Abstract
In a cross-sectional study, prevalence of ovine herpesvirus 2 (family: Herpesviridae, subfamily: Gammaherpesvirinae, genus Macavirus and species: Ovine herpesvirus 2) infection was estimated in sheep population of Karnataka state in India. Based on the three stage cluster sampling method, whole blood samples (356) of sheep were collected from 11 sheep-dense districts of the state. The samples were tested for presence of OvHV-2 genome by recommended hemi-nested polymerase chain reaction (PCR) test. The true prevalence of OvHV-2 infection in sheep population of Karnataka was 24.44 %. Of the 11 district surveyed, highest true prevalence of 42.42 % (CI 25.56–59.29) was found in Raichur followed by Tumkur (39.02 %, CI 24.09–53.96). Inverse distance weighted interpolation of prevalence indicated that OvHV-2 prevalence within a given district is not uniform and there are areas of varied prevalence. The nucleotide sequence of the 422 bp DNA fragment, amplified in PCR, matched 99 % with OvHV-2 reference sequence and other sequences reported from India. Grouping of OvHV-2 sequences obtained from Karnataka with those from Andhra Pradesh, Tamil Nadu and Jammu and Kashmir in the neighbour joining tree indicated a close relationship among the OvHV-2s circulating in India. This is the first study in the country where systematic screening of sheep population of a state for the presence of OvHV-2 infection has been carried out, which indicated a widespread prevalence calling for an urgent need for policy measures to prevent economic losses due to the disease in susceptible cattle and buffalo species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant catarrhal fever (MCF) is usually a fatal disease of bovids, cervids and certain other susceptible ruminant species. The disease has occasionally been reported in domestic pigs [16]. MCF primarily affects the lymphoid organs, the respiratory and alimentary tracts and is characterised by high fever, inappetence, ocular and nasal discharge, kerato-conjunctivitis with corneal opacity and enlargement of peripheral lymph nodes. Death may occur within few days or several weeks after the onset of clinical signs [27].
Globally, two endemic forms of MCF with distinct geographical distributions exist [27]. Alcelaphine herpesvirus 1 (family: Herpesviridae, subfamily: Gammaherpesvirinae, genus Macavirus and species: Alcelaphine herpesvirus 1) (AlHV-1), which is prevalent in Africa, is wide-spread among wildebeest populations and causes the wildebeest-associated MCF (WA-MCF) in cattle and other species [24]. The other form is caused by ovine herpesvirus 2 (family: Herpesviridae, subfamily: Gammaherpesvirinae, genus: Macavirus and species: Ovine herpesvirus 2) (OvHV-2), which infects domestic sheep asymptomatically [5] and causes sheep-associated MCF (SA-MCF) in large ruminants like cattle, bison, deer, water buffalo and occasionally swine [16, 27, 29]. SA-MCF has been reported worldwide, including North and South America [25, 26], Europe [33, 39], the Middle East [3], Asia [4, 9], Africa [5, 24] and New Zealand [10].
Although MCF has been reported worldwide, the information on the existence of the disease in India is scanty and reported at varied intervals [31]. The first confirmed case based on histopathology was reported in the year 1975 from North Indian state of Punjab [24], where 31 buffaloes and one cattle died of the disease. While this is the first authenticated report of MCF, there were unconfirmed reports of the disease in India as early as 1941 (Annual Report IVRI, 1941–1942). A report of clinical evidence in a Jersey Sindhi crossbred bull was also published in 1969 [9]. Almost three decades later, the occurrence of SA-MCF in cattle and sheep was reported from another North Indian state of Jammu and Kashmir [37, 38]. The presence of OvHV-2 in South India became evident, when screening of 612 samples from sheep and goats with some respiratory disease, showed that 17 of these were positive [1]. Recently, OvHV-2 was detected in a captive bison, which died of MCF like symptoms at Bannerghatta National Park, Bengaluru [30]. Detailed investigations revealed the occurrences of clinical cases of MCF in cattle in the areas, surrounding the park. [32]. Very recently, a large outbreak of MCF in cattle has been described from another South Indian state of Andhra Pradesh [11]. At present, it is still not clear whether the recent surge in the cases of MCF in India is the result of true emergence of the virus or merely due to the increased awareness about this disease. Keeping in view of the above, the present study was undertaken to generate baseline information about the extent of OvHV-2 infection in sheep population in India beginning with Karnataka state, so that appropriate control measures can be devised to prevent transmission of OvHV-2 to cattle and other susceptible species.
