Abstract
Internal reference genes that are stably expressed are essential for normalization in comparative expression analyses. However, gene expression varies significantly among species, organisms, tissues, developmental stages, stresses, and treatments. Therefore, identification of stably expressed reference genes in developmental endosperm of bread wheat is important for expression analysis of endosperm genes. As the first study to systematically screen for reference genes across different developmental stages of wheat endosperm, nine genes were selected from among 76 relatively stable genes based on high-throughput RNA sequencing data. The expression stability of these candidate genes and five traditional reference genes was assessed by real-time quantitative PCR combined with three independent algorithms: geNorm, NormFinder, and BestKeeper. The results showed that ATG8d was the most stable gene during wheat endosperm development, followed by Ta54227, while the housekeeping gene GAPDH, commonly used as an internal reference, was the least stable. ATG8d and Ta54227 together formed the optimal combination of reference genes. Comparative expression analysis of glutenin genes indicated that credible quantification could be achieved by normalization against ATG8d in developmental endosperm. The stably expressed gene characterized here can act as a proper internal reference for expression analysis of wheat endosperm genes, especially nutrient- and nutrient synthesis–related genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Internal reference genes are commonly used as internal controls in northern blotting, RNase protection assays, conventional semi-quantitative PCR, and real-time quantitative PCR (RT-qPCR) for normalization in gene expression analyses (Paolacci et al. 2009). Widely used housekeeping genes (HKGs), such as 18S rRNA, actin, tubulin, polyubiquitin, and GAPDH, are involved in basic cellular processes and therefore are assumed to have stable and uniform expression. However, HKGs are not always reliable controls, as their expression levels vary among species, organs, tissues, developmental stages, stresses, and treatments, as reviewed by Joseph et al. (2018). Therefore, it is essential to identify reliable reference genes that are stably expressed under given conditions.
Wheat is one of the three major crops worldwide. Its endosperm stores most of the seed nutrients (> 10% protein, 2.4% lipids, and 79% carbohydrates) and serves as an important food resource for over one-third of the global human population (Vasil 2007). Several key genes involved in starch synthesis (reviewed by Guzmán and Alvarez 2016) and transcriptional control of storage protein genes (Dong et al. 2007; Guo et al. 2015) have been identified and functionally studied in wheat. However, the genetic regulation of nutrient synthesis, processing, trafficking, and accumulation that accompanies endosperm development is an unknown and complex process. Identification of stably expressed genes would benefit studies on gene expression and provide valuable information for understanding the underlying mechanisms regulating the material storage process and wheat endosperm development. Paolacci et al. (2009) performed a cross-search for stably expressed genes in the UniGene and TIGR databases and, by using RT-qPCR combined with computational analysis, verified that Ta54227 (cell division control protein, AAA-superfamily of ATPases), Ta2291 (ADP-ribosylation factor), and Ta2776 (RNase L inhibitor-like protein) are expressed more stably in various wheat tissues and development stages than traditional HKGs (tHKGs) such as encoding α-tubulin, β-tubulin, Ubiquitin, Actin, GAPDH, ribosomal protein, and histone. Wu et al. (2015) verified that Ta2291 and EF (encoding elongation factor) were the two most stable genes among nine reference genes selected from previous reports, and can be applied to normalization of quantitative analyses as single or combined reference genes during wheat kernel development. Although several internal reference genes for various wheat kernel developmental stages have been identified based on high-throughput RNA sequencing (RNA-Seq) data and bioinformatics analysis (Paolacci et al. 2009; Wu et al. 2015), no reference genes have been reported for wheat endosperm, particularly across its different developmental stages.
The rapid development of RNA-Seq technology provides new opportunities for screening for the most stable reference genes based on the whole transcriptome data. In RNA-Seq analysis, the expected fragments per kilobase of transcript per million fragments sequenced (FPKM) is the most common method for estimating the level of gene expression (Trapnell et al. 2010). Recently, several potential stably expressed genes were screened by analyzing and comparing FPKM values, followed by further validation by RT-qPCR combined with computational analysis, in a variety of tissues and developmental stages in plants (Fernández-Aparicio et al. 2013; Stanton et al. 2017; Liu et al. 2018). By identifying candidates screened from RNA-Seq and microarray data, Lin et al. (2014) verified that a hypothetical protein-coding gene can be used as an internal reference due to its stable expression across 26 maize lines under different nitrogen treatments. Similarly, of six candidate genes selected from the transcriptome atlas, Fb15 was identified as the most stable gene in developing rice endosperms, both under normal and high-temperature culture conditions (Xu et al. 2015).
