Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) nucleocapsid protein (NP) is the immunodominant region of PRRSV viral proteins. Non-structural protein 2 (Nsp2) and its hypervariable region play an essential role in the differential diagnosis of PRRSV. Western blot and immunofluorescence assay (IFA) analyses found that 2 out of 18 monoclonal antibodies (MAbs) recognized the NP and that 5 of 11 MAbs recognized Nsp2–120aa. IFA data demonstrated that 2 MAbs raised against the NP have a positive reaction to PRRSV; either HP-PRRSV, classic PRRSV or the vaccine strain at 1:100 dilution. Two MAbs raise against Nsp2–120aa also react positively with the classic PRRSV nor HP-PRRSV, but not with the PRRSV vaccine strain TJM-F92. Epitope mapping using truncated proteins identified a novel Nsp2–120aa epitope. In addition, we show that MAb BR/PNsp2-2A20 recognizes a 20 amino acid peptide (707) GRFEFLPKMILETPPPHPCG (727) of Nsp2. Based on our findings, we propose that MAb BR/PNsp2-2A20, raised against Nsp2-120aa of PRRSV, as a candidate specific diagnostic MAb for differentiation of the PRRSV virulent strains infected pig from vaccine strain TJM-F92 inoculated ones. The MAbs developed here have potential for use in diagnostic and research tools, including immunofluorescence assay, enzyme-linked immunosorbent assay and Western blotting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine reproductive and respiratory syndrome (PRRS), causing late-term reproductive failure and severe pneumonia in neonatal pigs, continues to have an economically significant impact on the swine industry worldwide. Porcine reproductive and respiratory syndrome virus (PRRSV) is an enveloped single stranded positive-sense RNA virus classified in the order Nidovirales, the family Arteriviridae, and the genus Arterivirus [3, 6]. The viral genomic RNA contains at least nine open reading frames (ORFs)-ORF1a, ORF1b, ORFs 2 (2a and 2b), ORF3 and 4 and ORF5s (5a and 5b) to 7, as well as the 5′ and 3′ untranslated regions [2, 11, 18, 26]. ORF1a and ORF1b generate at least 14 nonstructural proteins (Nsps) by autocatalytic processing. ORF1a encodes Nsp1α, Nsp1β, Nsp2–Nsp6, Nsp7α, Nsp7β and Nsp8, and ORF1b encodes Nsp9–Nsp12 [4, 9, 14, 21]. Among the nonstructural proteins of PRRSV, Nsp2 is the largest part of the cleavage product of the replicative protein. The Nsp2 protein of PRRSV can be divided into three major domains: the conserved N-terminal cysteine proteinase domain (PLP2/OTU), a hydrophobic transmembrane region at the C terminus; a large hypervariable domain (HV); and a transmembrane domain (TM), which are located between the former two domains [5, 10, 22, 23]. Porcine reproductive and respiratory syndrome virus (PRRSV) was divided into North American and European genotypes [1]. The highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) that emerged in 2006, shares an identical discontinuous deletion of 30 amino acids (aa) in Nsp2 [13, 15, 29]. The TJM-F92 vaccine strain derived from the HP-PRRSV TJ strain by passage 92 times in the African green monkey kidney epithelial cell line Marc-145 has an additional 120-aa deletion in Nsp2 [24, 25].The additional 120-aa deletion in Nsp2 (aa 628–747) located between the HV1 and HV2 regions in a relatively conserved region of Nsp2 [12].
The nucleotide protein (NP), encoded by the PRRSV ORF7 gene is considered to be one of the most conserved PRRSV proteins [17]. In addition, NP has rich antigenic determinants, which induce production of long-time antibodies after the PRRSV infects cells. NP has good immunogenicity and reaction activity [8, 20, 27, 28].
