
It is shown in Figs. 4a–c, 5a–c, and 6a–c that the models perform for Pearson's correlation coefficient, fractal connectivity, and non-fractal connectivity based on the combination of connectivity measures and age groups, respectively, where (a) represents the performance metrics for the 6- to 11-year-old age group; (b) represents the performance metrics for the 11 to 18-year-old age group; and (c) represents the performance metrics for the 6- to 18-year-old age group, respectively, for each FC measure.
Based on Pearson's correlation-based datasets, DenseNet201 distinguished the age group 6–11 years most accurately, with accuracy, sensitivity, precision, and F1 scores of 72.19, 84.62%, 71.63%, and 77.59%. Similarly, the age group 6 to 11 years achieved impressive results in fractal connectivity with accuracy, sensitivity, precision, and F1 score of 69.30%, 65.28%, 80.25%, and 72%, respectively. In the case of non-fractal connectivity, the 6–11 age group remained on top with 76.25% accuracy, 77.09% sensitivity, 79.77% precision, and 78.41% F1 score. MobileNetV2 was used to train this non-fractal connectivity dataset. The non-fractal connectivity measure with DenseNet201 achieved a higher accuracy in distinguishing the ASD based on the rs-fMRI derived from 6 to 11 years old subjects.
According to classifiers and connectivity measures, participants between 6 and 11 years of age produced the best results. Non-fractal connectivity scored best across all age groups and both classifiers when compared to Pearson's correlation and fractal connectivity. In terms of classifier performance, both MobileNetV2 and DenseNet201 performed well across the different datasets tested. In the 11–18 age group, the fractal connectivity measure produced a maximum classification rate of 74.47%, a sensitivity of 68.71%, a prediction of 78.36%, and an F1-score of 73.72% in MobileNetv2. DenseNet201 network also achieved a maximum mean classification rate of 70.59%, sensitivity of 90.26%, precision of 66.86%, and F1-score of 76.82% using non-fractal connectivity features. Based on the age-based ASD classification, the proposed non-fractal connectivity measure produces higher accuracy than state-of-the-art methods reported in the literature and with other features and networks used in this study.
Similar classification pipelines were used for participant cohorts sub-grouped by severity scores (ADOS). The results for different combinations of FC measures and ADOS scores can be visualized in Figs. 7a, b, 8a, b, and 9a, b for Pearson’s correlation, fractal connectivity, and non-fractal connectivity, respectively, where (a) represent the performance metrics for cohorts with ADOS scores less than or equal to 11; (b) represent the performance metrics for cohorts with ADOS scores greater than 11, respectively.
Using DenseNet201, high-severity datasets (ADOS scores over 11) performed better in Pearson's correlation, achieving accuracy, sensitivity, precision, and F1 scores of 72.61%, 60.73%, 87.21%, and 71.60%, respectively. With MobileNetV2, high-severity groups outperformed low-severity groups, obtaining 73.52% accuracy, 58.01% sensitivity, 81.72% precision, and an F1 score of 67.85%. On severity-specific datasets, nonfractal measures performed better than fractal measures in classifying ASD from TD with an accuracy of 83.45%. In high severity (ADOS score of more than 11) datasets, it achieved a sensitivity of 87.3%, a precision of 79.13%, and an F1 score of 83.01% using the DenseNet201 classifier. Based on severity, the high severity group, i.e., those with ADOS scores greater than 11 outperformed the low severity group on all counts. It is again non-fractal connectivity that performs best, and both classifiers produce comparable results when combined with different datasets.
4 Discussion
4.1 Effect of Functional Connectivity methods
In this study, Pearson's correlation coefficient method was used to compare fractal and non-fractal measures of connectivity. These results are presented in Figs. 3a–c, 4a–c, and 5a–c. According to our findings, the non-fractal approach outperforms the fractal approach for identifying differences in neural connectivity between individuals with ASD and those with TD. A non-fractal analysis of the BOLD signal in both space and time provides more meaningful information about the heterogeneity present in the input data than a linear correlation method like PCC. Furthermore, Pearson's correlation performed better than datasets based on fractal connectivity. Interestingly, these results are consistent with what was previously reported in studies that compared the effectiveness of these diagnostic methods in diagnosing autism spectrum disorders [34] as well as Alzheimer's disease [33]. According to the proposed pipeline, 76.25% accuracy was achieved with nonfractal connectivity, 72.19% accuracy with Pearson's correlation, and 69.30% accuracy with fractal connectivity. According to Sadiq et al. [34], their non-fractal FC was best by a margin of 5.65% to 7.74% when compared with Pearson's correlation-based approach. Based on the support vector machine (SVM) classifier, Naseem et al. [33] also reported an accuracy of 83.3% for the non-fractal connectivity of the ADNI dataset, outperforming fractal connectivity and Pearson's correlation by 16.4% and 17.2%, respectively. According to a previous study using fractals on fMRI datasets, individuals with autism spectrum disorders experience a significant reduction in the complexity of the signals in specific brain regions, including the amygdala, vermis, basin ganglion, and hippocampus [29].