In this study, nine genes stably expressed across ten periods of Chinese Spring (CS) wheat endosperm development were selected as candidate reference genes based on transcriptome data. RT-qPCR combined with computational analysis of eight new candidate reference genes and five commonly used traditional reference genes showed that ATG8d exhibited the most stable expression levels throughout endosperm development, and together with Ta54227 formed the most stable gene combination. ATG8d can be used as a reference gene in research of gene expression analysis in developing wheat endosperm.
Materials and methods
Plant materials
The bread wheat Triticum aestivum cv. CS was grown in the greenhouse of the Chinese Academy of Agricultural Sciences in Beijing, in 2017. Seeds were collected individually from the central part of the spikes from three tillers at 5, 8, 11, 14, 17, 20, 23, 26, 29, and 32 days post anthesis (DPA), immediately frozen in liquid nitrogen, and then stored at − 80 °C for total RNA extraction and high-throughput RNA-Seq.
RNA extraction and complementary DNA (cDNA) synthesis
Embryos were carefully removed from seeds using a scalpel blade. Total RNA was extracted from endosperms following the TRIzol method (Li et al. 2010). Total RNA integrity was monitored on the 1% agarose gel electrophoresis and its quantity was spectrophotometrically adjusted using a NanoDrop ND-2000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA) for cDNA synthesis with a PrimeScriptTM RT Reagent Kit (TaKaRa, Dalian, Japan). After treatment with gDNA Eraser to eliminate genomic DNA, equal amount of RNA taken from each sample was subjected to reverse transcription for cDNA synthesis. The quality of all cDNA samples was examined by amplifying the coding of GAPDH and ATG8d genes. The primer sequences were listed as follows: GAPDH-F, ATGGCTCCGATCAAGATCG; GAPDH-R, TTACTTGGAGCTGTGCATGTGAC; ATG8d-F, CTGGAAAGGAGGCAAGCTGA; ATG8d-R, CGAAAGTGTTCTCGCCACTG.
RNA-Seq and transcriptome analysis
Libraries for 30 samples collected from 5, 8, 11, 14, 17, 20, 23, 26, 29, and 32 DPA (three replicates for each stage) were prepared according to the method described by Hu et al. (2015) and sequenced (150-bp paired-end reads) on an Illumina HiSeq platform. Paired-end reads from raw sequence data were imported into CLC Genomics Workbench v7.0.4 (CLC Bio, Aarhus, Denmark; presently Qiagen, Germantown, MD, USA). Trimmed reads were mapped against the CS reference gDNA database (RefSeq v1.0; International Wheat Genome Sequencing Consortium) using HISAT2 v2.0.4 with default parameters (Kim et al. 2015). Mapped reads were counted using the HTSeq v0.6.1 Python package (parameters: −m union, −s no) (Anders et al. 2015). All RNA-Seq data have been deposited at the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra/) under accession number PRJNA545291.
Screening of candidate reference genes from transcriptome data of developing endosperms
An in silico approach was applied to screen for genes with stable expression levels among different developmental periods of wheat endosperm. The gene transcription level in each period was represented as FPKM. To select stable genes, we (i) eliminated genes that were not expressed during all developmental periods; (ii) evaluated the mean FPKM, coefficient of variation (CV), and relative range (RR) for each gene; and (iii) further filtered out potential references that contained more than three copies or two types of alternative splicing (AS). Additionally, the expression stability of the five most frequently used reference genes in wheat or related species was also evaluated based on our transcriptome data.
