Abstract
Since its inception more than a decade ago, gene silencing mediated by double-stranded small interfering RNA (siRNA) has been widely investigated as a potential therapeutic approach for a variety of diseases. However, the use of siRNA is hampered by its rapid degradation and poor cellular uptake in vitro and in vivo. Recently, peptide-based carriers have been applied to siRNA delivery, as an alternative to the traditional delivery systems. Here, a histidine-containing amphipathic amino acid pairing peptide, C6M3, which can form complexes with siRNA, was used as a new siRNA delivery system. This peptide exhibited a high affinity for siRNA and ability to efficiently deliver siRNA into the cells. The interaction of C6M3 with siRNA was investigated to determine the loading capacity of C6M3 at different peptide/siRNA molar ratios. At C6M3/siRNA molar ratio of 10/1, siRNA molecules were entirely associated with C6M3 as indicated by a gel electrophoretic assay and further confirmed by zeta potential analysis. The particle size distribution of the C6M3-siRNA complexes was studied using dynamic light scattering, which showed an intensity-based size distribution peaked approximately at 100 nm in RNase-free water and 220 nm in the Opti-MEM medium. C6M3 adopted a helical secondary structure in RNase-free water and became more so after forming complexes with siRNA. The interaction of siRNA with C6M3 is an entropy-driven spontaneous process, as determined by isothermal titration calorimetry (ITC) study. The efficiency of cellular uptake of the siRNA complexes at different C6M3/siRNA molar ratios was evaluated, and the results showed that C6M3 promoted efficient cellular uptake of siRNA into cells. Furthermore, a significant level of GAPDH gene silencing efficiency (69%) was achieved in CHO-K1 cells, with minimal cytotoxicity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The term RNA interference (RNAi) was first coined after the discovery that double-stranded RNAs (dsRNAs) can trigger silencing of complementary messenger RNA (mRNA) sequences in the nematode Caenorhabditis elegans [1]. Since the discovery of RNAi, there has been an explosion of interest in using this technology for basic and applied research, including analysis of signaling pathways and gene functions, as well as in developing a therapeutic method to target disease-related mRNAs in the cytoplasm of cells [1–7]. In spite of the high therapeutic potential of small interfering RNAs (siRNAs), their negative charge has limited their ability to penetrate cell membranes, and naked siRNAs are easily degraded by nucleases [8].
To date, a number of materials have been used to develop siRNA delivery systems, including virus vectors, liposomes, lipoplexes, polymers, and peptides [9, 10]. Among them, cell-penetrating peptides (CPPs) have been utilized for intracellular delivery of a variety of macromolecules, including proteins, peptide nucleic acids (PNA), and siRNAs [11].
CPPs can be conjugated to siRNA through covalent cross-links such as disulfide bonds. The disulfide bonds are cleaved in cells, releasing the siRNA into the cytoplasm, where the RNA-induced silencing complex (RISC) forms [11]. CPPs and siRNA can also form non-covalent complexes through electrostatic interactions. At certain molar or charge ratios, siRNA and CPPs form positively charged complexes that are translocated across the plasma membrane. The advantages of this methodology include the low cost of the reagents and the easy preparation of CPP-siRNA complexes [12, 13].
CPP-siRNA complexes can enter cells either through endocytotic pathways or by directly crossing the cell membrane [14]. Amino acid pairing peptides are a new class of peptides, which are recently developed as drug/gene delivery systems [15–17]. There are two main domains in an amino acid pairing peptide: one is the amino acid pairing domain—responsible for peptide self assembly—and the other is the cell permeation domain—a functional group for cell penetration or a group of cationic amino acids [18]. In our previous study, we reported that a new amphipathic, amino acid pairing peptide, C6, formed complexes with siRNAs in a non-covalent manner and protected the siRNA from degradation [19]. Although its cellular uptake was highly efficient, we found that its gene silencing activity was not significantly high (not reported). Therefore, it was concluded that after entering cells, the C6-siRNA complexes were trapped in endosomes [19], which decreased their gene silencing efficiency. Strategies to induce the membrane disruption are required. Tryptophan (W), because of its aromaticity, flat rigid shape, and π electronic structure, has a strong preference at the membrane interface and acts as an anchor to the cell membrane [20–23], thus enhances the peptide-membrane interaction. Histidine proves to enhance the endosomal membrane disruption through proton sponge effect [24–26]. Therefore, C6 (Ac-RLLRLLLRLWRRLLRLLR-NH2) was modified to C6M3 (Ac-RLWHLLWRLWRRLHRLLR-NH2), where histidine and a higher amount of tryptophan were incorporated. In this study, the physicochemical properties, cytotoxicity, cellular uptake, and gene silencing efficiency of C6M3-siRNA complexes were investigated.