Materials and methods
Study area
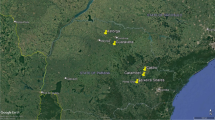
The survey was restricted to Karnataka state, which lies between 11°30′ North and 18°30′ North latitudes 74° East and 78°30′ East longitudes. It has a total land area of 191,791 km2 and stands eighth in terms of size in the country. The state also has second largest sheep population (9,339,897 as per 2012 livestock census) and has thirty districts with varying sheep density. In general, sheep population is concentrated in eastern half of the state (Fig. 1).
Sampling
Before the survey, a sampling frame containing hobli-wise (hobli = revenue villages formed by cluster of hamlets) population of sheep was prepared based on the 2007 livestock census. The data for the above purpose was obtained from the website of Department of Animal Husbandry Dairying and Fisheries, Government of India. As no previous knowledge of OvHV-2 prevalence in India was available, it was decided to collect minimum number samples (n = 300) required to detect a rare event [17] and accordingly three stage cluster sampling, keeping district as primary cluster, was done by allocating the samples proportionately to the sheep population. Briefly, sampling was done in 11 districts, which were selected based on sheep density and these districts accounted for nearly 72 % of the total sheep population of the state. The sheep density in these districts, barring Bijapur district, varied from 60 sheep/sq km to 118 sheep/sq km. At the second stage, 2 taluks (sub-district) within each district, having highest sheep populations were selected. Finally, in the third stage, 2 hoblies (cluster of hamlets) within each taluks having highest sheep populations were selected. In all, sampling was done in 44 hoblies (74 hamlets), 22 taluks and 11 districts and 356 samples were collected. The survey was conducted during a period of 4 months from November 2012 to February 2013. The details of sampling locations are shown in Fig. 1.
Samples, DNA extraction and PCR
Blood samples were drawn in heparinated blood vaccutainers (BD, USA) from the jugular vein of sheep and shipped on ice to the laboratory for further testing. DNA extraction from peripheral blood leukocytes (PBL) and hemi-nested PCR were performed as described earlier [2, 32].
Nucleotide sequencing and sequence analysis
The PCR amplicons of 422 bp from two of the positive samples, one from northern and another from southern part of the state, were sequenced by Sanger’s dideoxy method in an automatic DNA sequencer (ABI 3130, USA). Briefly, the two amplicons were cloned in pGEM-T easy plasmid vector (Promega, USA) as per manufacturer’s protocol. Recombinant plasmids were purified using DNA purification kit (Promega, USA).The plasmids were sequenced using Big Dye Terminator (V 3.1) cycle sequencing kit (Applied Biosystems) and T7 and SP6 universal primers (Promega, USA). Sequences were aligned using the Clustal X, Ver. 1.8.3 programme [36] and edited manually using GeneDoc [21]. Nucleotide divergence/similarity was calculated using Megalign module of Lasergene package (DNASTAR Inc., USA). The transition/transversion (Ts/Tv) rate was estimated from the dataset using TreePuzzle, Ver. 5.2 [34], and was found to be 1. The bootstrap values were calculated with the modules SEQBOOT, DNADIST, NEIGHBOUR and CONSENSE of the PHYLIP Ver. 3.59 package [7]. The phylogenetic trees, calculated by using the neighbour-joining method, were computed with the DNADIST, NEIGHBOUR modules with the same parameters. Trees were visualised using the programme, TREEVIEW Ver. 1.6.6 [22].
Mapping and spatial interpolation
Location mapping and spatial interpolation were done using Quantum GIS software (QGIS) version 2.0.1-Dufour (OSGeo, Beaverton, OR, USA). For spatial interpolation of prevalence data inverse distance weighting (IDW) method was used.
Results
Genomic detection of OvHV-2
Of the 356 PBL DNA obtained from blood samples collected from 11 sheep dense-districts of Karnataka a total of 87 samples were recorded as positive for OvHV-2 by hemi-nested PCR, which yielded two separate fragments of 422 bp and 238 bp in the first and second stage PCR respectively (Fig. 2a, b). No amplification was noticed in negative control. In certain samples chosen randomly, the specificity of the PCR products was determined by RE digestion with RSa I. In addition, PCR products, one each from South and North Karnataka were cloned and sequenced, which again confirmed the specificity of the products. The samples were considered positive only when they were positive in both the PCRs. The prevalence of OvHV-2 infection in sheep population of Karnataka was estimated by using ‘WinEpi’ software (developed by University of Spain) at a confidence interval (CI) of 95 %, and was found to be 24.44 % (CI 19.97–28.90).