4.2 Significance of Age
In the present study, participants with ASD or TD who were 6 to 11 years old were able to classify more accurately than those who were 11 to 18 years old or those who were 6 to 18 years old. In this study, we found a significant relationship between age and FC differences in individuals with ASD. It could be because early childhood has a significant amount of neural development that is faster than that of children and adolescents, as well as FC patterns that are substantially different from those observed in children and adolescents. Deep learning algorithms can easily identify abnormal FC patterns, which can be used to diagnose diseases. Haghighat et al. [69] conducted a study to build ASD diagnostic models for three different age groups: children, adolescents, and adults. According to the results of the study, children with ASD are well discriminated from the TD (Accuracy: 95.23%), followed by adults (Accuracy: 83.33%) and adolescents (Accuracy: 78.57%). As well, Subbaraju et al. [7] found similar results when they examined ASD and TD diagnoses in gender-specific adolescents and adults (Accuracy: 85.4% for subjects under 18 years of age; Accuracy: 78.6% for subjects over 18 years of age). According to Eslami et al. [39], diagnostic classification models for males aged under 15 performed better (Accuracy: 82%) than the entire dataset used for their analysis (Accuracy: 68%). According to Table 3, the performance of the present work is compared with existing studies based on age. Compared to the existing work, our work has utilized both age, and symptoms severity of the same dataset for ASD classification. In addition, our present has considered three different age groups and utilized the functional connectivity measures based on PCC, FrC, and NFrC in contrast with other works. In summary, the differences in FC observed among individuals with autism suggest that there are underlying neurobiological differences contributing to the disorder's symptoms. In addition to FC changes observed in different age groups, these developmental changes may also result in changes in brain function in individuals with autism spectrum disorders.
4.3 Significance of ADOS Score
We used DenseNet201 to evaluate participant cohorts with ADOS scores over 11 and achieved an accuracy of 83.45%. According to Reiter et al. [10], high-severity ASD subjects were also classified similarly based on heterogeneous datasets. It may be that the higher severity score is characterized by significant under- and over-connectivity in the neural architecture, which is easily captured by FC measures. According to Haweel et al. [71]), task-based fMRI can accurately categorize ASD subjects into mild, moderate, and severe categories. In addition, classification accuracy was improved in mild versus severe (81%) compared to mild versus moderate (80%) when tested on ADOS scores. According to the results, the best classification of ASD was moderate versus severe (77%) when comparing the fMRI data with the ADOS scores [72]. A severity-based model is important for making diagnoses of autism spectrum disorders based on the results of the study.
4.4 Effect of Classifiers
In our study, MobileNetV2 and DenseNet201 are equally effective when analyzing three different sets of data. MobileNetV2 achieved the highest accuracy of 76.25% in the same age group, while DenseNet201 achieved the highest accuracy of 74.68%. A high classification accuracy for DenseNet201 has been demonstrated in both sMRI [73] and fMRI [47] studies. Based on Ahmed et al.'s [49] research, MobileNetV2 is 76.5% accurate at diagnosing autism spectrum disorder (ASD).
4.5 Significance of Proposed Methodology in ASD Diagnosis
In the study, 30 combinations of pipelines were examined, each with three ages, two severity groups, three FC methods, and two classification models. Based on the results of the analysis, the processing pipeline that uses age-specific data (6–11 years old), non-fractal FC, and MobileNetV2 had the best classification accuracy (76.25%). Among severity-specific datasets (ADOS scores greater than 11), non-fractal FC and DenseNet201 produced the highest accuracy, 83.45%. These pipelines outperformed Pearson and fractal FC models as well as other deep learning models. Several studies have used age-specific datasets [7, 39, 69], severity-specific datasets [10], fractal and nonfractal methods [29, 33], along with deep learning algorithms [47, 73]. It is the first time this pipeline has been used in a study.