Materials and methods
Materials
Peptide C6M3 (Ac-RLWHLLWRLWRRLHRLLR-NH2, MW = 2621 g/mol) was synthesized by CanPeptide Inc. (Quebec, CA) with a purity of >95%. siRNA targeting the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene and Cy3-labeled GAPDH siRNA were purchased from Ambion (Silencer™ GAPDH siRNA kit). The siRNA targeting sequence for eGFP, GCGACGUAAACGGCCACAAGU, was purchased from Dharmacon. The antisense sequence is ACUUGUGGCCGUUUACGUCGC, and the sense sequence is GACGUAAACGGCCACAAGUUC. The negative control siRNA, which sets the threshold, is crucial for determining the transfection efficiency of a siRNA. The negative control siRNA used here was purchased from Ambion. Cell Counting Kit-8 (CCK-8) was used for cell viability test and was purchased from Dojindo Molecular Technologies.
Methods
Cell line
Chinese hamster ovary cells (CHO-K1) purchased from American Type Culture Collection (ATCC CCL-61) were grown in F-12K medium (Thermo Scientific, Ontario, CA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, Ontario, CA). The cells were incubated at 37 °C in 5% CO2.
Preparation of siRNA complexes
For the cell treatments, the peptide/siRNA complexes were prepared in Opti-MEM (Invitrogen, CA, USA). For the physicochemical characterization, the complexes were prepared in RNase-free water. The complexes were generally incubated at room temperature for 20 min immediately before transfection/measurements.
Agarose gel shift assay
Samples containing 300 ng of eGFP siRNA were electrophoresed on a 1.2% w/v agarose gel in 1× TBE at 55 V for 1 h. The ethidium bromide-stained siRNA was visualized on a UV transilluminator equipped with a camera.
Isothermal titration calorimetry
The isothermal titration calorimetry experiments were conducted using a Nano-ITC calorimeter (TA Instruments). A 250 μM C6M3 peptide solution and a 10 μM siRNA solution were both prepared in RNase-free water. All of the samples were degassed in a degassing station (TA Instruments) prior to the experiments. RNase-free water was placed in the ITC reference cell. For each titration, 2 μl of the peptide in a pipette rotating at 250 rpm was injected into the siRNA solution in the sample cell of the calorimeter, which was equilibrated to 25 °C, with an interval of 300 s between injections. The heat of dilution was measured by titrating the C6M3 solution into RNase-free water and was later subtracted from the sample measurement. The data were analyzed using NanoAnalyze software v.2.3.0.
Dynamic light scattering and zeta potential
Particle size measurements were conducted at 25 °C in transparent ZEN0040-disposable microcuvette cells (40 μl) using a Zetasizer Nano ZS (Malvern, UK) after the samples were allowed to stabilize for 20 min at peptide-siRNA molar ratios of 10/1, 20/1, or 40/1 with a final siRNA concentration of 100 nM in RNase-free water. At molar ratio 40/1, the particle size of peptide-siRNA complexes was also measured after incubation for 40 and 90 min, respectively. After the incubation of peptide-siRNA complexes (molar ratio 40/1) for 20 min at 25 °C, the sample was further incubated at 37 °C and the particle sizes were measured after incubation for 30 and 60 min, respectively. In order to study the effect of Opti-MEM culture medium on the particle size, the peptide-siRNA complexes (molar ratio 40/1) were prepared in Opti-MEM and incubated for 20 min at 25 °C following with size measurements. The zeta potentials of the peptide-siRNA complexes were measured using a Clear DTS1070-zeta dip cell.