Real-time quantitative PCR
The specific RT-qPCR primers for selected candidates were designed using the Primer3Plus online tool (http://www.primer3plus.com/cgi-bin/dev/primer3plus.cgi); primer sequences are listed in Tables S1 and S2. RT-qPCR was performed using the TB Green™ Premix Ex Taq™ Kit (TaKaRa) on a CFX96 Real-Time system (Bio-Rad, Hercules, CA, USA). The 20-μL reactions contained 10 μL TB Green™ Premix Ex Taq™ (Tli RNaseH Plus), 0.1 μM each primer, and 2 μL cDNA. The PCR program was as follows: pre-denaturation at 95 °C for 3 min followed by 40 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 15 s, and extension at 72 °C for 10 s. The products were heat-denatured over a temperature gradient of 0.1 °C/s from 72 °C to 95 °C.
Expression stability analysis of potential reference genes
Three Microsoft Excel-based tools, geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004), and BestKeeper (Pfaffl et al. 2004), were used to evaluate the expression stability of candidates. Relying on sequential pair-wise comparisons, geNorm calculates the stability value M and the variation value V, by which the stability of each gene and the number of optimal reference genes are determined. Similar to geNorm, NormFinder ranks individual candidates according to their stability value. BestKeeper determines the optimal references by employing pair-wise correlation analysis of all pairs of candidates. The Ct values of genes obtained by RT-qPCR were converted into their relative expression levels following the formula Q = EΔCt; the relative expression levels were imported into geNorm and NormFinder. BestKeeper provides the standard deviation (SD), CV, and Pearson correlation coefficient (r) based on cycle threshold (Ct) values.
Results
Screening of potential reference genes stably expressed in developing wheat endosperm
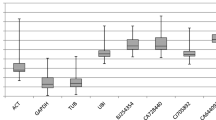
A total of 132,126 genes were expressed during CS endosperm development based on RNA-Seq analysis. Examination of the FPKM distribution by violin plot revealed that lots of genes expressed at a low level in each stage, especially in the last two stages (Fig. S1). Genes with low expression abundance are unsuitable as reference genes (Wu et al. 2015). We used FPKM > 50 as the cut-off value for candidate selection because the most frequently used reference genes with proper expression intensity, such as GAPDH, ubiquitins, actin, and histone, exhibited FPKM > 50 in our transcriptome data. Furthermore, any candidate genes with more than two isoforms or a CV > 10% were excluded in this study. After screening, 76 genes satisfied the above criteria (Table S3). By comparing FPKM values among candidate genes, several genes showed low CV values but high expression variance, indicating that either low- or high-level expression occurred during at least one of the ten development periods. Therefore, the relative range was used to filter out unstably expressed genes. Finally, the top seven genes were selected as candidate reference genes according to their expression stability and intensity, and the number of isoforms (Fig. 1, Table S3). In addition, irrespective of their expression levels, two genes, AL1 and PSMB3, expressed across all 10 development periods, showed the least variation due to their low CV values (CV < 5%; Tables S3, S4). We also selected five traditional reference genes among those most commonly used as internal controls in expression experiments of wheat or related species for comparative analysis of expression variance (Tables S3, S5). Interestingly, none of the five genes appeared in the list of 76 candidates.
Expression levels of new candidate and traditional reference genes across ten developmental stages basing on RNA-Seq data. Each FPKM value is a mean of triplicate replicates. The median (horizontal line), upper and lower quartiles (box), and maximum and minimum values (whiskers) are shown for each gene
Expression level analysis of new candidate genes and traditional reference genes across ten endosperm development periods by RT-qPCR
Prior to performing cDNA synthesis, the integrity of total RNA was analyzed using 1.0% agarose gel electrophoresis (Fig. S2). Each sample showed clear 28S (upper) band and 18S (lower) band, which indicated that RNA samples extracted from 10 stages were intact and appropriate for transcript level analysis. The quality of all cDNA samples was also examined by evaluating the expression of the GAPDH and ATG8d genes. Two specific products with the predicted molecular size, representative of the complete GAPDH coding sequence (1014 bp) and a partial ATG8d coding sequence (316 bp), were successfully amplified from each cDNA sample (Fig. S3), confirming the viability of all cDNA samples.