4.6 Limitations and Future Work
It is evident that the proposed methodology is effective in diagnosing ASD; however, some limitations exist in this pipeline. The training dataset contains just seven sites, which is relatively small. As a result of the stringent inclusion criteria used in this study, the remaining sites in the ABIDE database were not considered for analysis. Due to the limited availability of data for this age group in the ABIDE database, we did not include participants over 18 years old in our analysis. Data cross-validation was not possible due to the computational complexity of the model training. As part of the testing dataset, augmented images were included. There is the possibility of expanding this study to include more participants. In future work, feature ranking and selection could be included to ensure the classifier includes only relevant features. In addition, unsupervised clustering algorithms can be used to reduce heterogeneity in datasets by identifying underlying patterns. Based on robust and promising patterns in clustered datasets, ASD and TD could be better classified as diagnostic conditions.
5 Conclusion
In this study, FC matrices were computed based on fMRI data and three different connectivity measures, including Pearson correlation, fractal connectivity, and non-fractal connectivity, to examine how age, severity, and FC measures affect ASD classification. The matrices were converted into heat maps and used to train MobileNetV2 and DenseNet201. The age-specific dataset with participants between the ages of 6 and 11 years performed the best across all three types of connectivity in comparison to the datasets with 11 to 18-year-olds and 6 to 18-year-olds. The MobileNetV2 network scored the highest in accuracy, sensitivity, precision, and F1-score, with 76.25%, 77.09%, 79.77%, and 78.41%, respectively. Using DenseNet201, high-severity datasets with ADOS scores above 11 achieved 83.45% accuracy, 87.3% sensitivity, 79.13% precision, and an F1-score of 83.01% compared to low-severity datasets. Researchers found that a dataset with reduced heterogeneity and specific extraction of short-term temporal memory dependencies from time-series data was more accurate when fed to a deep network. Furthermore, the findings suggest that inconsistency in neuroanatomical reports of ASD may be due to differences in age or symptom severity in the study cohorts, highlighting the importance of considering the effects of age, symptom severity, biological factors, and methodology in future studies of ASD's underlying neural mechanisms.
References
Heinsfeld, A.S.; Franco, A.R.; Craddock, R.C.; Buchweitz, A.; Meneguzzi, F.: Identification of autism spectrum disorder using deep learning and the ABIDE dataset. NeuroImage: Clin. 17, 16–23 (2018)
Hull, J.V.; Dokovna, L.B.; Jacokes, Z.J.; Torgerson, C.M.; Irimia, A.; Van Horn, J.D.: GENDAAR research consortium corrigendum: resting-state functional connectivity in autism spectrum disorders: a review. Front. Psych. 9, 268 (2018)
Hashem, S.; Nisar, S.; Bhat, A.A.; Yadav, S.K.; Azeem, M.W.; Bagga, P.; Fakhro, K.; Reddy, R.; Frenneaux, M.P.; Haris, M.: Genetics of structural and functional brain changes in autism spectrum disorder. Transl. Psych. 10(1), 229 (2020)
Solmi, M.; Song, M.; Yon, D.K.; Lee, S.W.; Fombonne, E.; Kim, M.S.; Park, S.; Lee, M.H.; Hwang, J.; Keller, R.; Koyanagi, A.: Incidence, prevalence, and global burden of autism spectrum disorder from 1990 to 2019 across 204 countries. Mol. Psych. 29, 1–9 (2022)
Maximo, J.O.; Cadena, E.J.; Kana, R.K.: The implications of brain connectivity in the neuropsychology of autism. Neuropsychol. Rev. 24, 16–31 (2014)
Lombardo, M.V.; Lai, M.C.; Baron-Cohen, S.: Big data approaches to decomposing heterogeneity across the autism spectrum. Mol. Psych. 24(10), 1435–1450 (2019)
Subbaraju, V.; Suresh, M.B.; Sundaram, S.; Narasimhan, S.: Identifying differences in brain activities and an accurate detection of autism spectrum disorder using resting state functional-magnetic resonance imaging: a spatial filtering approach. Med. Image Anal. 35, 375–389 (2017)
Iidaka, T.: Resting state functional magnetic resonance imaging and neural network classified autism and control. Cortex 63, 55–67 (2015)
Plitt, M.; Barnes, K.A.; Martin, A.: Functional connectivity classification of autism identifies highly predictive brain features but falls short of biomarker standards. NeuroImage Clin. 7, 359–366 (2015)
Reiter, M.A.; Jahedi, A.; Fredo, A.J.; Fishman, I.; Bailey, B.; Müller, R.A.: Performance of machine learning classification models of autism using resting-state fMRI is contingent on sample heterogeneity. Neural Comput. Appl. 33, 3299–3310 (2021)
Kim, S.; Moon, H.S.; Vo, T.T.; Kim, C.H.; Im, G.H.; Lee, S.; Choi, M.; Kim, S.G.: Whole-brain mapping of effective connectivity by fMRI with cortex-wide patterned optogenetics. Neuron. (2022)
Liégeois, F.; Elward R.: Functional magnetic resonance imaging. InHandbook of clinical neurology 2020 Jan 1 (Vol. 174, pp. 265–275). Elsevier.