a PCR product (422 bp) from tegument region amplified from blood sample of positive sheep (lane 1, 2, 4, 5 and 7) positive control (lane 8), negative control (lane 9). b Nested PCR product (238 bp) from the positive blood samples (lane 1, 2, 3, 5 and 6) positive control (lane 7), negative control (lane 8)
Among the various districts screened, prevalence was highest in Raichur district (42.42 %, CI 25.56–59.29) followed by Tumkur (39.02 %, CI 24.09–53.96) and was lowest in Mandya district (14.29 %, CI 0.00–29.25) (Table 1). Similarly, comparison of prevalence among the taluks (sub districts) showed highest prevalence in Raichur taluk (54.55 %, CI 25.12–83.97), and lowest in Maddur (0 %, CI 0.00–34.81) and Shorapur taluks (0 %, CI 0.00–20.58) (Table 1). The observed true prevalence in young (<1 year of age) and adult animals (>1 year of age) was 17.48 % (CI 10.14–24.81) and 27.27 % (CI 21.78–32.76), respectively.
Mapping and spatial interpolation
In order to understand how the prevalence was distributed spatially within the surveyed districts, IDW interpolation technique was used to create surface maps from the prevalence data at sampled locations/taluks/districts and is shown in the Fig. 3a–c. From the three figures, it can be seen that location based-IDW map (Fig. 3a) produced many areas showing bull’s eye effect. Changing the distance coefficient P and increasing the cell size did not improve the results. Map produced using taluk centroids (Fig. 3b) produced smoother map, results of which resembled those obtained from location based interpolation map. The map produced using district centroid (Fig. 3c) produced an unrealistic surface map. Since the map, produced using the taluk centroid, matched the prevalence data closely (lower root mean square error), we used this map (Fig. 3b) for further analysis. In the Fig. 3b, it can be seen that, there are areas of higher prevalence as well as lower prevalence within a district. For example, in the eastern part of the Raichur district OvHV-2 prevalence is very high (about 60 %), while in northwest region, the prevalence is low (0–13 %). Similarly, in central part of Bellary district prevalence of OvHV-2 is very low, while in southern part it is about 40 %.
Nucleotide sequencing and analysis
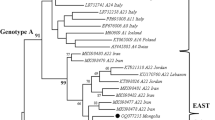
For understanding the genetic relatedness of OvHV-2s circulating in Karnataka, cloned second stage PCR products, one each from Southern and Northern Karnataka were nt sequenced (Gen Bank Accession No: KF303529 and KF303530) and aligned with few others from NCBI Genbank. Comparison of nt sequences indicated that both the products have identical sequences and share sequence homology of 99.6 % with one of the OvHV-2 sequence obtained earlier from a Bison died of SA-MCF. The genetic closeness was further indicated by their grouping in the neighbour joining (NJ) tree (Fig. 4). Alignment of nt sequences of tegument region with OvHV-2s from India, Brazil, Turkey, Europe and South Africa showed that these are about 0–2.4 % divergent from each other. The grouping pattern in the NJ tree implies that OvHV-2s recovered from world over from variety of species are closely related (Fig. 4).
Phylogenetic analysis of the specific 422 bp segment of the tegument gene of OvHV-2 sequences determined in the present study (shaded) and other sequences from India, Brazil, Turkey, Europe and South Africa. Neighbour-joining tree showing grouping of various OvHV-2 sequences reveal genetic closeness among Indian sequences and divergence of 0–2.4 % from other countries. Only boot strap values above 50 are indicated
Discussion
In the present study, level of OvHV-2 infection in the sheep of Karnataka state, India, was assessed using hemi-nested PCR. Of the 356 blood samples screened 87 were found positive for OvHV-2, with an overall true prevalence of 24.44 %, which is comparatively lower than that reported elsewhere; for example, in a hemi-nested PCR and CI ELISA based study to determine the level of SA-MCF infection in 144 randomly selected healthy adult sheep from 5 states of USA [12], 143 sheep were found positive for OvHV-2. Similarly, Mirangi and Kang’ee [18], who have used hemi-nested PCR for surveying the prevalence of OvHV-2 in both endemic and non endemic regions of WA-MCF in Kenya, have reported the presence of OvHV-2 in 90.4 % (161/178) of sheep. These authors have also noted that rates of infection did not differ between WA-MCF-endemic (89 %) and WA-MCF-free areas (92 %). The prevalence of OvHV-2 in other parts of India is currently not known and one available report from Kashmir valley [38] indicated a prevalence of 84.8 % (28/33) in sheep. However, this study was restricted to small group of animals within a small study area and may not be representative of the state. The lower prevalence obtained in our study probably justified the earlier held notion that SA-MCF is not a common disease and deserved the tag of an exotic disease until recently. However, further studies are needed to understand the OvHV-2 prevalence in other parts of the country, as regions/areas with higher prevalence cannot be ruled out.