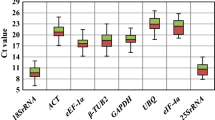
The constructed standard curve of Polyubi11L was based on less than five different concentrations with three times gradient dilution points (Table S1), which suggested that Polyubi11L was expressed at a low level during the development of wheat endosperm, and wasn’t suitable as a candidate reference gene in this experiment. The remaining eight candidate genes and five traditional reference genes were further analyzed across ten development periods. Specific primers are listed in Table S1. Gene-specific amplification by each primer pair was verified by a single dominant peak in the dissociation curve, and a single band with the predicted molecular size on a 2.0% agarose gel (Fig. S4). The amplification efficiency of each primer pair was 90–110% (Table S1). Based on the Ct values obtained from RT-qPCR, a box plot was generated to reflect the differences in gene expression level across the ten development periods (Fig. 2). Ct values ranged from 20 to 35, while most were between 25 and 30. Ubi-4 was the most abundantly transcribed, while ARF1 was the least abundant based on the number of cycles needed to reach the fluorescence threshold. We calculated the CVs of Ct values and found that Actin-7 showed the least variation, followed by ATG8d, SAR1a, and SKP1a, while GAPDH had the largest variation. Therefore, Actin-7, ATG8d, SAR1a, and SKP1a were determined to be relatively stable candidate genes, and GAPDH was the least stable.
Expression levels of new candidate and traditional reference genes across ten developmental stages basing on the RT-qPCR data. Each cycle threshold (Ct) value is a mean of triplicate technical replicates. The median (horizontal line), upper and lower quartiles (box), and maximum and minimum values (whiskers) are shown for each gene
Evaluation of expression stability of new candidate genes and traditional reference genes
geNorm analysis
The geNorm is the most commonly used software for verifying the expression stability of genes. geNorm calculates a stability value, M, as a criterion for assessing the average pair-wise variation of one target gene compared to other genes in a candidate gene list. It is generally accepted that a gene with lower M value, especially < 0.5, is considered to be a stably expressed gene (Dekkers et al. 2012; Wu et al. 2015). We used this application to rank all 13 genes according to their M values. The top three candidates, with M values < 0.5, were ATG8d, Ta54227, and PSMB3 (Fig. 3), among which the most stable genes were ATG8d and Ta54227. The bottom three candidates, GAPDH, SAR1a, and Actin-7, exhibited M values > 0.7, indicating their expression instability. In addition to M values, geNorm also evaluates the optimal gene numbers for multiple reference genes based on the ratio Vn/Vn + 1. The default V value of 0.15 is the pair-wise variation threshold, and additional reference genes are unnecessary when Vn/Vn + 1 drops below the threshold (Vandesompele et al. 2002). As shown in Fig. 4, the value of V2/V3 was 0.144, substantially below the threshold of 0.15, indicating that only ATG8d and Ta54227 were essential for normalization-based quantification during wheat endosperm development.
NormFinder analysis
Unlike geNorm, the NormFinder algorithm ranks the expression stability of candidate references based on a stability value derived from the synthetic calculation of inter- and intra-group expression variations, which can prevent co-regulated genes from being selected from the candidate list (Andersen et al. 2004). The stability value calculated by NormFinder is similar to the M value; namely, lower value indicates more stable gene expression. The results from NormFinder showed that ATG8d had the lowest value, followed by SKP1a, Ta54227, and Ubi-4 (Table 1), indicating that ATG8d was the most stably expressed among the 13 genes. Meanwhile, as expected, the traditional HKG GAPDH had the highest value in NormFinder. Consistent with the geNorm results, NormFinder predicted that ATG8d was the most stable gene while GAPDH was the least stable.
BestKeeper analysis
The BestKeeper algorithm calculates the SD, CV, and r and its P value based on the crossing point (Cp) value. The most stable reference genes defined by BestKeeper should have the lowest SD (usually < 1) and significantly higher correlations with other candidate genes (Pfaffl et al. 2004). BestKeeper was used to analyze the expression stability of the 13 candidates during wheat endosperm development (Table 2). Only the top two genes, Actin-7 and ATG8d, had an SD < 1 and a CV value lower than the other genes. We determined ATG8d to be the most stable because it showed stronger correlations with the remaining genes than Actin-7. Although SAR1a, Ta54227, Ta097800, Ta27922, PSMB3, ADP, and GAPDH had higher correlations with other candidate genes at significant levels, they showed unexpected SD values (> 1), especially GAPDH.
