Abstract
Cassava (Manihot esculenta Crantz) holds significant economic importance globally. Evaluating a diverse range of germplasm based on molecular characteristics not only enhances its preservation but also supports its utilization in breeding programs. In this study, we assessed genetic diversity and population structure among 155 cassava genotypes from Uganda using 5247 single nucleotide polymorphism (SNP) markers. Genotyping by sequencing (GBS) was employed for SNP discovery and to evaluate genetic diversity and population structure using the ADMIXTURE software. The cassava accessions comprised two populations: 49 accessions from Ugandan lines and 106 accessions resulting from crosses between South American and Ugandan lines. The average call rate of 96% was utilized to assess marker polymorphism. Polymorphic information content values of the markers ranged from 0.1 to 0.5 with an average of 0.4 which was moderately high. The principal component analysis (PCA) showed that the first two components captured ~ 24.2% of the genetic variation. The average genetic diversity was 0.3. The analysis of molecular variance (AMOVA) indicated that 66.02% and 33.98% of the total genetic variation occurred within accessions and between sub-populations, respectively. Five sub-populations were identified based on ADMIXTURE structure analysis (K = 5). Neighbor-joining tree and hierarchical clustering tree revealed the presence of three different groups which were primarily based on the source of the genotypes. The results suggested that there was considerable genetic variation among the cassava genotypes which is useful in cassava improvement and conservation efforts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cassava (Manihot esculenta Crantz) stands as one of the most important staple crops, providing sustenance and livelihood to millions of people globally, especially in tropical regions (Ceballos et al. 2020). This crop serves as the primary source of calories and income for small-scale farmers with its starchy roots providing carbohydrates and leaves offering vitamins, proteins, and minerals (Bayata 2019). Cassava’s unique ability to thrive in marginal ecologies with low soil fertility and rainfall makes it a crucial player in global agriculture, food security, and economic growth in such ecologies (Ngongo et al. 2022).
Advancements in breeding programs heavily rely on gaining a deeper understanding of the genetic diversity within populations which functions as a repository of diverse genes with significant potential for enhancing productivity and adapting to both abiotic and biotic stress (Adu et al. 2021). The relevance is particularly pronounced in the context of ongoing climate change and global warming as genetic variation serves as a foundation upon which breeding efforts are built, providing the raw material for developing improved varieties. Variations in traits such as yield, disease resistance, drought tolerance, and nutritional content are essential for enhancing cassava resilience and nutritional value, making them fundamental prerequisites for sustainable cassava cultivation, crop improvement, and conservation. The analysis of genetic diversity and population structure plays a crucial role in understanding the historical patterns of natural selection and the genetic connections among different accessions (Luo et al. 2019).
In cassava research, genetic diversity has been extensively explored through the utilization of diverse morphological and molecular markers, encompassing various germplasm accessions and traits. Morphological markers were used to study genetic diversity and relationships among elite cassava cultivars in Ghana (Sivan et al. 2023) and Benin (Agre et al. 2016). However, the use of morphological markers has been limited for breeding purposes due to its vulnerability to environmental influence and low polymorphism. In contrast, molecular markers represent efficient and precise tools for unveiling and estimating genetic diversity and ascertaining the population structure within plant populations (Pour-Aboughadareh et al. 2022).
Several molecular markers, such as SSR (Adjebeng-Danquah et al., 2020), DArT (Adu et al. 2021), and ISSR (Sandra et al. 2019), have been utilized to assess genetic variation in cassava accessions. SNP markers have also been employed to evaluate the diversity of whiteflies (Wosula et al. 2017), cassava mosaic begomovirus (Aimone et al. 2021), and genetic information within and between populations. In recent studies, SNP markers and SilicoDArt were used to identify 47% of distinct cassava genotypes represented by 87 accessions from the CIAT cassava GenBank (Carvajal-Yepes et al. 2023). A study aimed at determining diversity within local landraces in Burundi identified six clusters and pairs of duplicates (Pierre et al. 2022). Another study reported high diversity for CIAT germplasm, while low diversity was reported for IITA and East, South, and Central Africa (Ferguson et al. 2019a). The use of SNP markers was chosen to understand the genetic diversity within and between populations because of their abundance, stability, polymorphism, and compatibility with automation.