Scanning electron microscopy
SEM was used to characterize the morphology of C6M3-siRNA complexes at a molar ratio of 20/1 with siRNA concentration of 100 nM. Right after incubation for 20 min, 40 μl samples were allowed to dry at room temperature over substrates. The samples were coated with gold (10 nm in thickness) before taking images. The SEM images were taken with a LEO FESEM 1530 field emission SEM.
Circular dichroism spectroscopy
Spectra at 250 to 190 nm with a spectral resolution and pitch of 1 nm and scan speed of 200 nm/min were recorded using a J-810 spectropolarimeter (Jasco, USA). Increasing amounts of siRNA were added to the peptide solution (30 μM), to obtain different molar ratios. The samples were transferred into 1-mm-long quartz cells and maintained at 25 °C. The spectra presented here are the average of three measurements.
Cytotoxicity of peptide-siRNA complexes
CHO-K1 cells were seeded at 5000 cells/well in clear, flat-bottomed, 96-well plates (Costar) 24 h before treatment. After washing the cells, the peptide-siRNA complexes or control samples prepared in 100 μl of Opti-MEM were added to the wells and incubated for 4 h. Thereafter, 50 μl of 30% serum containing medium was added, and the cytotoxicity of the relevant reagents was determined by the CCK-8 assay after 48 h. The reagent was added according to the manufacturer’s instructions, and the absorbance at 570 nm was measured with a plate reader (BMG, FLUOstar OPTIMA, Germany). The background absorbance of the multiwell plates at 690 nm was determined and subtracted from the 570-nm measurement. The results obtained from triplicate wells were averaged and normalized to the value obtained from non-treated samples.
Cellular uptake
In this research, siRNA is labeled with Cy3 at the 5′-end of the sense strand and peptide C6M3 is not labeled. CHO-K1 cells were seeded in a 24-well plate 24 h before treatment. The cells were incubated with C6M3-Cy3-labeled GAPDH siRNA complexes at a molar ratio of 20/1 for 4 h. Thereafter, the cells were washed three times with PBS and then washed with heparin (10 U/ml) following the method reported before [27] to remove extracellularly bound C6M3-siRNA complexes. After the thorough wash, the cells were fixed by 4% paraformaldehyde (PFA). The nucleus of the cell was stained by DAPI. The image showing the intracellular localization of Cy3-labeled siRNA was taken with a fluorescence microscope.
The cellular uptake of Cy3-labeled siRNA was quantified using flow cytometry (BD Biosciences, BD FACS Vantage SE Cell Sorter, USA). CHO-K1 cells (50,000) were incubated in 24-well plates for 24 h. The medium was replaced with Opti-MEM (Invitrogen) containing complexes or control samples with a final siRNA concentration of 100 nM. After incubating for 4 h, the cells were rinsed with PBS and then washed with heparin (10 U/ml). After washing the cells, trypsin-EDTA was added to detach them from the plate. The cells were re-suspended in 4% PFA and placed in fluorescence-activated cell sorting (FACS) tubes for analysis.
In vitro gene silencing efficiency
CHO-K1 cells were seeded at 35,000 cells/well in 24-well plates and incubated for 24 h. Next, the cells were incubated with various complex formulations in Opti-MEM at 37 °C for 4 h. Then, the cells were incubated for an additional 48 h in complete culture medium. siRNA concentration was 100 nM.
For qRT-PCR analysis, the total RNA was extracted from the treated cells using the SV Total RNA Isolation System (Promega, CA, USA). A Nanodrop (Nanodrop spectrophotometer ND-1000, Thermo Scientific, Ottawa, CA) was used to determine the RNA concentrations. The RNA samples were reverse-transcribed into complementary DNA (cDNA) using a Bio-Rad iScript cDNA synthesis kit according to the manufacturer’s protocol. After the cDNA was synthesized, PCR was performed with Brilliant II Fast SYBR Green QPCR Master Mix (Agilent Technologies, Wilmington, DE, USA) using an Mx3005PTM real-time PCR system (Agilent Technologies). The sequences of the primers used for the mouse GAPDH gene are 5′-TTGCTGTTGAAGTCGCAGGAG-3′ and 5′-TGTGTCCGTCGTGGATCTGA-3′ (Sigma, Oakville, Ontario, Canada). Cyclophilin, a housekeeping gene, was used as an internal control to normalize the GAPDH gene expression. Mouse cyclophilin mRNA amplified using the following primers: 5′-AGGGTTTCTCCACTTCGATCTTGC-3′ and 5′-AGATGGCACAGGAGGAAAGAGCAT-3′ (Sigma, Oakville, Ontario, CA).