Evaluation of the reference gene ATG8d
Glutenins are seed storage proteins that play a key role in determination of nutrient quality and bread-making quality (Shewry and Halford 2002). As low-molecular-weight glutenin subunits (LMW-GSs) are the major constituent of glutenin (D’Ovidio and Masci 2004), four LMW-GS genes were selected for expression pattern construction based on both RT-qPCR and RNA-Seq data; the primers are listed in Table S2. The expression dynamics of the four LMW-GS genes were normalized against reference gene ATG8d or GAPDH to calculate relative expression levels. After normalization with ATG8d, all four genes were expressed at lower levels at 5, 29, and 32 DPA, but at higher levels from 14 to 23 DPA (Fig. 5a). Samples from 5, 11–20, and 29–32 DPA are representative of the late undifferentiated tissue stage, middle grain filling stage, and late grain filling stage, respectively, corresponding to the initial, peak, and end points of glutenin synthesis (Evers and Millar 2002; Liu et al. 2012; Hurkman et al. 2013). LMWGS2 transcript abundance continuously increased before 14 DPA, but gradually decreased during subsequent development, consistent with the results obtained from RNA-Seq data (Fig. 5b). The expression patterns of the other genes were largely in agreement with the RNA-Seq results, with minor differences (e.g., a slight increase or decrease at certain points). Specifically, the levels of LMWGS1 and LMWGS3 increased until peaking at 17 DPA, while LMWGS4 peaked at 11 DPA. In contrast, each glutenin gene obtained an obviously different expression pattern from that constructed by RNA-Seq data when normalization against GAPDH (Fig. 5c). The universal consistency in expression patterns constructed from RT-qPCR with ATG8d as the reference and RNA-Seq data indicated that ATG8d is a reliable reference gene and can be applied for normalization in developing wheat endosperm.
Discussion
The use of an appropriate reference gene for proper interpretation of gene expression data emerged as a significant concern in Northern blotting, RNase protection assay, conventional semi-quantitative PCR, and real-time quantitative PCR experiments. In earlier studies, it was difficult to identify stably expressed genes due to the lack of whole-genome and transcriptome data, and most internal references were derived from HKGs involved in basic cellular metabolism. However, the most commonly used HKGs exhibit substantial variability under different experimental conditions (Suzuki et al. 2000; Brunner et al. 2004; Dheda et al. 2004; Czechowski et al. 2005; Paolacci et al. 2009; Wu et al. 2015). Genes unrelated to housekeeping activity, even those with unknown functions, can also be used for normalization purposes by virtue of their stable expression patterns (Czechowski et al. 2005). Therefore, it is reasonable that any gene stably expressed under the given condition can be used as a reference gene (Joseph et al. 2018).
The recent advances in high-throughput technologies permit direct screening for stable reference genes at the whole-genome level. RNA-Seq has many advantages, and is an attractive whole-transcriptome method for the screening of stably expressed genes (Stanton et al. 2017). Based on RNA-Seq data and experimental validation, appropriate, stable genes have been identified as internal references applicable under specific experimental situations (Fernández-Aparicio et al. 2013; Stanton et al. 2017; Liu et al. 2018). Ta54227 was screened from microarray data, and verified to outperform tHKGs, such as GAPDH, ribosomal protein, and ubiquitin encoding genes, in different tissues and developmental stages of wheat (Paolacci et al. 2009). In this study, we systematically and quantitatively evaluated five traditional references and eight stable genes obtained from screening whole-transcriptome RNA-Seq data from ten developmental periods of wheat endosperm. Of the 13 genes, ATG8d was the most stably expressed throughout endosperm development, followed by Ta54227, while GAPDH was least stable. The previously reported Ta54227 ranked behind the ATG8d as the second most suitable internal reference gene, demonstrating that whole transcriptome analysis (RNA-Seq) in combination with RT-qPCR and computational analysis is a very robust approach for discovering novel internal reference genes in developing wheat tissues.