In recent decades, highly specific monoclonal antibodies (MAbs) have been exploited to provide reliable diagnoses for many diseases. The MAbs have monovalent affinity due to their binding to the same epitope, and can specially recognize antigenic. MAbs are important tool in biochemistry, molecular biology and medicine. MAbs are also very useful for identification and antigenic characterization of pathogens in animal disease diagnosis. Here we produced MAbs against PRRSV NP and PRRSV Nsp2-120aa. The MAbs against PRRSV NP can be used to diagnose PRRS, and can identify all of the North American genotype PRRSV strains. The MAbs raised against Nsp2-120aa can be used to differentiate PRRSV infection from the vaccine inoculation of TJM-F92 in pigs.
Materials and methods
Cells, animals, viruses and plasmids
E. coli strains (DH5α)and BL21 (DE3) were purchased from TRANSGEN BIOTECH. Marc-145 cells and Mouse myeloma cells (Sp2/0) were purchased from ATCC. Marc-145 cells were cultured in MEM medium (Gibco, USA), 1% solution of antibiotics (streptomycin and penicillin—Sangon Biotech), and 10% fetal bovine serum (FBS—Gibco). Sp2/0 cells were maintained in IMDM medium (Invitrogen, USA) with 15% FBS. Cells were cultured at 37 °C and 5% CO2. Balb/c mice were obtained from the Animal Centre of Peking University Health Science Center. PRRSV JL-04/12 was a HP-PRRSV isolated in 2012. PRRSV TJM-F92 was a commercial vaccine strain. We made use of a series of prokaryotic expression plasmids constructed by Liu et al. [16]: Nsp2 (628–727), Nsp2 (628–707), Nsp2 (628–707), Nsp2 (648–747), Nsp2 (668–747), Nsp2 (688–747) were. The expression plasmids pGEX-NP and pGEX-dNsp2, expressing PRRSV NP and Nsp2-120aa respectively, were constructed by our research group using standard cloning methods.
Bacterial expression and purification of N and Nsp2-120aa recombinant protein
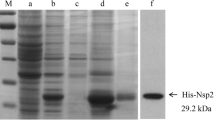
pGEX-NP and pGEX-dNsp2 were transformed into E. coli BL-21 cells and expression of the NP and Nsp2-120aa induced with 0.1 mmol/L isopropyl-β-d-thiogalactoside (IPTG). Expression of the NP and Nsp2-120aa proteins was confirmed by sodium dodecyl sulfate polyacrylamide gelelectrophoresis (SDS-PAGE) and western blotting. The proteins were purified using a B-PER (tm) GST Purification Kit (Thermo, USA) according to the manufacturer’s instructions.
Immunization of the animals
Four mice were inoculated on day zero with two subcutaneous doses of 0.05 mL containing 60 μg concentrated NP or Nsp2-120aa, diluted 1:1 in Freund’s Incomplete Adjuvant into the footpad of the hind legs. Three subsequent booster inoculations were made at half doses at 14 day intervals. Antibody titers were determined by enzyme-linked immunosorbent assay (ELISA) from 10 to 36 days after the first booster. The mice with the highest antibody titers were chosen for cell fusion. A fourth booster immunization was given to the chosen mice with 50 µg of pathogen by intraperitoneal injection 3 days before cell fusion.
Production of monoclonal antibodies from hybridoma cell
Cell fusion was performed three days after the last immunization. Mouse myeloma Sp2/0 cells were cultured and propagated in RPMI-1640 culture medium (Gibco) and 10% fetal bovine serum (FBS) (Gibco). First, collecting growth well Sp2/0 cells, and cells transferred into a 50 ml centrifuge tube. The mice were euthanasia and soaked for 5 min in 75% alcohol. The thymocyte were isolated from the immunized mouse and mixed with the Sp2/0 cells at a ratio of 5:1 in IMDM free FBS, 2 ml of HAT was added, followed by centrifugation at 1500 rad/min for5 min. The supernatant was discarded and cells gently resuspended with IMDM free FBS for washing. Then, 1 ml of PEG was slowly added to the cells under 37 °C warm water and stood for 1 min. Ten milliliters of IMDM free FBS was added within 2 min (2 ml initially, then a further 8 ml), followed by centrifugation at 1000 rad/min for 5 min. The cells were carefully resuspended in 10 ml FBS after pouring away the supernatant and then added to the prepared thymus cells. Sterile semi-solid medium (25 ml) was added and divided into culture dishes. The cells were cultured in a hot and humid incubator.