Chen, K.; Azeez, A.; Chen, D.Y.; Biswal, B.B.: Resting-state functional connectivity: signal origins and analytic methods. Neuroimag. Clin. 30(1), 15–23 (2020)
Dugré, J.R.; Potvin, S.: Altered functional connectivity of the amygdala across variants of callous-unemotional traits: a resting-state fMRI study in children and adolescents. J. Psych. Res. 2023.
Krishnamurthy, K.; Yeung, M.K.; Chan, A.S.; Han, Y.M.: Effortful control and prefrontal cortex functioning in children with autism spectrum disorder: an fNIRS study. Brain Sci. 10(11), 880 (2020)
Guo, X.; Duan, X.; Chen, H.; He, C.; Xiao, J.; Han, S.; Fan, Y.S.; Guo, J.; Chen, H.: Altered inter-and intrahemispheric functional connectivity dynamics in autistic children. Hum. Brain Mapp. 41(2), 419–428 (2020)
Long, Z.; Duan, X.; Mantini, D., et al.: Alteration of functional connectivity in autism spectrum disorder: effect of age and anatomical distance. Sci. Rep. 6, 26527 (2016). https://doi.org/10.1038/srep26527
Uddin, L.Q.; Supekar, K.; Lynch, C.J., et al.: Salience network-based classification and prediction of symptom severity in children with autism. JAMA Psych. 70(8), 869–879 (2013). https://doi.org/10.1001/jamapsychiatry.2013.104
Supekar, K.; de Los, A.C.; Ryali, S.; Cao, K.; Ma, T.; Menon, V.: Deep learning identifies robust gender differences in functional brain organization and their dissociable links to clinical symptoms in autism. Br. J. Psych. (2022). https://doi.org/10.1192/bjp.2022.13
Liu, X.; Huang, H.: Alterations of functional connectivities associated with autism spectrum disorder symptom severity: a multi-site study using multivariate pattern analysis. Sci. Rep. 10, 4330 (2020). https://doi.org/10.1038/s41598-020-60702-2
Ko, C.; Lim, J.; Hong, J.; Hong, S.; Park, Y.R.: Development and validation of a joint attention-based deep learning system for detection and symptom severity assessment of autism spectrum disorder. JAMA Netw. Open 6(5), e2315174 (2023). https://doi.org/10.1001/jamanetworkopen.2023.15174
Chaitra; Vijaya, P.A.: Machine learning based comparison of Pearson’s and partial correlation measures to quantify functional connectivity in the human brain. IJNBS 6(3), 23–30 (2018)
Wee, C.Y.; Yang, S.; Yap, P.T.; Shen, D.: Alzheimer’s disease neuroimaging Initiative. Sparse temporally dynamic resting-state functional connectivity networks for early MCI identification. Brain Imag. Behav. 10, 342–356 (2016)
Berto, S.; Treacher, A.H.; Caglayan, E.; Luo, D.; Haney, J.R.; Gandal, M.J.; Geschwind, D.H.; Montillo, A.A.; Konopka, G.: Association between resting-state functional brain connectivity and gene expression is altered in autism spectrum disorder. Nat. Commun. 13(1), 3328 (2022)
Sun, C.; Yang, F.; Wang, C.; Wang, Z.; Zhang, Y.; Ming, D.; Du, J.: Mutual information-based brain network analysis in post-stroke patients with different levels of depression. Front. Hum. Neurosci. 12, 285 (2018)
Ronicko, J.F.; Thomas, J.; Thangavel, P.; Koneru, V.; Langs, G.; Dauwels, J.: Diagnostic classification of autism using resting-state fMRI data improves with full correlation functional brain connectivity compared to partial correlation. J. Neurosci. Methods 345, 108884 (2020)
Mohanty, R.; Sethares, W.A.; Nair, V.A.; Prabhakaran, V.: Rethinking measures of functional connectivity via feature extraction. Sci. Rep. 10(1), 1298 (2020)
You, W.; Achard, S.; Stadler, J.; Brückner, B.; Seiffert, U.; Fractal analysis of resting state functional connectivity of the brain. In The 2012 International Joint Conference on Neural Networks (IJCNN) 2012 Jun 10 (pp. 1–8). IEEE.