Statistical analysis
Results were expressed as mean values ± SD. Data were analyzed by two-tailed t test, and only P values <0.05 were considered statistically significant.
Results
Physicochemical characterization of siRNA complexes
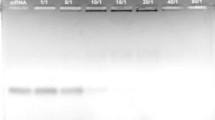
The binding of siRNA with C6M3 was investigated using agarose gel electrophoresis. A 21-mer siRNA that targets the eGFP gene was used in this experiment. C6M3-siRNA complexes prepared using a series of molar ratios were investigated. As shown in Fig. 1, lane 1, siRNA alone easily migrated through the gel due to its small size and negative charge. Due to the larger size and positive charge of the cationic C6M3-siRNA complexes, their migration through the gel was impeded. At a molar ratio of 10/1, the siRNA molecules were all bound to C6M3 because no free siRNA was detected in the agarose gel.
Binding ability of siRNA to C6M3 studied by agarose gel shift assay. The formed C6M3-siRNA complexes, stained with ethidium bromide, were investigated by electrophoresis on agarose gel (1.2% w/v). siRNAs, targeting eGFP genes, were complexed with C6M3 at a series of molar ratios from 1/1 to 80/1. Lane 1 was siRNA control, and lanes 2–8 indicated correlated molar ratios. The amount of siRNA was 300 ng/well
The interactions between C6M3 and siRNA were further studied by ITC, while the siRNA (in RNase-free water) was titrated with C6M3 at 25 °C. As shown in Fig. 2a, the heat exchange during the titration of siRNA by C6M3 was detected by the machine and output as raw data. By fitting the raw ITC data to a single-site model (Fig. 2b), the thermodynamic parameters of the interaction are obtained and listed in Table 1.
Calorimetric titration of siRNA with C6M3 at room temperature in RNase-free water. a Corrected thermogram of calorimetric titration of siRNA with C6M3. b Binding analysis of siRNA with C6M3 by fitting the raw data with an independent model. C6M3 concentration was 250 μM, and siRNA concentration was 10 μM
The obtained molar stoichiometry of 7.0 was the same as the theoretical value of 7, considering that the C6M3 peptide contains six positively charged arginine residues and there are 21 pairs of negatively charged nucleotides in the siRNA molecule. With an enthalpy of −16.1 kJ/mol, a ΔS of 117.0 J/(mol K), and an entropy of −34.9 kJ/mol, the binding was predominantly entropy-driven. Moreover, the Gibbs free energy calculated using the equation ΔG = ΔH − TΔS was −51.0 kJ/mol, indicating that the C6M3 peptide and siRNA formed complexes spontaneously [27–29].
Table 2 shows the detailed results of the size and charge characterization of C6M3-siRNA formulations evaluated in this study. As shown in Table 2, the particle size decreased over the increase of molar ratios. The possible reason is that at higher molar ratios, the nanoparticles were more condensed [30]. The size of C6M3-siRNA complexes remained in the range of 70–100 nm. At a molar ratio of 10/1, the C6M3-siRNA complexes have an approximately neutral zeta potential. Increasing the molar ratios, C6M3-siRNA complexes exhibited a more positive surface charge due to the addition of more cationic peptides. In order to increase the affinity of C6M3-siRNA complexes with cell membranes, we usually use molar ratios higher than 10/1 for the cell transfection experiments. The size range and positive surface charge of C6M3-siRNA complexes indicated that the complexes most possibly entered cells through endocytosis [31, 32].