The study aims to address the historical challenges posed by Cassava Mosaic Disease (CMD) and Cassava Brown Disease (CBSD) to cassava production by utilizing the resistant cassava populations to elucidate the genetic characteristics, population structure, and gene flow patterns. Therefore, the objectives of this study are (1) to unravel allelic diversity and heterozygosity levels, (2) to explore population structure, (3) to detect potential admixture, and (4) to identify gene flow patterns. The ultimate goal includes contributing valuable information to cassava breeding programs in Uganda for the development of more resistant and disease-resistant varieties.
Materials and methods
Plant material and populations development
Two groups comprising a total of 155 cassava genotypes were used in the study. The first group consisted of 80 biparental genotypes derived from the parents MM06 128 and TME14, collected from existing germplasm at the National Agriculture Semi-Arid Research Resources Institute (NaSARRI) in Eastern Uganda. The second group included 75 biparental genotypes resulting from crosses between the parents COL40 and TME14 obtained from the International Institute of Tropical Agriculture (IITA) in Sendusu, Central Uganda. The parent COL40 originates from South America, TME14 from West Africa, and MM060128 from East Africa.
Genotyping
A total of 155 cassava leaf samples were collected from a single representative plant per accession. A representative plant was selected randomly from a plant with young top leaves, folded, and punched four leaf disks using a 5-mm puncher and placed in a 96 plate. The samples were oven dried, shipped in silica gel to Intertek, Australia Lab for DNA extraction. Genotyping-by-Sequencing and SNP calling were done for each sample using Diversity Array Technology sequencing (DArTseq™) genotyping platform at Bruce, ACT Australia (https://www.diversityarrays.com/technology-and-resources/dartreseq). This technology involves library construction through the DArTSeq complexity reduction method, which entails digestion of the genomic DNA using restriction enzymes followed by ligation of barcoded adaptors. The adaptor-ligated fragments were then amplified using polymerase chain reaction (PCR). The resultant PCR products of each sample were sequenced using Hiseq2500 (Illumina Inc. San Diego, CA, USA). The resultant identical sequences were collapsed into FASTQCOL from which the software package DArTsoft14 was used for markers discovery and scoring. The Single nucleotide polymorphism (SNP) markers were scored and converted to HapMap format after mapping them to the cassava (Manihot esculenta) reference genome v8.1 available in Phytozome (Goodstein et al. 2012).
Genotype data processing
The data in HapMap format was converted to variant call format (VCF) using TASSEL (Bradbury et al. 2007). The genotype data was filtered by removing SNPs with less than 80% call rate and less than 5% minor allele frequency (MAF) using VCFtools (Danecek et al. 2011). The filtered markers were used for subsequent analysis. The marker characteristics such as polymorphic information content (PIC), reproducibility, and call rate were determined in the dartR package of R (Gruber et al. 2018).
Population structure and diversity analysis of cassava genotypes
Population structure analysis and admixed ancestry were estimated using a model-based clustering method implemented in ADMIXTURE software (Alexander et al. 2009). To determine the actual number of populations, ten-fold cross-validation (CV) procedure for K1 to K10 was run in ADMIXTURE, and the K value with the lowest CV error was selected as the optimal number of sub-populations. Principal component analysis (PCA) was performed in dartR package of R (Gruber et al. 2018), and the first two principal components were plotted based on the sub-populations pre-determined by ADMIXTURE to visualize structure stratification.
The genetic diversity indices including observed heterozygosity (Ho), expected heterozygosity (He), and fixation index (Fst) for the sub-populations were calculated using adegenet package in R (Jombart 2008). Analysis of molecular variance (AMOVA) was determined using the poppr package of R (Kamvar et al. 2014). The genetic differentiation among the sub-populations identified in population structure analysis was assessed using the Nei’s pairwise fixation indices (Fst) using the hierfstat package in R (Goudet 2005).
Phylogenetic analysis
A neighbor-joining phylogenetic tree showing the different sub-populations from ADMIXTURE was constructed based on the Nei’s pairwise fixation indices (Fst) generated from hierfstat package of R (Goudet 2005). The relationship among individuals was shown by generating a Euclidian distance matrix in R (R Core Team [R Foundation for Statistical Computing], 2021) which was further subjected to hierarchical clustering with the Unweighted Pair-Group Method with Arithmetic Means (UPGMA). The resultant phylogenetic tree was exported in Newick format using the ape package in R (Paradis et al. 2004) for visualization and annotation in the interactive tree of life (iTOL) Version 6.8 (https://itol.embl.de/, accessed on 5th September, 2023) (Letunic & Bork 2016).