Dona, O.; Hall, G.B.; Noseworthy, M.D.: Temporal fractal analysis of the rs-BOLD signal identifies brain abnormalities in autism spectrum disorder. PLoS ONE 12(12), e0190081 (2017)
Campbell, O.; Vanderwal, T.; Weber, A.M.: Fractal-based analysis of fMRI BOLD signal during naturalistic viewing conditions. Front. Physiol. 12, 2465 (2022)
Campbell, O.L.; Weber, A.M.: Monofractal analysis of functional magnetic resonance imaging: an introductory review. Hum. Brain Mapp. 43(8), 2693–2706 (2022)
Ochab, J.K.; Wątorek, M.; Ceglarek, A.; Fafrowicz, M.; Lewandowska, K.; Marek, T.; Sikora-Wachowicz, B.; Oświęcimka, P.: Task-dependent fractal patterns of information processing in working memory. Sci. Rep. 1, 17866 (2022)
Sadiq, A.; Yahya, N.; Tang, T.B.; Hashim, H.; Naseem, I.: Wavelet-based fractal analysis of rs-fMRI for classification of alzheimer’s disease. Sensors 22(9), 3102 (2022)
Sadiq, A.; Al-Hiyali, M.I.; Yahya, N.; Tang, T.B.; Khan, D.M.: Non-oscillatory connectivity approach for classification of autism spectrum disorder subtypes using resting-state fMRI. IEEE Access 10, 14049–14061 (2022)
Liu, Y.; Xu, L.; Li, J.; Yu, J.; Yu, X.: Attentional connectivity-based prediction of autism using heterogeneous rs-fMRI data from CC200 atlas. Exp. Neurobiol. 29(1), 27–37 (2020). https://doi.org/10.5607/en.2020.29.1.27
Taban Eslami and Fahad Saeed. 2019. Auto-ASD-Network: A Technique Based on Deep Learning and Support Vector Machines for Diagnosing Autism Spectrum Disorder using fMRI Data. In Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics (BCB '19). Association for Computing Machinery, New York, NY, USA, 646–651. https://doi.org/10.1145/3307339.3343482
Qureshi, M.S.; Qureshi, M.B.; Asghar, J.; Alam, F.; Aljarbouh, A.: Prediction and analysis of autism spectrum disorder using machine learning techniques. J. Healthcare Eng. 4853800, 10 (2023). https://doi.org/10.1155/2023/4853800
Parlett-Pelleriti, C.M.; Stevens, E.; Dixon, D., et al.: Applications of unsupervised machine learning in autism spectrum disorder research: a review. Rev. J. Autism Dev. Disord. 10, 406–421 (2023). https://doi.org/10.1007/s40489-021-00299-y
Eslami, T.; Mirjalili, V.; Fong, A.; Laird, A.R.; Saeed, F.: ASD-DiagNet: a hybrid learning approach for detection of autism spectrum disorder using fMRI data. Front. Neuroinform. 13, 70 (2019)
Byeon, K.; Kwon, J.; Hong, J.; Park, H.: Artificial neural network inspired by neuroimaging connectivity: application in autism spectrum disorder. In2020 IEEE International Conference on Big Data and Smart Computing (BigComp) 2020 Feb 19 (pp. 575–578). IEEE
Dvornek, N.C.; Ventola, P.; Pelphrey, K.A.; Duncan, J.S.: Identifying autism from resting-state fMRI using long short-term memory networks. InMachine Learning in Medical Imaging: 8th International Workshop, MLMI 2017, Held in Conjunction with MICCAI 2017, Quebec City, QC, Canada, September 10, 2017, Proceedings 8 2017 (pp. 362–370). Springer International Publishing.