In our study, highest true prevalence of 42.42 % (CI 25.56–59.29) was found in Raichur district followed by Tumkur (39.02 %, CI 24.09–53.96), which is also reflected in interpolated surface map (Fig. 3b). It is pertinent to note that northern and southern boundaries of the former district are formed by two rivers leading to substantial irrigated land areas that attract migration of sheep from neighbouring districts as well as from neighbouring state (Andhra Pradesh) during the dry seasons, thereby providing a suitable environment for intermixing of flocks. It is to be noted that cases of MCF have been recently recorded in neighbouring Mahabubnagar and Karnool districts (Dr. Sudarshana Rao personal communication) of Andhra Pradesh state. Some districts like Bagalkot (25.58 %, CI 12.54–38.62), Chitradurga (25.86 %, CI 14.59–37.13) and Bellary (16.13 % CI 3.18–29.08) have moderate overall prevalence as compared to Raichur and however, distribution of prevalence within the district is not uniform; from the figure (Fig. 3b), it can seen that there are areas of high (>30 %), moderate (15–30 %) and low (<15 %) prevalence. It is to be noted that, Tumkur and Chitradurga districts have large sheep population and most of which are migratory, resulting in frequent intermixing of flocks there by facilitating the transmission of OvHV-2 among flocks.
Mandya and Yadgir districts although have lower overall prevalence (Table 1), have a substantial land mass with moderate prevalence (Fig. 3c). For example, taluks such as Krishnaraj Pet, Pandavapura in Mandya district are located within the Kaveri basin and have lot of irrigated lands, which attract sheep migration during dry season. Nagamangala taluk, which falls within migratory route of sheep has higher prevalence, as a result intermixing of the flocks, where as Mandya taluk, which does not fall in the migratory route has lower prevalence. Moderate prevalence in Yadgir taluk (Fig. 3c), which falls in the dry agroclimatic zone, could be due to close proximity to Raichur taluk, which has a high OvHV-2 prevalence and the resultant frequent intermixing with flocks from the said taluk.
In the present study, the prevalence of OvHV-2 infection in young animals has been shown to be lower than that of the adult animals. This is not surprising considering that the majority of lambs are not infected with OvHV-2 until after 2 months of age [13] and those lambs which are separated from infected sheep from an early age have been shown to remain uninfected as adults [14]. Sheep producers and zoo managers in the United States and in Europe have used this strategy to produce OvHV-2-free sheep [19]. The lower prevalence among younger animals in our study further confirms the above observation and higher level of infection in older animals indicates that these animals have acquired the infection as they grew older.
We have used sequences of ORF 75, which codes for viral tegument protein for constructing NJ tree. The same region has been earlier used by Baxter et al. [2] and Sausker and Dyer [28] for comparing OvHV-2 sequences from Europe and USA respectively. Comparison of both the OvHV-2 nt sequences determined in this study indicated that these are identical in the region compared and have grouped in the NJ tree with a OvHV-2 sequence that was obtained from a Bison died of SA-MCF earlier. Upon comparison of nt sequences of OvHV-2 tegument protein gene obtained from the sows and from three cases of sheep associated MCF in Finnish Cattle, Syrjälä et al. [35] have found these to be identical. They have found tegument protein gene region of OvHV-2 as highly conserved among European OvHV-2. Grouping of OvHV-2 sequences obtained from Karnataka with those from Andhra Pradesh, Tamil Nadu and Jammu and Kashmir in the present study further confirms the above observation (Fig. 4). Similar results were obtained by Dunowska et al. [6], who found glycoprotein B gene sequences from cattle, bison and sheep to be highly similar; however, higher variability in some sequences, so as to warrant a new subgroup among OvHV-2 sequences from North American bison and sheep has been reported [28].