Results
Characterization of SNP markers
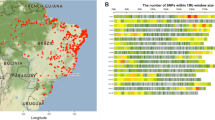
In this study, a comprehensive analysis of single nucleotide polymorphism (SNPs) within the cassava (M. esculenta) genome utilizing the reference genome v8.1 was conducted. Initially, a total of 12,841 SNPs was identified, and through the application of stringent filtration criteria (retaining markers with a minor allele frequency > 0.05 and a call rate > 80), 5247 SNPs were retained for further investigation. The outcome of this filtration process is visually represented in Fig. 1, where the average call rate for the genotypes was determined to be 96%, exhibiting a narrow range of 92% to 98% (Fig. 1A).
The distribution across chromosomes was analyzed, and it was noted that chromosome 1 contained the highest number of SNPs, amounting to 477, while chromosome 18 displayed the lowest count with 200 SNPs. The average SNP count per chromosome was calculated to be 292, providing a comprehensive overview of the genomic landscape (Fig. 1B).
Furthermore, the 5247 retained SNPs exhibited an average polymorphic information content (PIC) value of 0.4, indicating a moderate level of diversity. The PIC values ranged from 0.10 to 0.5, highlighting the variability in informativeness among the identified SNPs (Fig. 1C).
Population structure analysis of the cassava genotypes
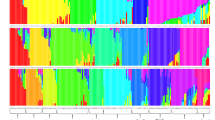
Population structure based on 5247 (MAF > 0.05 and 80% call rate) identified 5 sub-populations across the 155 cassava genotypes (Fig. 2). The 5 sub-populations (pop1, pop2, pop3, pop4, and pop5) were pre-defined by K value of five which showed the least cross-validation error in ADMIXTURE (Fig. 2A). Pop1 (10) and Pop2 (38) were composed of materials from a cross between a variety from Ibadan Nigeria, TME14 and Ugandan genotype MM160128. Most of the samples clustered in pop3 (75 genotypes) were from a cross between a Columbian variety COL40 and Ibadan, Nigeria variety TME14. Pop4 (18) was composed of materials from a cross between a Ugandan variety MM160128 and Ibadan, Nigeria variety TME14. The genotypes in pop5 (14) were highly admixed and were derived from a cross between Ugandan and Ibadan varieties, Variety TME14 and Variety MM060128, respectively. Significant portion of the genetic composition observed in individuals from population 5 (pop5) was derived from population 2 (pop2). Additionally, the genetic profile of population 3 (pop3) exhibited a genetic profile primarily derived from population 5 (popo5).
Population structure analysis of 155 cassava genotypes based on 5247 genome-wide SNP markers with K = 5. A Hierarchical organization of the genetic relatedness of the 155 cassava genotypes. Each bar represents a single genotype and the colored segments within each bar represents the proportional contribution of each sub- population to that accession. B A biplot of the first two principal components with colors based on the 5 sub-populations. C Neighbor-joining tree showing the genetic differentiation among the five sub-populations from ADMIXTURE analysis
In the investigation of genotypic relationship, principal component analysis (PCA) was employed as shown in Fig. 2B. The examination of the first two principal components, PC1 and PC2, revealed that they collectively accounted for approximately 24.2% of the total genetic variation. The resultant biplot of PC1 and PC2 exhibited a discernible clustering pattern among the samples, mirroring the trends observed in the structure analysis conducted through ADMIXTURE. This alignment with the source-based structure analysis underscored the reliability of the findings.
To further understand the genetic relationship among the subpopulations, a neighbor-joining tree based on the Nei’s pairwise fixation indices (Fst) was constructed (Fig. 2C). The analysis identified three major groups within the studied population. Group 1 which was made of pop1, pop3, and pop5 suggests a close genetic affinity among these subpopulations. In contrast, pop2 and pop4 each formed distinct clusters, indicating a genetic distinctiveness that sets apart from the previously mentioned Group 1. This tree-based approach provided a complimentary perspective, further enriching our understanding of the intricate genetic structure within the studied populations.
The genetic relationship among the individuals in the 5 sub-populations was determined by hierarchical clustering of the Euclidean distance matrix using UPGMA method. The analysis led to the clustering of individuals from the five sub-populations into three distinct major clades (Fig. 3): The first clade comprised of populations 1, 3, and 5 (pop1, pop3, and pop5); the second clade encompassed population 2 (pop2); and the third clade population 4 (pop4). This clustering was similar to that of the neighbor-joining phylogenetic tree in Fig. 2C above.