Table 3 shows that in RNase-free water, after incubation for 40 min at 25 °C, the particle size of C6M3-siRNA complexes (molar ratio 40/1) slightly increased. When the incubation time was increased to 90 min, the particle size increased to 140 nm, possibly due to the aggregation of nanoparticles. This data indicated that the favorable incubation time before treatment was between 20 and 40 min. Table 3 shows that after the peptide C6M3 and siRNA formed complexes at 25 °C, the particle size changed after incubation at 37 °C for 30 and 60 min, respectively. At 25 °C, C6M3-siRNA complexes (molar ratio 40/1) in RNase-free water displayed size distribution around 70 nm; at 37 °C, the particle size decreased to 50 nm (30 min). Considering that the C6M3-siRNA complex formation process is entropy driven (ITC data), increasing the temperature leads to the increase of hydrophobic interactions, which may increase the compactness of the particles. Additionally, at 37 °C, increasing the incubation time to 60 min, the particle size remained almost the same as that after incubation for 30 min, possibly because the strong hydrophobic interactions increase the probability of interparticle interactions and the increased compactness of the particles increases the net surface charge to achieve higher stability. At 25 °C, C6M3-siRNA complexes (molar ratio 40/1) prepared in Opti-MEM exhibited size distribution around 220 nm (Table 3), which was much higher than that prepared in RNase-free water. This difference is possibly caused by the ionic effect of components within the Opti-MEM.
In Fig. 3a, the white dots are the particles of C6M3-siRNA complexes, which display a spherical shape. Moreover, the size distribution of C6M3-siRNA complexes was consistent with the DLS results. The impact of siRNA on the secondary structure of C6M3 was evaluated using CD spectroscopy. As shown in Fig. 3b, C6M3 in water exhibited a maximum spectrum of approximately 190 nm and a minimum of approximately 208 nm, which together indicated a helical conformation. When a small amount of siRNA (molar ratio of 20/1) was added, a shift in the spectrum minimum from 207 to 210 nm and a second minimum spectrum of approximately 222 nm was observed. Adding more siRNAs to attain a molar ratio of 10/1 increased the absolute values in spectrum minima at 208 and 222 nm and the maximum around 190 nm. These results indicated that an increase in the α helical secondary structure of the peptide occurred upon the addition of siRNA. The more siRNA was added, the more helical content of C6M3 secondary structure adopted. The conformational changes of the peptide after forming complexes with siRNA confirm that once siRNA neutralized some of the positive charges of the peptide, the repulsion between these charges decreased. Therefore, it became much easier for the peptide to form typical α helical secondary structure that was reported to facilitate the cellular uptake of peptide-siRNA complexes [33].
a Morphology of C6M3-siRNA complexes at a molar ratio of 20/1, where siRNA concentration was 100 nM. b CD spectra of C6M3 alone and C6M3-siRNA at different molar ratios. MR molar ratio (peptide/siRNA). C6M3 concentration was fixed as 30 μM, and C6M3-siRNA complexes were formulated at molar ratios of 10/1 and 40/1
Cytotoxicity of C6M3-siRNA complexes
As shown in Fig. 4, the viability of cells treated with C6M3-siRNA complexes was more than 90%. However, cells treated with Lipofectamine 2000 showed significant toxicity, with cell viability of 57%. These results clearly show that peptide C6M3 induced much lower cytotoxicity than Lipofectamine 2000. Moreover, peptide alone at corresponding concentrations of different molar ratios achieved >90% cell viability.
Viability of CHO-K1 cells after 48-h treatment. N.T. non-treated, MR peptide/siRNA molar ratio, Lipo Lipofectamine 2000 (1 μl of Lipo/well). siNRA concentration was at 100 nM. *P value <0.05, the cell viability after GL1 treatment is statistically significant than that after Lipo treatment. Results are expressed as mean ± standard deviation. n = 3
Cellular uptake of siRNA complexes
CHO-K1 cells were treated with the C6M3-Cy3-labeled siRNA complexes to study the intracellular localization and cellular uptake of siRNA. When provided in complexes with a peptide/siRNA molar ratio of 20/1 (Fig. 5), the siRNA was localized to regions in close proximity to the nuclear membrane. The siRNAs were distributed in a non-homogeneous pattern at the periphery of the nucleus, indicating the possibility of endocytotic delivery [34]. The efficiency of C6M3 to deliver siRNA into CHO-K1 cells was evaluated using FACS. As shown in Fig. 6, the treatment of CHO-K1 cells with Cy3-labeled GAPDH siRNA alone achieved almost no uptake, implying that siRNA without an efficient carrier was not able to enter the intracellular environment. However, the efficiency of cellular uptake of the siRNA significantly increased with the increase in the C6M3 peptide/siRNA molar ratio from 10/1 to 40/1. In addition, at molar ratios of 40/1, C6M3-Cy3-labeled siRNA complexes entered significantly more cells than Lipofectamine 2000-siRNA complexes, suggesting the pronounced delivery efficiency of C6M3.