Characterization of monoclonal antibodies
Monoclonal antibody (Mab) isotypes were determined by enzyme linked immunosorbent assay (ELISA) using a mouse isotyping primary antibody (Southern Biotech) and various subtype secondary antibodies (Southern Biotech). PBS with 2% BSA and 3% sucrose was used as a blocking solution. Ten microliters of 30% H2O2 and 0.2 ml H2O with 15 mg/ml diammonium salt were added into 10 ml of sodium citrate-hydrochloric acid buffer as color-substrate solution.
Reactivity of MAbs with PRRSV Screening test—indirect immunofluorescence
The chosen MAbs were screened using indirect immunofluorescence (IFA). High pathogenic PRRSV JL-04/12 and vaccine strain TJM-F92 were inoculated in Marc-145 cells. After 48 h, the cells were fixed with 70% ethyl alcohol. The hybridoma supernatant of NP and Nsp2-120aa were used as primary antibody and goat anti-mouse IgG Labeled HRP as secondary antibody. The antibody incubation times were 45 min at 37 °C. The results were observed under fluorescence microscope (Leca, Japan).
Western blotting confirm the binding activity of MAbs and PRRSV
To verify the reactivity against recombinant proteins and PRRSV, the supernatants of the obtained stable cell clones were tested by western blotting, using recombinant proteins and PRRSV JL-04/12 and TJM-F92 cell cultural lysates as antigens. The antigens were resolved by 12% SDS-PAGE and transferred onto 0.22 μm PVDF membrane (Millipore, USA)at room temperature and 5 mA for 2 h. The membrane was blocked with 5% skimmed milk in PBS-T overnight at 4 °C. The culture supernatants of the desired hybridoma cells were added as the primary antibodies. The horseradish peroxidase (HRP) conjugated goat anti-mouse IgG antibody (Sigma, USA) served as the secondary antibody, whose HRP activity was detected using an Enhanced ECL chemilumine scence detection kit (concentrated) (Vazyme, China).
Peptide identification of MAbs of Nsp2-120aa
A series of prokaryotic expression plasmids [Nsp2 (628–727), Nsp2 (628–707), Nsp2 (628–707), Nsp2 (648–747), Nsp2 (668–747) and Nsp2 (688–747)] were transfected into Marc-145 cells. At 36 h after transformation, the cells were fixed with 70% ethyl alcohol. Then IFA was done to figure out which plasmid will act with the chosen MAbs. So we can verified the corresponding sequence of MAb.
Purification of MAbs
BALb/c mice were injected with 0.5 ml incomplete Freund’s adjuvant in the abdominal cavity 1 week in advance. Then, the thrive hybridoma were centrifuged and the culture medium discarded. Cells were resuspended in PBS or incomplete medium and injected into the abdominal cavity of the mice. Seven to 10 days later, ascitic fluid was sucked from the enlarged abdomen with a syringe and centrifuged to remove impurities (e.g., cells, protein and fat). The MAbs were purified using a Protein A IgG Purification Kit (Thermo Fisher Scientific, USA) following the manufacturer’s instructions. We were able to recover MAbs with relatively high purity and good antibody activity.
Affinity determination of MAbs
We determined the affinity constant of the MAbs using competition ELISA. The antigen was coated with (citrate-buffered saline) CBS at 2 µg/ml and the hybridoma supernatant of the MAbs diluted from 1:200 to 1:204,800. The antigens were gradient diluted two times. The diluted MAbs were mixed with an equal volume of the corresponding diluted antigens and incubating for 20 h at room temperature. The mixture was added into the wells of a 96-well plate and incubated for 30 min at 37 °C. After three washes with PBS-T, then added secondary antibody diluted to 1:10,000, and incubated for 30 min at 37 °C. Following a further three washes, the substrate TMB was added and incubated for 10 min. The action was stopped with 0.5 M H2SO4. The optical density (OD) was detected using a BIO-RAD Microplate reader. The affinity constant was calculated using the formula (150,000 × A)/original concentration of the MAb.
