Abstract
Leaf beetles (Coleoptera: Chrysomelidae) constitute one of the most species-rich insect families, and live exclusively on leaves or other plant parts. Early histological works described the presence of symbiotic bacteria in gut-associated symbiotic organs of some chrysomelid species, but their microbiological nature has been poorly characterized except for those associated with reed beetles of the subfamily Donaciinae. Here we investigated symbiotic bacteria of the leaf beetle Bromius obscurus (L.) belonging to the subfamily Eumolpinae. Specific bacterial 16S ribosomal RNA and gyrB gene sequences were consistently obtained from the symbiotic organs, which radially surround the foregut-midgut junction, of all adult males and females examined. In adult females, the same sequences were also obtained from a pair of genital accessory organs, which are presumably for vertical symbiont transmission. Whole mount in situ hybridization specifically detected the symbiont in the gut symbiotic organs endocellularly and also in the female genital accessory organs extracellularly. In the gut symbiotic organs, the endocellular symbiont cells were small and rosette-like or aggregated and granule-like, whereas in the female genital organs the extracellular symbiont cells were of a condensed form. Molecular phylogenetic analysis showed that the symbiont of B. obscurus constitutes a distinct lineage in the Gammaproteobacteria. Molecular evolutionary analysis has identified significantly accelerated molecular evolution and a highly adenine–thymine-biased nucleotide composition of the symbiont genes, presumably reflecting reductive evolution of the symbiont genome. These results suggest an intimate and stable host-symbiont association in B. obscurus, in which the symbiont may play some important, though hitherto unknown, biological roles in its herbivorous insect host.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf beetles (Coleoptera: Chrysomelidae), with some 50,000 described species, constitute one of the most species-rich insect groups in the world. Both adults and larvae of chrysomelids exclusively feed on leaves and other parts of various plants, and consequently, many species are recognized as notorious agricultural pests including the Colorado potato beetle Leptinotarsa decemlineata (Say) and the Western corn rootworm Diabrotica virgifera LeConte (Gray et al. 2009; Hare 1990; Hunt et al. 2007; Jolivet et al. 1988, 1994; Jolivet and Cox 1996).
Many insects are associated with symbiotic microorganisms, which confer either beneficial or detrimental effects on their host’s biology such as nutrient provisioning (Douglas 2009; Moran et al. 2008), food production (Currie 2001; Klepzig and Six 2004), food digestion (Brune 2014; Ohkuma 2003), defense against natural enemies (Clay 2014; Oliver et al. 2014), tolerance to environmental stresses (Montllor et al. 2001; Rodriguez et al. 2008), modifying the food plant range (Hosokawa et al. 2007; Tsuchida et al. 2004), detoxifying noxious chemicals (Bosch and Welte 2016; Kikuchi et al. 2012), manipulating sex differentiation and reproduction (Hurst and Frost 2015; Werren et al. 2008), etc. Previous studies have identified a variety of facultative microbial associates of leaf beetles such as Wolbachia, Arsenophonus and Cardinium, and other diverse assemblage of gut and external bacteria (Clark et al. 2001; Chung et al. 2013; Keller et al. 2004; Kelley and Dobler 2011; Kondo et al. 2002, 2011; Krawczyk et al. 2015; Montagna et al. 2015; Muratoglu et al. 2011; Roehrdanz et al. 2006; Roehrdanz and Levine 2007; Roehrdanz and Wichmann 2013; Takano et al. 2017; Werren et al. 1995), although their biological effects on their chrysomelid hosts have been poorly characterized in general, except for some Wolbachia and other bacterial strains interfering with the reproduction of their host insects by causing cytoplasmic incompatibility (Keller et al. 2004; Kondo et al. 2002; Takano et al. 2017) and several gut bacteria of L. decemlineata suppressing plant defense responses induced by the insect’s infestation (Chung et al. 2013). Notably, early histological works in the 1930s described conspicuous gut-associated symbiotic organs and dense bacterial populations therein in several groups of the Chrysomelidae: reed beetles of the subfamily Donaciinae, tortoise beetles of the subfamily Cassidinae, and brown-and-black beetles of the subfamily Eumolpinae (Stammer 1935, 1936). However, the microbiological nature of these symbiotic bacteria has been poorly characterized for decades, except for those associated with reed beetles of the subfamily Donaciinae (Kleinschmidt and Kölsch 2011; Kölsch et al. 2009; Kölsch and Pedersen 2010).