The fluorescence microscopy image of CHO-K1 cells. The red fluorescence indicated Cy3-labeled siRNA; blue fluorescence represented DAPI-stained nuclei. The lower panel was non-treated (NT) cells, used as a control. The upper panel was treated with C6M3-siRNA complexes at molar ratio 20/1, and siNRA concentration was at 100 nM
Cellular uptake of Cy3-labeled siRNA. Non-treated (N.T.) sample was negative control; Lipofectamine 2000 (Lipo) was positive control (1 μl of Lipo/well). MR molar ratio (peptide/siRNA). siRNA concentration was at 100 nM. *P value <0.05; the difference of cellular uptake efficiency of siRNA delivered by C6M3 at a molar ratio of 40/1 and Lipofectamine 2000 is significant. Results are expressed as mean ± standard deviation. n = 3
In vitro transfection
The efficiency of GAPDH gene silencing by C6M3-siRNA complexes at the mRNA level was investigated using qRT-PCR. Lipofectamine 2000 was used as the positive control. As shown in Fig. 7, the Lipofectamine 2000-siRNA complexes induced the silencing of GAPDH gene by 84%, and the C6M3-siRNA complexes at 40/1 achieved 69% GAPDH gene silencing efficiency. The significant silencing efficiency of GAPDH gene induced by C6M3-siRNA complexes implied that the C6M3 peptide could effectively deliver bioactive siRNA into the cytosol and induce specific gene silencing.
GAPDH gene silencing in CHO-K1 cells. GAPDH siRNA concentration was 100 nM. NT non-treated sample, Lipo Lipofectamine 2000 (1 μl of Lipo/well), MR peptide/siRNA molar ratio. *P value <0.05, the gene silencing efficiency at a molar ratio of 40/1 and 60/1 is significantly different from Lipo-siRNA treatment. Results are expressed as mean ± standard deviation. n = 3
Discussion
Here, we evaluated the ability of C6M3 to form complexes with siRNA and deliver siRNA to the cytosol with concomitant silencing of GAPDH expression. Based on the data of ITC and zeta potential, the complex formation is not only involving electrostatic force but also hydrophobic interaction. To investigate whether this complex formation could promote internalization of the peptide-siRNA complexes, FACS, a quantitative method using fluorescently labeled siRNA, was performed. Interestingly, with the increase of molar ratios from 10/1 to 40/1, the uptake efficiency of siRNA increased. The possible reasons are that at higher molar ratios, the peptide-siRNA complexes display higher positive charge resulting in stronger affinity with the negatively charged cell membrane and higher contents of tryptophan enhancing the peptide-membrane interaction.
The modification of C6 has resulted in the following changes. With the same siRNA concentration, siRNA molecules were entirely associated with C6M3 at a molar ratio of 10/1, while a very small amount of free siRNA was observed when interacting with C6 at a molar ratio of 10/1. This result is further confirmed by the zeta potential data. At a molar ratio of 10/1, C6M3-siRNA complexes exhibited a slight positive surface charge (0.4 mV), while C6-siRNA complexes obtained a negative surface charge (−13.1 mV). Moreover, C6M3-siRNA complexes showed smaller particle sizes (70–100 nm) than C6-siRNA complexes (170–260 nm). The ITC data showed that both C6 and C6M3 could spontaneously form complexes with siRNA. However, a higher value of Gibbs free energy was observed when titrating siRNA with C6M3 (−50 kJ/mol) than with C6 (−40 kJ/mol), suggesting the interaction of siRNA and C6M3 is highly favorable. In water, C6M3 adopted helical secondary structure, while C6 adopted random coil structure. This conformational change may facilitate C6M3 forming complexes with siRNA more efficiently with relatively small particle sizes due to the defined helical secondary structure.