Results
Production and characterization of the MAbs against PRRSV NP or Nsp2-120aa
The mouse with higher antibody titer were chosen and splenocytes were sterile collected and fused with sp2/0 cells. Thirty-two clones of NP (of 930 wells tested) and 23 clones of Nsp2-120aa (of 930 wells tested) tested positive by ELISA. In order to further determine the positive clones, further ELISA using PRRSV NP (for MAb of NP) or Nsp2-120aa (for MAb of Nsp2-120aa) and GST as antigens were performed. Eighteen hybridomas of NP and 11 hybridomas of Nsp2-120aa were selected. NP MAbs were characterized as 7 IgG2b and 8 IgG1 of the kappa light chain. Nsp2-120aa MAbs were characterized as 6 IgG2a, 1 IgG2b and 4 IgG1. The clone data are listed in supplementary tables 1 and 2.
Detection of interaction of MAbs and PRRSV by IFA
To examine if the obtained MAbs against PRRSV NP and Nsp2-120aa could react against PRRSV, HP-PRRSV JL-04/12 was infected into Marc-145 cells for IFA, with the MAbs as the primary antibody. These IFA data show that 2 of 18 the NP MAbs (# 13 and 29) and 5 of the 11 Nsp2-120aa MAbs (# 8, 13, 14, 15, 17) reacted positively with PRRSV (part of data showed in Fig. 1b). By IFA, we detected cells with strong green fluorescent both between NP MAbs and HP-PRRSV JL-04/12 and Nsp2-120aa MAbs and HP-PRRSV JL-04/12. This fluorescence localizes to different regions of the cells with different MAbs. NP MAbs showed strong cytoplasm staining in PRRSV infected MARC-145 cells in an immunofluorescence assay. Whereas Nsp2-120aa MAbs produced strong perinuclear staining and some cytoplasm staining.
a The schematic diagram of Nsp2-120aa overlapped truncated expression. The 120 amino acid of Nsp2 is a deleted region in Nsp2 of PRRSV vaccine strain TJM-F92 during the cell passages of its parental virus TJ strain. The 20 amino acids (aa707-727) are the polypeptide recognized by MAb BR/PNsp2-2A20. b The detection of MAbs reactivity with PRRSV by IFA. Marc-145 cells was inoculated with PRRSV JL-04/12 strain, fixed after 48 postinfection. IFA test was done with MAbs as primary antibody and HRP labeled anti-mice IgG as secondary antibody. The MAbs developed in present study can bind very well with PRRSV growing in Marc-145 cells
One epitope domain of Nsp2-120aa MAbs was determined
Mapping of the 6 six epitopes was performed using a panel of expressed polypeptides: Nsp2 (628–727), Nsp2 (628–707), Nsp2 (628–707), Nsp2 (648–747), Nsp2 (668–747) and Nsp2 (688–747) (Fig. 1a). The polypeptide Nsp2 (628–727) could be recognized by all 5 MAbs of Nsp2-120aa chosen in this study (Fig. 2a). Interestedly, four transiently expressed polypeptides: Nsp2 (628–727), Nsp2 (648–747), Nsp2 (668–747) and Nsp2 (688–747) were all recognized by MAb #13 of Nsp2-120aa (Fig. 2a, b).The overlapping epitope polypeptide sequence was (707) GRFEFLPKMILETPPPHPCG(727). We named this MAb BR/PNsp2-2A20.