The leaf beetle Bromius obscurus (Coleoptera: Chrysomelidae), known as the Western grape rootworm, infests the grapevine Vitis vinifera L. (Peacock 1992) and also lives on wild plants such as the fireweed Chamerion angustifolium (L.). It is widely distributed across the world, including North America, Europe, Russia, China, Korea, the Kuril Islands and Japan (Gruev 2004; Jolivet and Verma 2008; Xing-peng and Cheng-de 2007). Adult beetles feed on leaf tissue of grapevines by cutting distinctive slits of about 1-mm width, while larvae develop in soil and feed on small rootlets (Peacock 1992). In Japan, B. obscurus lives on the fireweed, the fuki Petasites japonicus (Siebold et. Zucc.) Maxim., the wild grape Ampelopsis glandulosa var. brevipedunculata (Wall.), and the aspen Populus tremula var. sieboldii (Miq.) (Ozono 2014). An early histological study described rosette- or granule-shaped symbiotic bacteria densely populating gut- or oviduct-associated symbiotic organs of B. obscurus (Stammer 1936). Recently, a bacterial 16S ribosomal RNA (rRNA) sequence was reported from a German specimen of B. obscurus (Kölsch and Synefiaridou 2012). In this study, in an attempt to integrate the old histological description into the recent molecular information, we characterized the symbiotic bacteria of B. obscurus collected in Japan using molecular phylogenetic, evolutionary and histological techniques.
Materials and methods
Sampling and DNA extraction
Adult insects of B. obscurus were collected from the fireweed Chamerion angustifolium (L.) at Manza-Kogen, Gumma, Japan, in July 2016 (Fig. 1a). We also attempted to collect larvae of B. obscurus from the rhizosphere of C. angustifolium, but in vain. The insects were preserved in an ultracold freezer at −80 °C until use. Gut symbiotic organs (Fig. 1b–d) and paired genital accessory organs (Fig. 1e) were dissected in 70% ethanol using fine tweezers. The isolated tissues were individually subjected to DNA extraction using QIAamp DNA Mini Kit or QIAamp DNA Micro Kit (Qiagen).
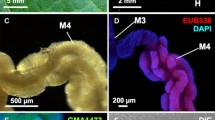
Symbiotic system of Bromius obscurus. a Adult insect. b Dissected alimentary tract of an adult female. Symbiotic organ consisting of finger-like lobes at the foregut (FG). Midgut (MG) junction is indicated by an arrow. c Enlarged image of the symbiotic organ. Arrows indicate the finger-like lobes. d Dissected FG-MG junction, to which each finger-like lobe is connected via a thin duct (arrowheads). e Enlarged image of the hindgut (HG)-ovary (OV) region, where female-specific genital accessory organs (arrowheads) are seen. f–l, n, o Symbiont localization visualized by whole-mount fluorescence in situ hybridization using the symbiont-specific probe Bro717. f Symbiont localization in the dissected alimentary tract of an adult female. Symbiont signals (red) are seen in the gut-associated symbiotic organs (arrow) and the female-specific genital accessory organs (arrowheads). g Symbiont localization in a dissected lobe of the gut-associated symbiotic organ. h–j Negative controls. h No-probe control. i Competition control. j RNase control. k Symbiont cells in the gut-associated symbiotic organ of an adult female. Note the peculiar rosette-like bacterial morphology within the host cytoplasm. l Symbiont cells in the gut-associated symbiotic organ of a different adult female. Note the peculiar aggregated granule-like bacterial morphology within the host cytoplasm. m Symbiont localization visualized by whole-mount fluorescence in situ hybridization using the general bacterial probe EUB338. n Symbiont localization in the female-specific genital accessory organ. o Symbiont cells in the inner cavity of the genital accessory organ of an adult female. Note that the extracellular symbiont cells are small and condensed in shape. g–o Symbiont 16S ribosomal RNA (rRNA) is visualized in red and DNA is stained in blue
DNA sequencing
Bacterial genes were amplified by polymerase chain reaction (PCR) using ExTaq DNA polymerase (Takara Bio) with the primers 16SA1 (5′-AGA GTT TGA TCM TGG CTC AG-3′) (Fukatsu and Nikoh 1998) and 1507R (5′′-TAC CTT GTT ACG ACT TCA CCC CAG-3′) (Sandström et al. 2001) for the 16S rRNA gene, and gyrBsymF (5′-TTA TCA TGA CWG TAT TAC ATG CWG G-3′) (Hosokawa et al. 2010) and gyrBsymR (5′-TCC AGC WGA ATC WCC TTC WAC-3′) (Hosokawa et al. 2010) for the gyrB gene. After checking successful amplification by electrophoresis on 1% agarose gels, each PCR product was purified using exonuclease I (New England Biolabs) and shrimp alkaline phosphatase (Takara Bio) at 37 °C for 15 min followed by 80 °C for 15 min. The purified PCR products were directly subjected to a sequencing reaction using BigDye Terminator version 3.1 Cycle Sequencing Kit (Applied Biosystems) and analyzed by a 3130xl Genetic Analyzer (Applied Biosystems). The internal primer 16SA2 (5′-GTG CCA GCA GCC GCG GTA ATA C-3′) (Fukatsu and Nikoh 1998) was also used for sequencing of bacterial 16S rRNA gene.