C6M3-siRNA complexes exhibited particle size ∼100 nm in water and ∼200 nm in Opti-MEM. Previous studies show that particles with the size range from 50 to 200 nm usually enter cells through caveolae and clathrin-mediated endocytotic pathway [35]. Fluorescence microscopy images of C6M3-Cy3-labeled siRNA complexes (Fig. 5) also indicated that the complexes entered cells possibly through endocytotic pathway [19]. Previous studies showed that histidine residues were incorporated in CPPs to enhance the endosomal escape of siRNAs [36, 37]. Due to the pH drop in endosome, the imidazole ring of histidine was protonated. The protonation induced an extensive inflow of water and ions into the endosomal environment, thereby resulting in disruption of the endosomal membrane and release of the entrapped components [24]. Taken together, the incorporation of tryptophan and histidine in C6M3 sequence led to over two fold increase of GAPDH silencing efficiency compared to that of C6-siRNA complexes (25%), confirmed by the qRT-PCR analysis.
Conclusions
C6M3, an 18-mer amphipathic peptide, formed complexes with siRNA exhibiting particle sizes from 50 to 220 nm. C6M3 adopted helical secondary structure in water and became more so upon binding to siRNA. Zeta potential and agarose gel experiments proved that siRNA molecules were completely associated with C6M3 at a molar ratio of 10/1. ITC data revealed that the stoichiometry between C6M3 and siRNA is 7/1, which is consistent with the theoretical value considering that each peptide consists of six positive charge residues (R) and siRNA contains 21 negative charge base pairs. In addition, FACS results revealed that higher molar ratios achieved better siRNA uptake efficiency because higher amount of peptides induced stronger interaction with the cell membrane. GAPDH gene silencing experiment and cell viability data showed that C6M3-siRNA complexes induced 69% silencing of GAPDH gene with minimal cytotoxicity (<10%). These data demonstrate that C6M3 is a promising carrier for siRNA delivery and the modification strategy is useful.
References
Kim WJ, Kim SW. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm Res. 2009;26:657–66.
Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–38.
Yao Y, Wang C, Varshney RR, Wang D-A. Antisense makes sense in engineered regenerative medicine. Pharm Res. 2009;26:263–75.
Saghir Akhtar IFB. Nonviral delivery of synthetic siRNAs in vivo. J Clin Inverstigation. 2007;117:3623–32.
Krebs MD, Alsberg E. Localized, targeted, and sustained siRNA delivery. Chemistry. 2011;17:3054–62.
Monaghan M, Pandit A. RNA interference therapy via functionalized scaffolds. Adv Drug Deliv Rev Elsevier BV. 2011;63:197–208.
Krebs MD, Jeon O, Alsberg E. Localized and sustained delivery of silencing RNA from macroscopic biopolymer hydrogels. J Am Chem Soc. 2009;131:9204–6.
Takahashi Y, Nishikawa M, Takakura Y. Nonviral vector-mediated RNA interference: its gene silencing characteristics and important factors to achieve RNAi-based gene therapy. Adv Drug Deliv Rev. 2009;61:760–6.
Zhen G, Hinton TM, Muir BW, Shi S, Tizard M, McLean KM, et al. Glycerol monooleate-based nanocarriers for siRNA delivery in vitro. Mol Pharm. 2012;9:2450–7.
Wang J, Lu Z, Wientjes MG, Au JL-S. Delivery of siRNA therapeutics: barriers and carriers. AAPS J. 2010;12:492–503.
Meade BR, Dowdy SF. Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev. 2007;59:134–40.
Deshayes S, Heitz A, Morris MC, Charnet P, Divita G, Heitz F. Insight into the mechanism of internalization of the cell-penetrating carrier peptide Pep-1 through conformational analysis. Biochemistry. 2004;43:1449–57.
Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–24.
Khalil I, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45.
Fung SY, Yang H, Sadatmousavi P, Sheng Y, Mamo T, Nazarian R, et al. Amino acid pairing for de novo design of self-assembling peptides and their drug delivery potential. Adv Funct Mater. 2011;21:2456–64.