MAbs binding activity with antigens or PRRSV by Western blotting
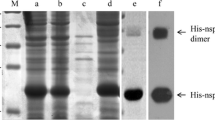
The recombinant proteins, HP-PRRSV JL-04/12 and PRRSV vaccine strain TJM-F92 were used as antigens during western blotting. The corresponding MAbs were used as the primary antibodies. Our data show that the MAbs against PRRSV NP strongly bind to the NP expressed by HP-PRRSV and the vaccine strain in Marc-145 cells (Fig. 3a). The corresponding MAbs of PRRSV Nsp2-120aa also had a strong binding activity with Nsp2-120aa. These data indicate that the MAbs recovered here were targeting the correct epitopes of the recombinant proteins. However, no apparent expected reaction band was seen when probing PRRSV strains with the MAbs raised against PRRSV Nsp2-120aa (Fig. 3b).
Western blot detection of reaction between recombinant expressed proteins, PRRSV and corresponding MAbs. a NP MAb No. 13. b Nsp2-120aa MAb No. 8. Lane 1: Recombinant expressed protein; Lane 2: Protein molecular weight ladder, WB Standard Marker 1.0 (HangZhou GoodHere Technology Co., Ltd. China, HangZhou); Lane 3: PRRSV JL-04/12; Lane 4: PRRSV TJM-F92; Lane 5: Marc-145 cells control
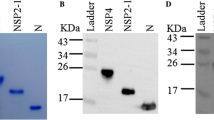
Purification and affinity determination of MAbs
We had purified the four MAbs against either PRRSV NP or Nsp2-120aa. The size is right and purity is good by SDS-PAGE detection (Fig. 4a, d). From the affinity curve, we obtained the affinity constant for each MAb. The affinity constant of #29 and #13 PRRSV NP MAbs are 8.99E + 09 and 7.93E + 09, respectively (Fig. 4b, c). The affinity constant of the No. 13 and No. 14 PRRSV Nsp-120aa MAbs were estimated as 2E + 06 and 5E + 09, respectively (Fig. 4e, f).
Discussion
MAbs directed against a single epitope should be homogeneous, highly specific and producible in unlimited quantities. MAbs generally exhibit exquisite specificity for the target antigen. In this study, we constructed are combinant expressed vector for the PRRSV NP and Nsp2-120aa. The recombinant proteins were successfully recovered from E.coli BL-21 and purified. The purified recombinant proteins were inoculated mice. After three rounds of screening by ELISA and IFA, we identified two NP MAbs (#13 and 29) and five Nsp2-120aa MAbs (#8, 13, 14, 15, 17). MAb subtype data indicated that we had obtained two G2b subtype NP MAbs and two G1 and 3 G2a subtype Nsp2-120aa MAbs. IFA data demonstrated that 2 MAbs raised against the NP have a positive reaction to PRRSV; either HP-PRRSV, classic PRRSV or the vaccine strain. All five Nsp2-120aa MAbs (#8, 13, 14, 15, 17) exhibited immunoreactivity with the denatured Nsp2-120aa recombinant protein in Western blotting and two MAbs produced strong cell membrane staining in PRRSV infected MARC-145 cells in an immunofluorescence assay. Two MAbs raise against Nsp2–120aa also react positively with the classic PRRSV nor HP-PRRSV, but not with the PRRSV vaccine strain TJM-F92.
All MAbs against NP and Nsp2-120aa showed good binding activity against the native PRRSV virion when analyzed by IFA, as well as clear reactivity with the corresponding recombinant proteins in western blotting. However, we did not find an reaction in Western blotting between MAbs of Nsp2-120aa and PRRSV JL-04/12 collected from Marc-145 cells. We propose that this result may be because of non-structural protein being expressed during the early period of virus replication, which is quickly degraded.
The epitope mapping of Nsp2-120aa was done by serial truncation of the Nsp2-120aa protein and identified Nsp-120aa epitope. Each truncation was transiently expressed in Marc-145 cells and detected with the developed MAbs by IFA. Finally, we identified an epitope recognized by MAb BR/PNsp2-2A20. The amino acid sequence is 20aa: (707) GRFEFLPKMILETPPPHPCG(727). Based on our findings, we propose that MAb BR/PNsp2-2A20, raised against Nsp2-120aa of PRRSV, as a candidate specific diagnostic MAb for differentiation of the PRRSV virulent strains infected pig from vaccine strain TJM-F92 inoculated ones.