Molecular phylogenetic and evolutionary analysis
Multiple alignments of the nucleotide sequences were generated using the program Clustal W (Thompson et al. 1994). The alignments were then inspected and corrected manually, and ambiguously aligned sites were removed. Phylogenetic analyses were conducted by the Bayesian and maximum-likelihood (ML) methods. The best-fit substitution model for each of the aligned sequences was selected using the program KAKUSAN version 4 (Tanabe 2011) on the basis of the Bayesian information criterion for the Bayesian analysis and Akaike’s information criterion for the ML analysis. Bayesian phylogenies were constructed using the program MrBayes version 3.2.6 (Ronquist et al. 2012) with substitution models selected by BIC4 (16S rRNA, GTR + G; gyrB, proportional model among codons, GTR + G for the first codon, HKY85 + G for the second codon, and GTR + G for the third codon). Two simultaneous, independent runs of the Metropolis-coupled Markov chain Monte Carlo (MCMC) method were performed for one million generations with trees sampled every 100th generation. Convergence of the MCMC procedure was assessed from effective sample size scores (all >100) using MrBayes and Tracer version 1.6 (Rambaut et al. 2014). The first 25% of the trees were discarded as burn-in, and the remaining trees were used to calculate the 50% majority rule consensus and to determine posterior probabilities for individual branches. ML phylogenies were inferred using the software Treefinder version March 2011 (Jobb 2011) based on models selected by AICc4 (16S rRNA, GTR + G; gyrB: separate model among codons, GTR + G for the first codon, GTR + G for the second codon, and GTR + G + I for the third codon). Bootstrap probabilities were determined by 100 replications. Relative rate tests were performed by the program RRTree (Robinson-Rechavi and Huchon 2000).
Histological procedures
Whole-mount fluorescence in situ hybridization targeting bacterial 16S rRNA was performed essentially as described previously (Koga et al. 2009). Dissected symbiotic organs were fixed in Carnoy’s solution (60% ethanol, 30% chloroform, 10% acetic acid) for 15 min and washed three times in 70% ethanol at room temperature. The dissected tissues were then washed three times in phosphate-buffered saline–Triton X (PBS–TX; 0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4, 0.3% Triton X-100) and washed twice in a hybridization buffer [20 mM TRIS–HCl (pH 8.0), 0.9 M NaCl, 0.01% SDS, 30% formamide]. For specifically targeting 16S rRNA of the symbiont of B. obscurus, the oligonucleotide probe Bro717 (5′-GTC GCT TTC GCC TCT GGT AT-3′) was labeled with the fluorochrome Alexa555 at the 5′-terminus. For universal detection of bacterial 16S rRNA, the probe EUB338 (5′-GCT GCC TCC CGT AGG AGT-3′) (Amann et al. 1990) whose 5′-terminus is labeled with Alexa555 was also used. The tissues were incubated in the hybridization buffer containing 50 nM of the probe and 4.5 µM of 4′,6-diamidino-2-phenylindole (Invitrogen) overnight at room temperature, washed thoroughly in PBS–TX, mounted in Slowfade antifade solution (Molecular Probes), and observed under an epifluorescence microscope (Axiophot; Carl Zeiss) and a laser scanning confocal microscope (LSCM Pascal5; Carl Zeiss). In situ hybridization controls included (1) no probe control (Fig. 1h), (2) competition control in which unlabeled Bro717 was added to hybridization buffer in excess to suppress the hybridization signals (Fig. 1i), (3) RNase digestion control in which the tissue samples were treated with RNase A prior to hybridization (Fig. 1j), and (4) a positive control with the general bacterial probe EUB338 (Fig. 1m).
For clearly visualizing the cellular construction of the symbiotic organs, dissected tissues were fixed in 4% paraformaldehyde in PBS (0.8% NaCl, 0.02% KCl, 0.115% Na2HPO4, 0.02% KH2PO4), washed twice in PBS, stained with Alexa Fluor 488 phalloidin (Molecular Probes) for visualizing cytoplasmic actin filaments, and counterstained with 4′,6-diamino-2-phenylindole for visualizing nuclear DNA.