Genetic diversity indices
The mean values for expected (He), observed (Ho), and unbiased expected heterozygosity (uHe) were 0.30, 0.32, and 0.31, respectively (Table 1). The gene diversity values represented by He ranged from 0.28 in pop1 to 0.31 in pop2. The Ho was between 0.3 (pop4) and 0.33 (pop2, pop3 and pop5).
The population fixation indices were used to estimate the genetic differentiation among the five sub-populations due to genetic structure (Table 2). Pop3 and pop4 showed the greatest genetic distance (Fst = 0.23), while pop1 and pop5 and pop2 and pop4 had the least genetic distance of Fst = 0.12.
Analysis of molecular variance (AMOVA) showed that the overall fixation index of the entire population was 0.13 (Table 3). The variation between sub-populations was ~ 34%, while variation within sub-populations was 66%. This finding suggests a higher level of diversity within individual accessions compared to variation observed between sub-populations indicating the presence of various genetic variants, and alleles within the accessions studied.
Discussion
In this study, a significant number of markers (5247 SNP) were generated from diverse collection of cassava genotypes which exhibited moderate polymorphism and revealed a moderate level of genetic diversity. These results were inconsistent with the results by Soro et al. (2023) and Ferguson et al. (2019b) that obtained lower number of SNPs; 36 SNPs from 184 cassava genotypes (Soro et al., 2023) and 1124 SNPs from 522 genotypes respectively. In addition, the contrary results were obtained in the study by Ogbonna et al. (2021) and Rabbi et al. (2017) that obtained higher 27,045 SNPs from 3354 genotypes and 72,279 genome-wide SNP markers from 672 cassava accessions respectively. These findings suggest that the number and distribution of markers across the chromosomes were linked to the level of genetic diversity (Ogbonna et al. 2021). The level of genetic diversity, as indicated by the number and distribution of SNP markers, directly influences the presence and distribution of disease resistance genes within cassava populations. The regions of the cassava genome that harbor a higher density of SNP often correspond to regions containing genes that are associated with disease resistance (Huang & Han 2014). This relationship enables breeders to identify and utilize diverse genetic resources for developing resistant varieties. It also emphasizes the importance of using SNP markers to identify and utilize genetic variants associated with disease resistance against the threatening diseases of CBSD and CMD in cassava, within crop breeding programs.
The average PIC value of SNPs markers in this study indicates a moderate level of informativeness with approximately 65% of the markers falling within the range of PIC values of 0.4–0.5. This level is higher compared to previous studies in cassava, where PIC values ranged from 0.18 to 0.26 (de Oliveira et al. 2014; Ogbonna et al. 2021; de Albuquerque et al. 2018). However, similar results were observed by Eltaher et al. (2018) with SNP markers. The higher PIC values of the SNP markers used in this study suggest that they are suitable for conducting genetic studies in cassava accessions. The PIC reflects the level of diversity captured by the markers, with higher values (0.4–0.5) indicating greater variability among the studied genotypes. In addressing CBSD and CMD challenges, the significance of using SNP with higher PIC values facilitates effective breeding strategies for developing disease-resistant varieties.
The population structure analysis identified five distinct sub-populations within the cassava panel, indicating significant differentiation among genotypes. The composition of each sub-population was strongly associated with the geographical origin of the materials and/or pedigree, given that all genotypes originated from biparental populations whose parents hailed from West Africa, East Africa, and Southern America. This kind of genetic divergence based on crop origin has been previously documented (Adjebeng-Danquah et al., 2020; Adu et al. 2021). Principal component analysis (PCA) further confirmed the genetic relationships among accessions in the diversity panel. The clustering of accessions in PCA biplots closely mirrored the results of structure analysis using ADMIXTURE, consistent with findings from previous studies (Adu et al. 2021; Soro et al., 2023). However, the total genetic variation explained by the first two principal components (PCs) was relatively lower in this study (~ 24.2%) compared to 38.2% and 48.13% in previous studies (Adu et al. 2021) and (Soro et al., 2023) respectively. This discrepancy may be attributed to the narrower genetic background of the population derived from biparental populations. Understanding population structure helps in selecting parental lines for hybridization, enhancing genetic recombination and introducing novel combinations of resistance alleles ultimately broadening the genetic base and enhancing the resilience of new varieties to disease pressure (Uba et al. 2021). In the current study, the population structure of cassava genotypes based on SNP data reveals clusters or groups of genotypes with similar genetic backgrounds. Certain genetic clusters may exhibit higher levels of resistance to cassava mosaic disease (CMD) and cassava brown streak disease (CBSD) due to shared genetic traits. Breeders can leverage this information strategically to incorporate disease-resistance traits into new varieties, thereby improving the overall sustainability and productivity of cassava cultivation.