Bayram, M.A.; İlyas, Ö.Z.; Temurtaş, F.: Deep learning methods for autism spectrum disorder diagnosis based on fMRI images. Sakarya Univ. J. Comput. Inf. Sci. 4(1), 142–155 (2021)
Niu, K.; Guo, J.; Pan, Y.; Gao, X.; Peng, X.; Li, N.; Li, H.: Multichannel deep attention neural networks for the classification of autism spectrum disorder using neuroimaging and personal characteristic data. Complexity 2020, 1–9 (2020)
Leming, M.; Górriz, J.M.; Suckling, J.: Ensemble deep learning on large, mixed-site fMRI datasets in autism and other tasks. Int. J. Neural Syst. 30(07), 2050012 (2020)
Wen, G.; Cao, P.; Bao, H.; Yang, W.; Zheng, T.; Zaiane, O.: MVS-GCN: A prior brain structure learning-guided multi-view graph convolution network for autism spectrum disorder diagnosis. Comput. Biol. Med. 142, 105239 (2022)
Jiang, H.; Cao, P.; Xu, M.; Yang, J.; Zaiane, O.: Hi-GCN: A hierarchical graph convolution network for graph embedding learning of brain network and brain disorders prediction. Comput. Biol. Med. 127, 104096 (2020)
Hiremath, Y.; Ismail, M.; Verma, R.; Antunes, J.; Tiwari, P.: Combining deep and hand-crafted MRI features for identifying sex-specific differences in autism spectrum disorder versus controls. InMedical Imaging 2020: Computer-Aided Diagnosis 2020 Mar 16 (Vol. 11314, pp. 445–451). SPIE.
Al-Hiyali, M.I.; Yahya, N.; Faye, I.; Khan, Z.: Autism spectrum disorder detection based on wavelet transform of bold FMRI signals using pre-trained convolution neural network. Int. J. Int. Eng. 13(5), 49–56 (2021)
Ahammed, M.S.; Niu, S.; Ahmed, M.R.; Dong, J.; Gao, X.; Chen, Y.: Darkasdnet: Classification of ASD on functional MRI using deep neural network. Front. Neuroinform. 15, 635657 (2021)
Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; Deen, B.: The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psych. 19(6), 659–667 (2014)
Nair, S.; Jao Keehn, R.J.; Berkebile, M.M.; Maximo, J.O.; Witkowska, N.; Müller, R.A.: Local resting state functional connectivity in autism: site and cohort variability and the effect of eye status. Brain Imag. Behav. 12(1), 168–179 (2018)
Power, J.D.; Mitra, A.; Laumann, T.O.; Snyder, A.Z.; Schlaggar, B.L.; Petersen, S.E.: Methods to detect, characterize, and remove motion artifact in resting state FMRI. Neuroimage 84, 320–341 (2014)
Cox, R.W.: AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29(3), 162–173 (1996)
Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.; Woolrich, M.W.; Smith, S.M.: Fsl. Neuroimage. 62(2), 782–790 (2012)
Teipel, S.J.; Wohlert, A.; Metzger, C.; Grimmer, T.; Sorg, C.; Ewers, M.; Meisenzahl, E.; Klöppel, S.; Borchardt, V.; Grothe, M.J.; Walter, M.: Multicenter stability of resting state FMRI in the detection of Alzheimer’s disease and amnestic MCI. NeuroImage Clin. 14, 183–194 (2017)
Kubanek, D.; Freeborn, T.; Koton, J.; Herencsar, N.: Evaluation of (1+ α) fractional-order approximated Butterworth high-pass and band-pass filter transfer functions. Elektronika ir Elektrotechnika. 24(2), 37–41 (2018)
Satterthwaite, T.D.; Elliott, M.A.; Gerraty, R.T.; Ruparel, K.; Loughead, J.; Calkins, M.E.; Eickhoff, S.B.; Hakonarson, H.; Gur, R.C.; Gur, R.E.; Wolf, D.H.: An improved framework for confound regression and filtering for control of motion artifact in the preprocessing of resting-state functional connectivity data. Neuroimage 64, 240–256 (2013)