Results and discussion
Bacterial 16S rRNA and gyrB gene sequences from gut symbiotic organs of B. obscurus
For all six males and three females we examined, the same 16S rRNA gene sequences, 1412 bp in size with a highly adenine–thymine (AT)-biased nucleotide composition at 56.4% (sequence accession number LC273302), were consistently obtained from the dissected finger-shaped organs radially surrounding the foregut-midgut junction (Fig. 1b–d, arrows), which were previously described as the main locations of the bacterial symbiont in B. obscurus (Stammer 1936). In a previous study (Kölsch and Synefiaridou 2012), a bacterial 16S rRNA gene sequence, 1090 bp in size with AT-biased nucleotide composition at 55.4% (accession number JQ805030), was reported from a specimen of B. obscurus collected in Germany. All 1070 nucleotide sites of the 1070-bp overlapping region between the sequences were identical. Furthermore, for all six males and three females we examined, the same gyrB gene sequences, 897 bp in size with extremely AT-biased nucleotide composition at 79.9% (sequence accession number LC273303), were consistently obtained from the dissected gut symbiotic organs. No other bacterial gene sequences were detected from the dissected organs, indicating that the specific bacterial species dominates the symbiotic organs although the possibility of coexisting minor bacterial associates cannot be ruled out. These results indicate that:
-
1.
A specific bacterial symbiont is associated with the gut symbiotic organ of B. obscurus.
-
2.
The same symbiont is fixed within the Japanese population and conserved across the Japanese and European populations, suggesting an intimate and stable host-symbiont association in B. obscurus.
-
3.
Judging from the extremely AT-biased nucleotide compositions of the symbiont genes, the symbiont genome might have experienced reductive genome evolution typical of long-lasting obligate endosymbiotic bacteria (Wernegreen 2002; Moran et al. 2008).
Localization of the bacterial symbiont in B. obscurus
Alimentary tract samples of two males and two females of B. obscurus were dissected, fixed and subjected to whole-mount fluorescence in situ hybridization using a specific probe targeting 16S rRNA of the bacterial symbiont. In all the samples, strong hybridization signals were detected in the gut symbiotic organs surrounding the foregut-midgut junction (Fig. 1f, arrow), confirming the principal symbiont location in vivo. In addition, specifically in the female samples, a pair of slender structures associated with the reproductive system exhibited remarkable, though less intense, hybridization signals (Fig. 1f, arrowheads). The delicate club-shaped organs were connected to the common oviduct and regarded as types of female-specific genital accessory organs (Fig. 1e), confirming the early histological observation of bacterial colonization in the female-specific organs of B. obscurus (Stammer 1936). PCR amplification and sequencing of bacterial 16S rRNA and gyrB genes from the accessory organs identified 1412- and 897-bp sequences, respectively, which were identical to those from the gut symbiotic organs. Close cytological inspection revealed that the symbiont cells are present in the cytoplasm of the gut symbiotic organs endocellularly (Fig. 1g, k, l), whereas the symbiont cells are localized in the inner cavity of the female genital accessory organs extracellularly (Fig. 1n, o). Each lobe of the symbiotic organ has a narrow inner cavity and is connected to the foregut-midgut junction through a thin duct (Fig. 2a–c). These observations suggest that:
-
1.
The gut symbiotic organs are the main symbiont location, where the substantial bacterial population plays some endocellular, though hitherto unknown, biological roles.
-
2.
The female genital accessory organs are, considering the anatomical configuration and the extracellular symbiont accumulation, likely to function as a symbiont transmission mechanism, which delivers a symbiont-containing secretion onto eggs upon oviposition, as observed in other symbiotic leaf beetles of the Donaciinae and Cassidinae (Stammer 1935, 1936).
-
3.
The same bacterial symbiont experiences both endocellular and extracellular conditions during the infection cycle in B. obscurus.
Fig. 2a–c Histological configuration of the finger-like lobe of the gut-associated symbiotic organ. a Dissected finger-like lobe connected to the foregut-midgut junction via a duct (arrow). b Enlarged image of the lobe-duct junction. c Cross section of the lobe-duct junction, in which a narrow inner cavity is seen at the center. In the confocal optical section images, green signals indicate cytoplasmic actin filaments, blue signals nuclear DNA
Morphological variation of the symbiont cells
In an early histological study, Stammer (1936) describes drastic morphological changes in the symbiont cells during the development of B. obscurus: in the gut symbiotic organs of the larvae, each symbiont cell is well developed and petal- or rosette-like, whereas in the gut symbiotic organs of adults, each symbiont cell is less developed and granule-like. In this study, we could not inspect larval samples, unfortunately, but we recognized conspicuous pleomorphism of the symbiont cells across tissues and samples of adult insects. In the gut symbiotic organs, the endocellular symbiont cells were small and rosette-like (Fig. 1k) or aggregated granule-like (Fig. 1l), whereas in the female genital accessory organs, the extracellular symbiont cells were of a more condensed form (Fig. 1o). These observations suggest a pleomorphic nature of the bacterial symbiont depending on the symbiotic conditions. The rosette-like amorphous shape of the symbiont cells is of particular interest in that:
-
1.