The NJ and hierarchical classified grouped the 155 cassava genotypes into three major groups revealing a shared gene pool within each cluster with Cluster 1 comprising of pop1, pop3, and pop5 whose cassava genotypes shared common parents. Additionally, the average observed heterozygosity (HO) was determined to be 0.32, aligning with findings from similar studies, which reported Ho of 0.33 (Ferguson et al. 2019b) and 0.32 (Ogbonna et al. 2021) respectively. Analysis of heterozygosity within distinct sub-populations significantly revealed diversity, suggesting potential hybridization and selective processes among the genotypes. This diversity underscores the adaptability and resilience of the species to environmental changes, diseases, and pests, as proposed by Goulet et al. (2017). Such genetic diversity serves as a valuable resource for breeders, offering to develop new varieties with enhanced traits and increased resistance to various stresses.
The differentiation among the groups was assessed using Fst index, which estimates genetic differentiation among groups. According to Wright (1968), Fst values are categorized as high (> 0.25), moderate (0.15–0.25), and low (< 0.05). This index serves as an estimate of genetic flow between sub-populations and, when considered alongside heterozygosity, can directly influence genetic differentiation. In this study, moderate Fst values (0.15–0.25) between certain population pairs (e.g., pop 1 and 3, pop 2 and 3, pop 3 and 4) indicate substantial genetic differentiation. These findings suggest the presence of distinct genetic clusters within cassava populations, which may harbor unique genetic variations, including traits related to disease resistance against CBSD and CMD. Moderate Fst values between population pairs highlight opportunities for targeted breeding efforts. Populations exhibiting moderate genetic differentiation can serve as potential sources of diverse genetic material for developing disease-resistant cassava varieties. The observed Fst values reflect the influence of geographical isolation and parental lineage on genetic differentiation. For instance, populations originating from geographically isolated regions (e.g., Africa and South America) show higher genetic differentiation (moderate Fst), indicating limited gene flow and potential genetic adaptation to local environments. Conversely, populations derived from the same parents (e.g., pop 2 and pop 5) exhibit lower differentiation (low Fst), suggesting recent common ancestry and potential sharing of genetic traits, including disease resistance genes. Breeders can leverage populations with moderate genetic differentiation to introduce novel genetic combinations and enhance disease resistance traits in new cassava varieties.
Conclusion
The SNP markers identified in this study exhibit high polymorphism and informativeness rendering them valuable for assessing diversity within cassava populations. These markers effectively classified cassava populations into sub-groups based on their allele variants. The moderate to high genetic diversity observed across the cassava populations suggests the presence of valuable alleles associated with desirable traits. Integrating these populations into cassava improvement efforts could facilitate the development of user-friendly breeding programs for farmers and stakeholders across the value chain. This research contributes to germplasm management and the establishment of core collections aimed at improving existing varieties.
In summary, the detailed distribution of SNPs across chromosomes and the assessment of polymorphic information content (PIC) significantly enhances our understanding of the genetic makeup of cassava. This groundwork sets the stage for future investigations into cassava diversity and its potential applications in breeding programs and genetic studies. The markers successfully differentiated cassava populations into distinct sub-groups based on their genetic variants. The observed range of moderate to high genetic diversity in genotypes underscores the presence of valuable alleles for desirable traits. The integration of these genotypes into cassava improvement efforts holds promise for developing enhanced varieties through breeding programs.