Wang M, Law M, Duhamel J, Chen P. Interaction of a self-assembling peptide with oligonucleotides: complexation and aggregation. Biophys J Elsevier. 2007;93:2477–90.
Sadatmousavi P, Soltani M, Nazarian R, Jafari M, Chen P. Self-assembling peptides: potential role in tumor targeting. Curr Pharm Biotechnol. 2011;12:1089–100.
Jafari M, Chen P. Peptide mediated siRNA delivery. Curr Top Med Chem. 2009;9:1088–97.
Jafari M, Xu W, Naahidi S, Chen B, Chen P. A new amphipathic, amino-acid-pairing (AAP) peptide as siRNA delivery carrier: physicochemical characterization and in vitro uptake. J Phys Chem B. 2012;116:13183–91.
Reithmeier R a F. Characterization and modeling of membrane proteins using sequence analysis. Curr Opin Struct Biol. 1995;5:491–500.
Von Heijne G. Membrane proteins: from sequence to structure. Annu Rev Biophys Biomol Struct. 1994;23:167–92.
Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RAF. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–8.
Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–8.
Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511.
Zimenkov Y, Dublin SN, Ni R, Tu RS, Breedveld V, Apkarian RP, et al. Rational design of a reversible pH-responsive switch for peptide self-assembly. J Am Chem Soc. 2006;128:6770–1.
Lee ES, Oh KT, Kim D, Youn YS, Bae YH. Tumor pH-responsive flower-like micelles of poly(l-lactic acid)-b-poly(ethylene glycol)-b-poly(l-histidine). J Control Release. 2007;123:19–26.
Ladbury JE, Chowdhry BZ. Sensing the heat: the application of isothermal titration calorimetry to thermodynamic studies of biomolecular interactions. Chem Biol. 1996;3:791–801.
Pierce MM, Raman CS, Nall BT. Isothermal titration calorimetry of protein-protein interactions. Methods. 1999;19:213–21.
Chen B, Xu W, Pan R, Chen P. Design and characterization of a new peptide vector for short interfering RNA delivery. J Nanobiotechnology. 2015;13:1–10.
Lehto T, Simonson OE, Mäger I, Ezzat K, Sork H, Copolovici D-M, et al. A peptide-based vector for efficient gene transfer in vitro and in vivo. Mol Ther Nature Publishing Group. 2011;19:1457–67.
Conner SD, Schmid SL. Regulated portals of entry into the cell. Nat. 2003;422:37–44.
Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8:848–66.
Pujals S, Fernández-Carneado J, López-Iglesias C, Kogan MJ, Giralt E. Mechanistic aspects of CPP-mediated intracellular drug delivery: relevance of CPP self-assembly. Biochim Biophys Acta. 2006;1758:264–79.
Veldhoen S, Laufer SD, Trampe A, Restle T. Cellular delivery of small interfering RNA by a non-covalently attached cell-penetrating peptide: quantitative analysis of uptake and biological effect. Nucleic Acids Res. 2006;34:6561–73.
Kolli S, Wong SP, Harbottle R, Johnston B, Thanou M, Miller AD. PH-triggered nanoparticle mediated delivery of siRNA to liver cells in vitro and in vivo. Bioconjug Chem. 2013;24:314–32.
Samir E-a, Henrik J. J, Potus L, Ulo L. Induction of splice correction by cell-penetrating peptide nucleic acids. J Gene Med. 2006;8:1262–73.
Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat Med. 2004;10:310–5.
Acknowledgements
Financial support from the Canadian Foundation for Innovation (CFI), the Natural Sciences and Engineering Research Council of Canada (NSERC), the Canadian Research Chairs (CRC) program, and Waterloo Institute of Nanotechnology (WIN) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We declare that the experiments comply with the current laws of Canada in which they were performed.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Chen, B., Yoo, K., Xu, W. et al. Characterization and evaluation of a peptide-based siRNA delivery system in vitro. Drug Deliv. and Transl. Res. 7, 507–515 (2017). https://doi.org/10.1007/s13346-017-0371-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13346-017-0371-x