Autophagy-related genes (ATGs) are a group of evolutionarily conserved genes involved in the entire process of autophagosomes in cells (Yorimitsu and Klionsky 2005). The lipid-conjugated ubiquitin-like protein ATG8 is a core machinery protein for the formation of autophagosomes, which are double-membrane vesicles responsible for the delivery of cytoplasmic material to lysosomes (Nakatogawa et al. 2007; Xie et al. 2008). Seo et al. (2016) identified 116 ATG8 genes from 18 different publicly available plant genome sequences, and found that their gene structures and protein domain sequences are conserved in plant lineages. Compared to other crops, such as rice (four ATG8 genes) and maize (five ATG8 genes), wheat possesses the largest number of ATG8 gene family members (13 ATG8 genes) (Seo et al. 2016; Yue et al. 2018). Of the 13 ATG8 genes, ATG8d and eight others are expressed specifically in the spike, far more than in other tissues, likely due to the high activity of cell division, metabolism, and senescence during spike development in wheat (Yue et al. 2018). In this study, ATG8d was identified as the most stably expressed gene in the developing wheat endosperm and could be used as an internal reference for expression analysis of endosperm genes. The expression stability of its orthologues in rice and maize, and their applicability for normalization require further study.
Conclusion
Overall, ATG8d, Ta54227, PSMB3, SKP1a, Ubi-4, and Actin-7 were evaluated as stably expressed genes during different development stages of CS endosperm based on three mathematical algorithms. Of these, ATG8d was the only gene ranked among the top two candidates in all three lists, exhibiting higher stability or lower variation than other genes. Additionally, Ta54227 was determined as stable by both geNorm and NormFinder. However, PSMB3, SKP1a, and Ubi-4 were determined as stable by only one of the three algorithms. Although Actin-7 was ranked in the top two by BestKeeper, its lower correlation coefficient and bottom ranks by geNorm and NormFinder indicated its instability. In contrast, GAPDH was ranked at the bottom by all three algorithms, showing that it was the least stable among the 13 candidate reference genes. Therefore, ATG8d was the most stable gene throughout the different development stages of CS endosperm, and together with Ta54227 formed the optimal gene combination of paired internal reference genes for accurate assessment of gene expression levels in wheat endosperm. Using ATG8d as an internal reference gene, credible expression patterns of LMW-GS genes could be achieved by RT-qPCR in CS endosperm development. The stably expressed gene characterized here can act as a proper internal reference for expression analysis of wheat endosperm genes.
References
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31(2):166–169
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64(15):5245–5250
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139(1):5–17
D’Ovidio R, Masci S (2004) The low-molecular-weight glutenin subunits of wheat gluten. J Cereal Sci 39:321–339
Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A (2004) Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37:112–119
Dong GQ, Ni ZF, Yao YY, Nie XL, Sun QX (2007) Wheat Dof transcription factor WPBF interacts with TaQM and activates transcription of an alpha-gliadin gene during wheat seed development. Plant Mol Biol 63:73–84
Evers T, Millar S (2002) Cereal grain structure and development: some implications for quality. J Cereal Sci 36:261–284
Fernández-Aparicio M, Huang K, Wafula EK, Honaas LA, Wickett NJ, Timko MP, Depamphilis CW, Yoder JI, Westwood JH (2013) Application of qRT-PCR and RNA-Seq analysis for the identification of housekeeping genes useful for normalization of gene expression values during Striga hermonthica development. Mol Biol Rep 40(4):3395–3407
Guo WW, Yang H, Liu YQ, Gao YJ, Ni ZF, Peng HR, Xin MM, Hu ZR, Sun QX, Yao YY (2015) The wheat transcription factor TaGAMyb recruits histone acetyltransferase and activates the expression of a high-molecular-weight glutenin subunit gene. Plant J 84:347–359
Guzmán C, Alvarez JB (2016) Wheat waxy proteins: polymorphism, molecular characterization and effects on starch properties. Theor Appl Genet 129(1):1–16
Hu LX, Li HY, Chen L, Lou YH, Amombo E, Fu JM (2015) RNA-seq for gene identification and transcript profiling in relation to root growth of bermudagrass (Cynodon dactylon) under salinity stress. BMC Genomics 16:575