References
Adjebeng-Danquah J, Manu-Aduening J, Asante IK, Agyare RY, Gracen V, Offei SK (2020) Genetic diversity and population structure analysis of Ghanaian and exotic cassava accessions using simple sequence repeat (SSR) markers. Heliyon 6(1):e03154. https://doi.org/10.1016/j.heliyon.2019.e03154
Adu BG, Akromah R, Amoah S, Nyadanu D, Yeboah A, Aboagye LM, Amoah RA, Owusu EG (2021) High-density DArT-based SilicoDArT and SNP markers for genetic diversity and population structure studies in cassava (Manihot esculenta Crantz). PLoS ONE 16(7):1–19. https://doi.org/10.1371/journal.pone.0255290
Agre AP, Gueye B, Adjatin A, Dansi M, Bathacharjee R, Rabbi IY, Gedil M (2016) Folk taxonomy and traditional management of cassava (Manihot esculenta Crantz) diversity in southern and central Benin. Int J Innov Sci Res 20(2):500–515
Aimone CD, Lavington E, Hoyer JS, Deppong DO, Mickelson-Young L, Jacobson A, Kennedy GG, Carbone I, Hanley-Bowdoin L, Duffy S (2021) Population diversity of cassava mosaic begomoviruses increases over the course of serial vegetative propagation. J Gen Virol 102(7) https://doi.org/10.1099/JGV.0.001622
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664. https://doi.org/10.1101/gr.094052.109
Bayata A (2019) Review on nutritional value of cassava for use as a staple food. Sci J Anal Chem 7(4):83. https://doi.org/10.11648/j.sjac.20190704.12
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635. https://doi.org/10.1093/bioinformatics/btm308
Carvajal-Yepes M, Ospina JA, Aranzales E, Velez-Tobon M, Correa Abondano M, Manrique-Carpintero NC, Wenzl P (2023) Identifying genetically redundant accessions in the world’s largest cassava collection. Front Plant Sci 14 https://doi.org/10.3389/fpls.2023.1338377
Ceballos H, Rojanaridpiched C, Phumichai C, Becerra LA, Kittipadakul P, Iglesias C, Gracen VE (2020) Excellence in cassava breeding: perspectives for the future. Crop Breed Genet Genom 2(2):e200008. https://doi.org/10.20900/cbgg20200008
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R (2011) The variant call format and VCFtools. Bioinformatics 27(15):2156–2158. https://doi.org/10.1093/bioinformatics/btr330
de Oliveira EJ, Ferreira CF, da Silva Santos V, de Jesus ON, Oliveira GAF, da Silva MS (2014) Potential of SNP markers for the characterization of Brazilian cassava germplasm. Theor Appl Genet 127(6):1423–1440. https://doi.org/10.1007/s00122-014-2309-8
de Albuquerque HYG, Carmo, CD, de Brito AC OliveiraEJ (2018) Genetic diversity of Manihot esculenta Crantz germplasm based on single-nucleotide polymorphism markers. Ann Appl Biol 173(3):271–284. https://doi.org/10.1111/aab.12460
Ferguson ME, Shah T, Kulakow P, Ceballos H (2019a) A global overview of cassava genetic diversity. PLoS ONE 14(11):1–16. https://doi.org/10.1371/journal.pone.0224763
Ferguson ME, Shah T, Kulakow P, Ceballos H (2019b) A global overview of cassava genetic diversity. PLoS ONE 14(11):e0224763
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(1):D1178–D1186. https://doi.org/10.1093/nar/gkr944
Goudet J (2005) hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes 5(1):184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour 18(3):691–699. https://doi.org/10.1111/1755-0998.12745
Huang X, Han B (2014) Natural variations and genome-wide association studies in crop plants. Annu Rev Plant Biol 65(1):531–551. https://doi.org/10.1146/annurev-arplant-050213-035715Annual Reviews Inc
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. https://doi.org/10.1093/nar/gkw290
Luo Z, Brock J, Dyer JM, Kutchan T, Schachtman D, Augustin M, Ge Y, Fahlgren N, Abdel-Haleem H (2019) Genetic diversity and population structure of a Camelina sativa spring panel. Front Plant Sci 10(February):1–12. https://doi.org/10.3389/fpls.2019.00184
Ngongo Y, Basuki T, Derosari B, Mau YS, Noerwijati K, Dasilva H, Sitorus A, Kotta NRE, Utomo WH, Wisnubroto EI (2022) The roles of cassava in marginal semi-arid farming in East Nusa Tenggara—Indonesia. Sustainability (switzerland) 14(9):1–22. https://doi.org/10.3390/su14095439
Ogbonna AC, Braatz de Andrade LR, Mueller LA, de Oliveira EJ, Bauchet GJ (2021) Comprehensive genotyping of a Brazilian cassava (Manihot esculenta Crantz) germplasm bank: insights into diversification and domestication. Theor Appl Genet 134(5):1343–1362. https://doi.org/10.1007/s00122-021-03775-5
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20(2):289–290. https://doi.org/10.1093/bioinformatics/btg412
Pierre N, Wamalwa LN, Muiru WM, Simon B, Kanju E, Ferguson ME, Ndavi MM, Tumwegamire S (2022) Genetic diversity of local and introduced cassava germplasm in Burundi using DArTseq molecular analyses. PLoS ONE 17(1):1–19. https://doi.org/10.1371/journal.pone.0256002
Pour-Aboughadareh A, Poczai P, Etminan A, Jadidi O, Kianersi F, Shooshtari L (2022) An analysis of genetic variability and population structure in wheat germplasm using microsatellite and gene-based markers. Plants 11(9):1–15. https://doi.org/10.3390/plants11091205
R Core Team (R Foundation for Statistical Computing) (2021) R: A Language and Environment for Statistical Computing.