It is generally accepted that serological tests are best suited for surveying MCF infection in asymptomatic carrier animals in the field. This is because virtually all reservoir hosts consistently develop antibodies after infection, which can be detected by any of the serological tests. Among the various serological tests, competitive inhibition enzyme linked immunosorbent assay (CI-ELISA), which uses the AlHV-1 antigen for detection of OvHV-2 antibodies has been the recommended method for surveillance and also for declaring individual as well as population freedom from infection [15]. The CI-ELISA, which uses a monoclonal antibody (MAb 15A) to an epitope conserved among MCF group of viruses, is reported to have high specificity [19]. The analytical sensitivity of this ELISA was later increased to four fold using the MAb directly conjugated with horse radish peroxidase [15]. An indirect ELISA, which uses crude AlHV-1 lysate with results comparable to CI-ELISA described by Fraser et al., [8] may be an another option for MCF serology. Unfortunately, CI-ELISA though commercialised is not readily available in many parts of the world, especially in developing countries. In this context, use of hemi-nested PCR was the only option available to us and was used for surveying the level of OvHV-2 infection in sheep. In view of this, the true prevalence obtained in this study could be slightly higher in actual sense due to fact that virus undergoes latency and may not be present at a detectable limit in blood all the time.
Finally, in this limited study to determine the prevalence of OvHV-2 infection in the state of Karnataka, we found that nearly one-fourth of sheep population carries the virus, which could act as a potential source of infection for the clinically susceptible species and also to the remaining reservoir population. This is the first study in the country where systematic screening of sheep population of a state for the presence of OvHV-2 infection has been carried out. Since the infection level is still at a comparatively lower level, enactment of a national policy in the form of a law to direct sheep farmers to segregate lambs at a young age and to raise them separately for developing OvHV-2 free sheep flock would be an ideal proposition. However, given the poor economic conditions and migratory nature of most of the sheep farmers, this might be difficult to accomplish.
References
Banumathi N, Sood R, Patil SS, Subramanian M, Pradhan HK. Genomic detection of ovine herpes virus-2 in sheep and goat in states of southern India. Indian J Anim Sci. 2008;78:13–6.
Baxter SIF, Pow I, Bridgen A, Reid HW. PCR detection of the sheep-associated agent of malignant catarrhal fever. Arch Virol. 1993;132:145–59.
Brenner J, Perl S, Lahav D, Garazi S, Oved Z, Shlosberg A, David A. An unusual outbreak of malignant catarrhal fever in a beef herd in Israel. J Vet Med B Infect Dis Vet Public Health. 2002;49:304–7.
Dabak M, Bulut H. Outbreak of malignant catarrhal fever in cattle in Turkey. Vet Rec. 2003;152:240–1.
De Kock G, Neitz WO. Sheep as a reservoir host of snotsiekte (or malignant catarrhal fever of cattle) in South Africa. S Afr J Sci. 1950;46:176–80.
Dunowska M, Letchworth GJ, Collins JK, DeMartini JC. Ovine herpesvirus-2 glycoprotein B sequences from tissues of ruminant malignant catarrhal fever cases and healthy sheep are highly conserved. J Gen Virol. 2001;82:2785–90.
Felsenstein J. Phylip: phylogeny inference package (version 3.5c). Cladistics. 1989;5:164–6.
Fraser SJ, Nettleton PF, Dutia BM, Haig DM, Russell GC. Development of an enzyme-linked immunosorbent assay for the detection of antibodies against malignant catarrhal fever viruses in cattle serum. Vet Microbiol. 2006;116:21–8.
Gnanaprakasam V. Malignant head catarrh—a case report. Indian Vet J. 1969;46:912–6.
James MP, Neilson FJ, Steward WJ. An epizootic of malignant catarrhal fever. 1. Clinical and pathological observations. N Z Vet J. 1975;23:9–12.
Kumar NV, Sreedevi B, Karthik A, Lakshmi SV, Reddy AG, Sreenivasulu D. Detection of OvHV-2 from an outbreak of sheep associated malignant catarrhal fever from crossbred cattle of Southern India. Vet Res Commun. 2014;. doi:10.1007/s11259-014-9612-3.
Li H, Shen DT, O’Toole D, Knowles DP, Gorham JR, Crawford TB. Investigation of sheep associated malignant catarrhal fever virus infection in ruminants by PCR and competitive inhibition enzyme-linked immunosorbent assay. J Clin Microbiol. 1995;33:2048–53.
Li H, Snowder G, O’Toole D, Crawford TB. Transmission of ovine herpesvirus 2 in lambs. J Clin Microbiol. 1998;36:223–6.
Li H, Snowder G, Crawford TB. Production of malignant catarrhal fever virus-free sheep. Vet Microbiol. 1999;65:167–72.