Hurkman WJ, Tanaka CK, Vensel WH, Thilmony R, Altenbach SB (2013) Comparative proteomic analysis of the effect of temperature and fertilizer on gliadin and glutenin accumulation in the developing endosperm and flour from Triticum aestivum L. cv. Butte 86. Proteome Sci 11(1):8
Joseph JT, Poolakkalody NJ, Shah JM (2018) Plant reference genes for development and stress response studies. J Biosci 43(1):173–187
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Li XH, Wang K, Wang SL, Gao LY, Xie XX, Hsam SLK, Zeller FJ, Yan YM (2010) Molecular characterization and comparative transcriptional analysis of LMW-m-type genes from wheat (Triticum aestivum L.) and Aegilops species. Theor Appl Genet 121(5):845–856
Lin F, Jiang L, Liu YH, Lv YD, Dai HX, Zhao H (2014) Genome-wide identification of housekeeping genes in maize. Plant Mol Biol 86:543
Liu W, Zhang YZ, Gao X, Wang K, Wang SL, Zhang Y, He ZH, Ma WJ, Yan YM (2012) Comparative proteome analysis of glutenin synthesis and accumulation in developing grains between superior and poor quality bread wheat cultivars. J Sci Food Agric 92(1):106–115
Liu J, Huang S, Niu X, Chen D, Chen Q, Tian L, Xiao FM, Liu YS (2018) Genome-wide identification and validation of new reference genes for transcript normalization in developmental and post-harvested fruits of Actinidia chinensis. Gene 645:1–6
Nakatogawa H, Ichimura Y, Ohsumi Y (2007) Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130(1):165–178
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515
Seo E, Woo J, Park E, Bertolani SJ, Siegel JB, Choi D, Dinesh-Kumar SP (2016) Comparative analyses of ubiquitin-like ATG8 and cysteine protease ATG4 autophagy genes in the plant lineage and cross-kingdom processing of ATG8 by ATG4. Autophagy 12(11):2054–2068
Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot 53:947–958
Stanton KA, Edger PP, Puzey JR, Kinser T, Cheng P, Vernon DM, Forsthoefel NR, Cooley AM (2017) A whole-transcriptome approach to evaluating reference genes for quantitative gene expression studies: a case study in Mimulus. G3-Genes Genom Genet 7(4):1085–1095
Suzuki T, Higgins PJ, Crawford DR (2000) Control selection for RNA quantitation. Biotechniques 29:332–337
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van BMJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):1–12
Vasil IK (2007) Molecular genetic improvement of cereals: transgenic wheat (Triticum aestivum L.). Plant Cell Rep 26:1133–1154
Wu D, Dong J, Yao YJ, Zhao WC, Gao X (2015) Identification and evaluation of endogenous control genes for use in quantitative RT-PCR during wheat (Triticum aestivum L.) grain filling. Genet Mol Res 14(3):10530–10542
Xie ZP, Nair U, Klionsky DJ (2008) Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19(8):3290–3298
Xu H, Bao JD, Dai JS, Li YQ, Zhu Y (2015) Genome-wide identification of new reference genes for qRT-PCR normalization under high temperature stress in rice endosperm. PLoS One 10(11):e0142015
Yorimitsu T, Klionsky DJ (2005) Autophagy: molecular machinery for self-eating. Cell Death Differ 12(S2):1542
Yue WJ, Nie XJ, Cui LC, Zhi YQ, Zhang T, Du XH, Song WN (2018) Genome-wide sequence and expressional analysis of autophagy gene family in bread wheat (Triticum aestivum L.). J Plant Physiol 229:7–21
Funding
This research was supported by grants from National Key R&D Program of China (2016ZX08009003-004), National Natural Science Foundation of China (31571652), the Natural Science Foundation of Beijing (6162002, 6122002), and the Youth Innovative Research Team of Capital Normal University.
Author information
Authors and Affiliations
Contributions
XHL conceived and designed the experiment. JYM, LC, YSG, LND, and SCH performed the experiment. JYM and LC analyzed the data. YXL and YMY provided technical assistance and scientific discussion. JYM and XHL wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by: Barbara Naganowska
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 1992 kb)
Rights and permissions
About this article
Cite this article
Mu, J., Chen, L., Gu, Y. et al. Genome-wide identification of internal reference genes for normalization of gene expression values during endosperm development in wheat. J Appl Genetics 60, 233–241 (2019). https://doi.org/10.1007/s13353-019-00503-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-019-00503-0