Such amorphous bacterial cells have been observed in diverse endosymbiotic bacteria of insects (Buchner 1965 for review; Buchner 1954; Fink 1952; Koch 1931 for specific examples).
-
2.
Similar amorphous bacterial cells have been often observed in endoparasitic/pathogenic bacteria with reduced genomes like Mycoplasma spp. (Allan et al. 2009).
-
3.
Such amorphous bacterial morphology, the so-called L-form, has been reported to occur when genes involved in cell wall synthesis and/or cell division are disrupted in bacteria (Leaver et al. 2009; Mercier et al. 2016).
-
4.
Therefore, the rosette-like amorphous shape of the symbiont cells may be relevant to the presumed symbiont genome erosion.
Evolution of the bacterial symbiont of B. obscurus
Molecular phylogenetic analysis based on the 16S rRNA gene sequences placed the bacterial symbionts of B. obscurus representing the Japanese and German populations as a distinct lineage in the Gammaproteobacteria (Fig. 3). The branch length of the symbiont lineage was remarkably elongated, suggesting an accelerated rate of molecular evolution. The relative rate test based on the 16S rRNA gene sequences confirmed significantly accelerated molecular evolution in the symbiont lineage: significantly and drastically faster than that of the free-living Escherichia coli, and even significantly faster than that of the genome-reduced aphid endosymbiont Buchnera aphidicola (Table 1). On the phylogenetic tree, the symbiont sequence from B. obscurus did not cluster with previously reported symbiont sequences from other chrysomelid leaf beetles such as Sagra femorata (Sagrinae), Macroplea mutica, Macroplea appendiculata, and Donacia semicuprea (Donaciinae) (Fig. 3). Molecular phylogenetic and evolutionary analyses based on the gyrB gene sequences exhibited similar evolutionary patterns (Fig. 4; Table 1). These results suggest independent evolutionary origins of these symbiotic bacteria in the Chrysomelidae.
Phylogenetic relationship of the symbiont of B. obscurus with representative Gammaproteobacteria based on 16S rRNA gene sequences. A Bayesian phylogeny inferred from 1441 aligned nucleotide sites is shown. Posterior probability values of the Bayesian analysis and bootstrap probability values of the maximum-likelihood analysis are depicted above and below each node (minus sign bootstrap values less than 50%), respectively. Sequence accession numbers are in brackets, followed by adenine–thymine content percentages. GS Gut symbiont, BS bacteriocyte symbiont, SS secondary symbiont
Phylogenetic relationship of the symbiont of B. obscurus with representative Gammaproteobacteria based on gyrB gene sequences. Bayesian phylogeny inferred from 906 aligned nucleotide sites is shown, as in Fig. 3
Conclusion and perspective
Here we unequivocally characterized the specific bacterial symbiont of the leaf beetle B. obscurus using molecular phylogenetic, evolutionary and histological approaches, thereby integrating the old histological description (Stammer 1936) and recent molecular information (Kölsch and Synefiaridou 2012) into a coherent picture. However, a number of biological aspects of the chrysomelid-bacterium symbiosis deserve further study. More detailed in vivo localization and infection dynamics of the symbiont in the life cycle of B. obscurus, in particular the vertical transmission process to the offspring and colonization process of the symbiotic organs, should be clarified using modern histological techniques. Considering the accelerated molecular evolution and the AT-biased nucleotide compositions of the symbiont genes, sequencing of the symbiont genome will provide clues to understanding the detailed process of reductive genome evolution and also presumable biological functions that the reduced genome is specialized for. A symbiont survey of other leaf beetles of the subfamily Eumolpinae, to which B. obscurus belongs, should lead to further symbiont discoveries and shed light on the host-symbiont coevolution in this insect group. The establishment of a rearing system for B. obscurus, the generation of aposymbiotic insects using antibiotic curing, and a fitness evaluation and physiological analysis of the insects are pivotal for understanding hitherto unknown symbiont functions in the leaf-eating insect group Chrysomelidae. Considering that many chrysomelids are notorious agricultural pests (Jolivet and Cox 1996; Jolivet et al. 1988) and some symbionts are involved in such pest-related traits as plant adaptation (Tsuchida et al. 2004), crop exploitation (Hosokawa et al. 2007) and pesticide resistance (Kikuchi et al. 2012), understanding symbiont roles in the Chrysomelidae would contribute to the control and management of these pest insects (Chung et al. 2013).