Gordon, E.M.; Laumann, T.O.; Adeyemo, B.; Huckins, J.F.; Kelley, W.M.; Petersen, S.E.: Generation and evaluation of a cortical area parcellation from resting-state correlations. Cereb. Cortex 26(1), 288–303 (2016)
Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; Albert, M.S.: An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31(3), 968–980 (2006)
Diedrichsen, J.; Balsters, J.H.; Flavell, J.; Cussans, E.; Ramnani, N.: A probabilistic MR atlas of the human cerebellum. Neuroimage 46(1), 39–46 (2009)
Nair, A.; Carper, R.A.; Abbott, A.E.; Chen, C.P.; Solders, S.; Nakutin, S.; Datko, M.C.; Fishman, I.; Müller, R.A.: Regional specificity of aberrant thalamocortical connectivity in autism. Hum. Brain Mapp. 36(11), 4497–4511 (2015)
Reiter, M.A.; Mash, L.E.; Linke, A.C.; Fong, C.H.; Fishman, I.; Müller, R.A.: Distinct patterns of atypical functional connectivity in lower-functioning autism. Biol Psych. Cogn. Neurosci. Neuroimag. 4(3), 251–259 (2019)
Kotu, V.; Deshpande, B.: Data science: concepts and practice. Morgan Kaufmann; 2018 Nov 27.
Song, C.; Havlin, S.; Makse, H.A.: Origins of fractality in the growth of complex networks. Nat. Phys. 2(4), 275–281 (2006)
Achard, S.; Bassett, D.S.; Meyer-Lindenberg, A.; Bullmore, E.: Fractal connectivity of long-memory networks. Phys. Rev. E 77(3), 036104 (2008)
Baillie, R.T.; Kapetanios, G.: On the estimation of short memory components in long memory time series models. Stud. Nonlinear Dyn. Econom. 20(4), 365–375 (2016)
Pradhan, A.; Srivastava, S.: Hierarchical extreme puzzle learning machine-based emotion recognition using multimodal physiological signals. Biomed. Signal Process. Control 83, 104624 (2023)
Kazeminejad, A.; Sotero, R.C.: Topological properties of resting-state fMRI functional networks improve machine learning-based autism classification. Front. Neurosci. 12, 1018 (2019)
Haghighat, H.; Mirzarezaee, M.; Araabi, B.N.; Khadem, A.: An age-dependent Connectivity-based computer aided diagnosis system for autism spectrum disorder using resting-state fMRI. Biomed. Signal Process. Control 71, 103108 (2022)
Vigneshwaran, S.; Suresh, S.; Mahanand, B.S.; Sundararajan, N.: ASD detection in males using MRI-an age-group based study. In2015 International Joint Conference on Neural Networks (IJCNN) 2015 Jul 12 (pp. 1–8). IEEE.
Haweel, R.; Dekhil, O.; Shalaby, A.; Mahmoud, A.; Ghazal, M.; Keynton, R.; Barnes, G.; El-Baz, A.: A machine learning approach for grading autism severity levels using task-based functional MRI. In2019 IEEE International Conference on Imaging Systems and Techniques (IST) 2019 Dec 9 (pp. 1–5). IEEE.
Haweel, R.; Shalaby, A.M.; Mahmoud, A.H.; Ghazal, M.; Seada, N.; Ghoniemy, S.; Casanova, M.; Barnes, G.N.; El-Baz, A.: A novel grading system for autism severity level using task-based functional MRI: a response to speech study. IEEE Access 9, 100570–100582 (2021)
Nomi, J.S.; Uddin, L.Q.: Developmental changes in large-scale network connectivity in autism. NeuroImage Clin. 7, 32–41 (2015)
Funding
This work was supported by the Science and Engineering Research Board through the Start-up Research Grant (SRG) scheme (SRG/2021/002289).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, V., Rakshe, C.T., Sengar, S.S. et al. Age- and Severity-Specific Deep Learning Models for Autism Spectrum Disorder Classification Using Functional Connectivity Measures. Arab J Sci Eng 49, 6847–6865 (2024). https://doi.org/10.1007/s13369-023-08560-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13369-023-08560-8