A previous research has shown thatVR2385-R, with a large spontaneous deletion of 145 aa of the Nsp2 gene, has an overlapping region in its deletion (aa 581–725) [19] with the TJM-F92 deleted 120aa (aa 628-747). We previously suggested the aa 727–747 of Nsp2 were most likely related to PRRSV’s replication ability [25]. Now, in this study, we had obtained a MAb against a peptide corresponding to another region (aa 707–727). This peptide is located between the HV1 and HV2 regions. Previous research has shown that Nsp2 has 18 B cell linear epitopes and that these are scattered along the Nsp2 amino acid sequence [7]. The peptide sequence (707) GRFEFLPKMILETPPPHPCG(727) recognized by MAb BR/PNsp2-2A20 is not included in the 18 B-cell linear epitopes. It is a novel epitope peptide found in this study.
In summary, the purified MAbs reacted well with recombinant proteins and PRRSV. The affinity constants of these MAbs were calculated by ELISA and all shown to have a high affinity for the antigens, except #13 MAb against Nsp2-120aa. These MAbs can now be used applied in the future development of diagnostic methods. Here we obtained two MAbs against PRRSV NP and five MAbs against PRRSV Nsp2-120aa. By IFA using transiently expressed truncated proteins, we identified the peptide targeted by the MAb BR/PNsp2-2A20. MAbs were purified in preparation for future experiment.
References
Allende R, Lewis TL, Lu Z, Rock DL, Kutish GF, Ali A, et al. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol. 1999;80(Pt 2):307–15.
Bautista EM, Meulenberg JJ, Choi CS, Molitor TW. Structural polypeptides of the American (VR-2332) strain of porcine reproductive and respiratory syndrome virus. Arch Virol. 1996;141:1357–65.
Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–33.
Chen Z, Lawson S, Sun Z, Zhou X, Guan X, Christopher-Hennings J, et al. Identification of two auto-cleavage products of nonstructural protein 1 (nsp1) in porcine reproductive and respiratory syndrome virus infected cells: Nsp1 function as interferon antagonist. Virology. 2010;398:87–97.
Chen Z, Zhou X, Lunney JK, Lawson S, Sun Z, Brown E, et al. Immunodominant epitopes in Nsp2 of porcine reproductive and respiratory syndrome virus are dispensable for replication, but play an important role in modulation of the host immune response. J Gen Virol. 2010;91:1047–57.
Conzelmann KK, Visser N, Van Woensel P, Thiel HJ. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329–39.
de Lima M, Pattnaik AK, Flores EF, Osorio FA. Serologic marker candidates identified among B-cell linear epitopes of Nsp2 and structural proteins of a North American strain of porcine reproductive and respiratory syndrome virus. Virology. 2006;353:410–21.
Doan DN, Dokland T. Cloning, expression, purification, crystallization and preliminary X-ray diffraction analysis of the structural domain of the nucleocapsid N protein from porcine reproductive and respiratory syndrome virus (PRRSV). Acta Crystallogr D Biol Crystallogr. 2003;59:1504–6.
Fang Y, Snijder EJ. The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154:61–76.
Han J, Rutherford MS, Faaberg KS. The porcine reproductive and respiratory syndrome virus Nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J Virol. 2009;83:9449–63.
Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol. 2011;92:1107–16.
Kim DY, Kaiser TJ, Horlen K, Keith ML, Taylor LP, Jolie R, et al. Insertion and deletion in a non-essential region of the nonstructural protein 2 (Nsp2) of porcine reproductive and respiratory syndrome (PRRS) virus: effects on virulence and immunogenicity. Virus Genes. 2009;38:118–28.
Li Y, Wang X, Bo K, Tang B, Yang B, Jiang W, Jiang P. Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J. 2007;174:577–84.
Li Y, Tas A, Snijder EJ, Fang Y. Identification of porcine reproductive and respiratory syndrome virus ORF1a-encoded non-structural proteins in virus-infected cells. J Gen Virol. 2012;93:829–39.