References
Allan EJ, Hoischen C, Gumpert J (2009) Chapter 1. Bacterial L-forms. Adv Appl Microbiol 68:1–39
Amann RI, Krumholz L, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770. doi:10.1128/jb.172.2.762-770.1990
Brune A (2014) Symbiotic digestion of lignocellulose in termite guts. Nat Rev Microbiol 12:168–180. doi:10.1038/nrmicro3182
Buchner P (1954) Studien an intrazellularen Symbionten. VIII. Die symbiontischen Einrichtungen der Bostrychiden (Apatiden). Z Morphol Ökol Tiere 42:550–633
Buchner P (1965) Endosymbiosis of animals with plant microorganims. Wiley, New York
Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW (2013) Herbivore exploits orally secreted bacteria to suppress plant defenses. Proc Natl Acad Sci USA 110:15728–15733. doi:10.1073/pnas.1308867110
Clark TL, Meinke LJ, Skoda SR, Foster JE (2001) Occurrence of Wolbachia in selected diabroticite (Coleoptera: Chrysomelidae) beetles. Ann Entomol Soc Am 94:877–885. doi:10.1603/0013-8746(2001)094[0877:OOWISD]2.0.CO;2
Clay K (2014) Defensive symbiosis: a microbial perspective. Funct Ecol 28:293–298. doi:10.1111/1365-2435.12258
Currie CR (2001) A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol 55:357–380. doi:10.1146/annurev.micro.55.1.357
Douglas AE (2009) The microbial dimension in insect nutritional ecology. Funct Ecol 23:38–47. doi:10.1111/j.1365-2435.2008.01442.x
Fink R (1952) Morphologische und physiologische Untersuchungen an den intrazellularen Symbionten von Pseudococcus citri Risso. Z Morphol Ökol Tiere 41:78–146
Fukatsu T, Nikoh N (1998) Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera). Appl Environ Microbiol 64:3599–3606
Gray ME, Sappington TW, Miller NJ, Moeser J, Bohn MO (2009) Adaptation and invasiveness of Western corn rootworm: intensifying research on a worsening pest. Annu Rev Entomol 54:303–321. doi:10.1146/annurev.ento.54.110807.090434
Gruev BA (2004) The leaf beetles (Insecta: Coleoptera: Chrysomeridae) of the Sredna Gora Mountains (Bulgaria). Fauna and zoogeography. Trav Sci Univ Plovdiv Animalia 40:77–96
Hare JD (1990) Ecology and management of the Colorado potato beetle. Annu Rev Entomol 35:81–100
Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T (2007) Obligate symbiont involved in pest status of host insect. Proc R Soc B 274:1979–1984. doi:10.1098/rspb.2007.0620
Hosokawa T, Kikuchi Y, Nikoh N, Meng X-Y, Hironaka M, Fukatsu T (2010) Phylogenetic position and peculiar genetic traits of a midgut bacterial symbiont of the stinkbug Parastrachia japonensis. Appl Environ Microbiol 76:4130–4135. doi:10.1128/AEM.00616-10
Hunt T, Bergsten J, Levkanicova Z, Papadopoulou A et al (2007) A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 318:1913–1916. doi:10.1126/science.1146954
Hurst GDD, Frost CL (2015) Reproductive parasitism: maternally inherited symbionts in a biparental world. Cold Spring Harbor Perspect Biol. doi:10.1101/cshperspect.a017699
Jobb G (2011) Treefinder, version March 2011. http://www.treefinder.de. Accessed 3 Mar 2017
Jolivet PHA, Cox ML (1996) Chrysomelidae biology. Kugler, Amsterdam
Jolivet P, Verma KK (2008) Eumolpinae—a widely distributed and much diversified subfamily of leaf beetles (Coleoptera, Chrysomelidae). Terr Arthropod Rev 1:3–37. doi:10.1163/187498308X345424
Jolivet P, Petitpierre E, Hsiao TH (eds) (1988) Biology of Chrysomelidae. Series Entomologica, vol 42. Springer, Netherlands
Jolivet PH, Cox ML, Petitpierre E (eds) (1994) Novel aspects of the biology of Chrysomelidae. Series Entomologica, vol 50. Springer, Netherlands
Keller GP, Windsor DM, Saucedo JM, Werrin JH (2004) Reproductive effects and geographical distributions of two Wolbachia strains infecting the Neotropical beetle, Chelymorpha alternans Boh. (Chrysomelidae, Cassidinae). Mol Ecol 13:2405–2420. doi:10.1111/j.1365-294X.2004.02213.x
Kelley ST, Dobler S (2011) Comparative analysis of microbial diversity in Longitarsus flea beetles (Coleoptera: Chrysomelidae). Genetica 139:541–550. doi:10.1007/s10709-010-9498-0
Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T (2012) Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA 109:8618–8622. doi:10.1073/pnas.1200231109
Kleinschmidt B, Kölsch G (2011) Adopting bacteria in order to adapt to water-how reed beetles colonized the wetlands (Coleoptera, Chrysomelidae, Donaciinae). Insects 2:540–554. doi:10.3390/insects2040540
Klepzig KD, Six DL (2004) Bark beetle-fungal symbiosis: context dependency in complex associations. Symbiosis 37:189–205
Koch A (1931) Die symbiose von Oryzaephilus surinamensis L. (Cucujidae, Coleoptera). Z Morphol Ökol Tiere 23:389–424
Koga R, Tsuchida T, Fukatsu T (2009) Quenching autofluorescence of insect tissues for in situ detecton of endosymbionts. Appl Entomol Zool 44:281–291. doi:10.1303/aez.2009.281
Kölsch G, Pedersen BV (2010) Can the tight co-speciation between reed beetles (Col., Chrysomelidae, Donaciinae) and their bacterial endosymbionts, which provide cocoon material, clarify the deeper phylogeny of the hosts? Mol Phylogenet Evol 54:810–821. doi:10.1016/j.ympev.2009.10.038
Kölsch G, Synefiaridou D (2012) Shared ancestry of symbionts? Sagrinae and Donaciinae (Coleoptera, Chrysomelidae) harbor similar bacteria. Insects 3:473–491. doi:10.3390/insects3020473
Kölsch G, Matz-Grund C, Pedersen BV (2009) Ultrastructural and molecular characterization of endosymbionts of the reed beetle genus Macroplea (Chrysomelidae, Donaciinae), and proposal of “Candidatus Macropleicola appendiculatae” and “Candidatus Macropleicola muticae”. Can J Microbiol 55:1250–1260. doi:10.1139/W09-085
Kondo N, Ijichi N, Shimada M, Fukatsu T (2002) Prevailing triple infection with Wolbachia in Callosobruchus chinensis (Coleoptera: Bruchidae). Mol Ecol 11:167–180. doi:10.1046/j.0962-1083.2001.01432.x
Kondo NI, Tuda M, Toquenaga Y, Lan Y-C, Buranapanichpan S, Horng S-B, Shimada M, Fukatsu T (2011) Wolbachia infections in world populations of bean beetles (Coleoptera: Chrysomelidae: Bruchinae) infesting cultivated and wild legumes. Zool Sci 28:501–508. doi:10.2108/zsj.28.501
Krawczyk K, Szymańczyk M, Obrępalska-Stęplowska A (2015) Prevalence of endosymbionts in Polish populations of Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). J Insect Sci 15:1–6. doi:10.1093/jisesa/iev085
Leaver M, Domínguez-Cuevas P, Coxhead JM, Daniel RA, Errington J (2009) Life without a wall or division machine in Bacillus subtilis. Nature 457:849–853. doi:10.1038/nature07742
Mercier R, Kawai Y, Errington J (2016) Wall proficient E. coli capable of sustained growth in the absence of the Z-ring division machine. Nature Microbiol 1:16091. doi:10.1038/NMICROBIOL.2016.91
Montagna M, Gómez-Zurita J, Giorgi A, Epis S, Lozzia G, Bandi C (2015) Metamicrobiomics in herbivore beetles of the genus Cryptocephalus (Chrysomelidae): toward the understanding of ecological determinants in insect symbiosis. Insect Sci 22:340–352. doi:10.1111/1744-7917.12143
Montllor CB, Maxmen A, Purcell AH (2001) Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol 27:189–195. doi:10.1046/j.1365-2311.2002.00393.x
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. doi:10.1146/annurev.genet.41.110306.130119
Muratoglu H, Demirbag Z, Sezen K (2011) The first investigation of the diversity of bacteria associated with Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Biologia 66:288–293. doi:10.2478/s11756-011-0021-6
Ohkuma M (2003) Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl Microbiol Biotechnol 61:1–9. doi:10.1007/s00253-002-1189-z
Oliver KM, Smith AH, Russell JA (2014) Defensive symbiosis in the real world—advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol 28:341–355. doi:10.1111/1365-2435.12133
Ozono A (2014) The handbook of the leaf beetle. Bun-ichi, Tokyo, Japan (in Japanese)
Peacock W (1992) Western grape rootworm. In: Flaherty DL (ed) Grape pest management. University of California Division of Agriculture and Natural Resources Publication 3343, 2nd edn, Oakland, pp 239–240