Rabbi IY, Udoh LI, Wolfe M, Parkes EY, Gedil MA, Dixon A, Ramu P, Jannink J-L, Kulakow P (2017) Genome-wide association mapping of correlated traits in cassava: dry matter and total carotenoid content. The Plant Genome 10(3) https://doi.org/10.3835/plantgenome2016.09.0094
Sandra DJA, Ricardo FCM, da Carlos ASL, Cláudia FF, Vanderlei da SS, Pascoal AM (2019) Genetic structure of cassava populations (Manihot esculenta Crantz) from Angola assessed through (ISSR) markers. Afr J Biotech 18(7):144–154. https://doi.org/10.5897/ajb2018.16720
Sivan S, Arya K, Sheela MN, Revathi BS, Prakash Krishnan BS, Muthusamy K (2023) Genetic diversity analysis of Indian Cassava (Manihot esculenta Crantz) accessions using morphological and molecular markers. S Afr J Bot 161:347–357. https://doi.org/10.1016/J.SAJB.2023.08.027
Soro M, Pita JS, Somé K, Otron DH, Yéo E, Mutuku JM, Néya JB, Tiendrébéogo F, Koné D (2023) Genomic analysis and identification of potential duplicate accessions in Burkina Faso cassava germplasm based on single nucleotide polymorphism. Front Sustain Food Syst 7(June):1–15. https://doi.org/10.3389/fsufs.2023.1202015
Uba CU, Oselebe HO, Tesfaye AA, Abtew WG (2021) Genetic diversity and population structure analysis of bambara groundnut (Vigna subterrenea L) landraces using DArT SNP markers. PLoS ONE 16(7):e0253600. https://doi.org/10.1371/journal.pone.0253600
Wosula EN, Chen W, Fei Z, Legg JP (2017) GBE unravelling the genetic diversity among Cassava Bemisia tabaci whiteflies using NextRAD Sequencing. Genome Biol Evol 9(11):2958–2973. https://doi.org/10.1093/gbe/evx219
Acknowledgements
The research grant was provided by NextGen cassava Project to Makerere Regional Centre for Crop improvement (MaRCCI) for the research and training of the first author. The International Institute for Tropical Agriculture (IITA) Uganda hosted field experiments at their experimental field stations and provided field assistance and INTERTEK Australia Lab for sequencing the cassava genotypes.
Funding
Funding for this work was provided by Cornell University through a sub-award agreement (N0.84941–11056 between TARI and Cornell University through Next Generation Cassava Breeding Project).
Author information
Authors and Affiliations
Contributions
The authors made the following contributions to the study: Conceptualization was led by KLS, IK, and WK; study design involved KLS, EK, and IK; data collection was carried out by KLS; analysis and investigation were conducted by KLS and WK; the writing of the draft and final manuscript was primarily the responsibility of KLS, with contributions from EAA, PR, IOD, EK, DM, and IK; the final manuscript was reviewed and approved by all authors; funding acquisition was managed by RE, HK, EK, and ISK; and supervision was provided by PR, EK, and ISK.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Disclaimer
The funders had no role in the design of the study, data collection, analysis, or interpretation of the results or in the writing of the manuscript.
Additional information
Communicated by: Izabela Pawłowicz
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sichalwe, K.L., Kayondo, I., Edema, R. et al. Genetic diversity and population structure of Uganda cassava germplasm. J Appl Genetics (2024). https://doi.org/10.1007/s13353-024-00892-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13353-024-00892-x