Li Z, Leng X, Qi Q, Wang F, Wen Y, Tan B, et al. Complete genome sequence of a highly pathogenic porcine reproductive and respiratory syndrome virus NM1 strain from Northern China. J Virol. 2012;86:13863–4.
Liu Y, Wang FX, Wen YJ, Li ZG, Liu X, Sun N, et al. Effect of nonstructural protein 2 hypervariable regions in the replication of porcine reproductive and respiratory syndrome virus in marc-145 cells. Intervirology. 2015;58:288–96.
Meulenberg JJ, Petersen-den Besten A, de Kluyver EP, Moormann RJ, Schaaper WM, Wensvoort G. Characterization of structural proteins of Lelystad virus. Adv Exp Med Biol. 1995;380:271–6.
Meulenberg JJ, Petersen den Besten A, de Kluyver E, van Nieuwstadt A, Wensvoort G, Moormann RJ. Molecular characterization of Lelystad virus. Vet Microbiol. 1997;55:197–202.
Ni YY, Huang YW, Cao D, Opriessnig T, Meng XJ. Establishment of a DNA-launched infectious clone for a highly pneumovirulent strain of type 2 porcine reproductive and respiratory syndrome virus: identification and in vitro and in vivo characterization of a large spontaneous deletion in the nsp2 region. Virus Res. 2011;160:264–73.
Ren X, Wang M, Yin J, Li G. Phages harboring specific peptides that recognize the N protein of the porcine reproductive and respiratory syndrome virus distinguish the virus from other viruses. J Clin Microbiol. 2010;48:1875–81.
Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol. 1998;79(Pt 5):961–79.
Sun Z, Chen Z, Lawson SR, Fang Y. The cysteine protease domain of porcine reproductive and respiratory syndrome virus nonstructural protein 2 possesses deubiquitinating and interferon antagonism functions. J Virol. 2010;84:7832–46.
Sun Y, Han M, Kim C, Calvert JG, Yoo D. Interplay between interferon-mediated innate immunity and porcine reproductive and respiratory syndrome virus. Viruses. 2012;4:424–46.
Wang F-X, Yang B-C, Wen Y-J, Liu Z, Leng X, Shi X-C, et al. Genomic characterization of porcine reproductive and respiratory syndrome virus TJM vaccine strain. Int Res J Microbiol. 2012;3:191–201.
Wang FX, Song N, Chen LZ, Cheng SP, Wu H, Wen YJ. Non-structural protein 2 of the porcine reproductive and respiratory syndrome (PRRS) virus: a crucial protein in viral pathogenesis, immunity and diagnosis. Res Vet Sci. 2013;95:1–7.
Wu WH, Fang Y, Farwell R, Steffen-Bien M, Rowland RR, Christopher-Hennings J, Nelson EA. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology. 2001;287:183–91.
Yu D, Han Z, Xu J, Shao Y, Li H, Kong X, Liu S. A novel B-cell epitope of avian infectious bronchitis virus N protein. Viral Immunol. 2010;23:189–99.
Zhou YJ, An TQ, Liu JX, Qiu HJ, Wang YF, Tong GZ. Identification of a conserved epitope cluster in the N protein of porcine reproductive and respiratory syndrome virus. Viral Immunol. 2006;19:383–90.
Zhou L, Zhang J, Zeng J, Yin S, Li Y, Zheng L, et al. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J Virol. 2009;83:5156–67.
Acknowledgements
This work was supported by the Special Fund for Agro-scientific Research in the Public Interest (201303042) and Key Technology R&D Program of Jilin Province (20140204073NY).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, FX., Yang, Y., Liu, X. et al. Development of monoclonal antibody for differentiating porcine reproductive and respiratory syndrome virus and identification of a novel non-structural protein 2 epitope peptide. VirusDis. 28, 408–415 (2017). https://doi.org/10.1007/s13337-017-0400-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-017-0400-x