Rambaut A, Suchard MA, Xie W, Drummond AJ (2014) Tracer version 1.6. http://beast.bio.ed.ac.uk/Tracer. Accessed 3 Mar 2017
Robinson-Rechavi M, Huchon D (2000) RRTree: relative-rate tests between groups of sequences on a phylogenetic tree. Bioinformatics 16:296–297. doi:10.1093/bioinformatics/16.3.296
Rodriguez RJ, Henson J, Van Volkenburgh E et al (2008) Stress tolerance in plants via habitat-adapted symbiosis. ISME J 2:404–416. doi:10.1038/ismej.2007.106
Roehrdanz RL, Levine E (2007) Wolbachia bacterial infections linked to mitochondrial DNA reproductive isolation among populations of northern corn rootworm (Coleoptera: Chrysomelidae). Ann Entomol Soc Am 100:522–531. doi:10.1603/0013-8746(2007)100[522:WBILTM]2.0.CO;2
Roehrdanz RL, Wichmann SGS (2013) Wolbachia wsp gene clones detect the distribution of Wolbachia variants and wsp hypervariable regions among individuals of a multistrain infected population of Diabrotica barberi (Coleoptera: Chrysomelidae). Ann Entomol Soc Am 106:329–338. doi:10.1603/AN12118
Roehrdanz R, Olson D, Bourchier R, Sears S, Cortilet A, Fauske G (2006) Mitochondrial DNA diversity and Wolbachia infection in the flea beetle Aphthona nigriscutis (Coleoptera: Chrysomelidae): an introduced biocontrol agent for leafy spurge. Biol Control 37:1–8. doi:10.1016/j.biocontrol.2005.12.004
Ronquist F, Teslenko M, Mark P et al (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi:10.1093/sysbio/sys029
Sandström JP, Russell JA, White JP, Moran NA (2001) Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. doi:10.1046/j.1365-294X.2001.01189.x
Stammer HJ (1935) Studien an Symbiosen zwischen Käfern und Mikroorganismen. I. Die Symbiose der Donaciinen (Coleopt. Chrysomel.). Z Morphol Ökol Tiere 29:585–608
Stammer HJ (1936) Studien an Symbiosen zwischen Käfern und Mikroorganismen. II. Die Symbiose des Bromius obscurus L. und der Cassida-Arten (Coleopt. Chrysomel.). Z Morphol Ökol Tiere 30:682–697
Takano S, Tuda M, Takasu K, Furuya N, Imamura Y, Kim S, Tashiro K, Iiyama K, Tavares M, Amaral AC (2017) Unique clade of alphaproteobacterial endosymbionts induces complete cytoplasmic incompatibility in the coconut beetle. Proc Natl Acad Sci USA 114:6110–6115. doi:10.1073/pnas.1618094114
Tanabe AS (2011) Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional, and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour 11:914–921. doi:10.1111/j.1755-0998.2011.03021.x
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1093/nar/22.22.4673
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989. doi:10.1126/science.1094611
van den Bosch TJM, Welte CU (2016) Detoxifying symbionts in agriculturally important pest insects. Microbiol Biotechnol 10:531–540. doi:10.1111/1751-7915.12483
Wernegreen JJ (2002) Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet 3:850–861. doi:10.1038/nrg931
Werren JH, Windsor D, Guo L (1995) Distribution of Wolbachia among Neotropical arthropods. Proc R Soc B 262:197–204. doi:10.1098/rspb.1995.0196
Werren JH, Baldo L, Clark MK (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. doi:10.1038/nrmicro1969
Xing-peng L, Cheng-de L (2007) Checklist of Eumolpidae (Coleoptera, Chrysomeloidea) from Mao’ershan region including one new record species from China. J For Res 18:65–68. doi:10.1007/s11676-007-0012-1
Acknowledgements
We thank Hiroshi Ikeda for helping with the fieldwork and for advice on the phylogenetic analysis. This study was supported by a JSPS Postdoctoral Fellowship for Young Scientists to K. F., and also by JSPS Kakenhi grants to K. F. (no. 16J40021) and T. F. (no. JP25221107).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Fukumori, K., Koga, R., Nikoh, N. et al. Symbiotic bacteria associated with gut symbiotic organs and female genital accessory organs of the leaf beetle Bromius obscurus (Coleoptera: Chrysomelidae). Appl Entomol Zool 52, 589–598 (2017). https://doi.org/10.1007/s13355-017-0513-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13355-017-0513-0