Li H, McGuire TC, Muller-Doblies UU, Crawford TB. A simpler, more sensitive competitive inhibition enzyme-linked immunosorbent assay for detection of antibody to malignant catarrhal fever viruses. J Vet Diagn Invest. 2001;13:361–4.
Loken T, Aleksandersen M, Reid H, Pow I. Malignant catarrhal fever caused by ovine herpesvirus-2 in pigs in Norway. Vet Rec. 1998;43:464–7.
Machin D, Campbell M, Fayers P, Pinol A. Sample size tables for clinical studies. 2nd ed. Blackwell Science 1997. IBSN 0-86542-870-0:34-135.
Mirangi PK, Kang’ee FM. Diagnosis of malignant catarrhal fever using the polymerase chain reaction. Vet Rec. 1999;145:558–9.
Müller-Doblies UU, Li H, Hauser B, Adler H, Ackermann M. Field validation of laboratory tests for clinical diagnosis of sheep-associated malignant catarrhal fever. J Clin Microbiol. 1998;36:2970–2.
Müller-Doblies UU, Egli J, Hauser B, Li H, Strasser M, Ehrensperger F, Braun U, Ackermann M. Malignant catarrhal fever in Switzerland. Epidemiol Schweiz Arch Tierheilkd. 2001;143:173–83.
Nicholas KB, Nicholas HB. GeneDoc: analysis and visualization of genetic variation. 1997. (distributed by the authors; http://www.psc.edu/biomed/genedoc).
Page RDM. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–8.
Parihar NS, Rajya BS, Gill BS. Occurrence of malignant catarrhal fever in India. Indian Vet J. 1975;52:857–9.
Plowright W, Ferris RD, Scott GR. Blue wildebeest and the aetiological agent of bovine malignant catarrhal fever. Nature. 1960;188:1167–9.
Rech RR, Schild AL, Driemeier D, Garmatz SL, Oliveira FN, Riet-Correa F, Barros CSL. Malignant catarrhal fever in cattle in Rio Grande do Sul, Brazil: epidemiology, clinical signs and pathology. Pesq Vet Bras. 2005;25:97–105.
Reid SW, Robinson BN. Malignant catarrhal fever in a 5-month-old calf. Can Vet J. 1987;28:489.
Russell GC, Stewart JP, Haig DM. Malignant catarrhal fever: a review. Vet J. 2009;179:324–35.
Sausker EA, Dyer NW. Polymerase chain reaction and DNA sequencing for detection of ovine herpesvirus 2 in American bison (Bison bison). J Vet Diagn Invest. 2002;14:40–6.
Singh G, Singh B, Gupta PP, Hothi DS. Epizootiological observations on malignant catarrhal fever and transmission of the disease in buffalo calves (Bubalus bubalis). Acta Vet Brno. 1979;48:95–103.
Sood R, Kumar M, Bhatia S, Pateriya AK, Khandia R, Kulkarni DD, Kumar MS, Venkatesha MD. Ovine herpesvirus type 2 infection in captive bison in India. Vet Rec. 2012;170:654.
Sood R, Hemadri D, Bhatia S. Sheep associated malignant catarrhal fever: an emerging disease of bovids in India. Indian J Virol. 2013;24:321–31.
Sood R, Khandia R, Bhatia S, Hemadri D, Kumar M, Patil SS, Pateriya AK, Siddiqui A, Kumar MS, Venkatesha MD, Kulkarni DD. Detection and molecular characterization of naturally transmitted sheep associated malignant catarrhal fever in cattle in India. Trop Anim Health Prod. 2014;46:1037–43.
Straver PJ, Jvan Bekkum JG. Isolation of malignant catarrhal fever virus from a European bison (Bos bonasus) in a zoological garden. Res Vet Sci. 1979;26:165–71.
Strimmer K, Von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–9.
Syrjälä P, Saarinen H, Laine T, Kokkonen T, Veijalainen P. Malignant catarrhal fever in pigs and a genetic comparison of porcine and ruminant virus isolates in Finland. Vet Rec. 2006;159:406–9.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82.
Wani SA, Bhat MA, Samanta I, Buchoo BA, Ishaq SM, Pandit F, Shah GUD, Buchh AS. Clinical, serological and molecular evidence of sheep associated malignant catarrhal fever. Vet Rec. 2004;155:242–4.
Wani SA, Samanta I, Pandit F, Buchoo BA, Bhat MA. Molecular epidemiology of ovine herpesvirus type2 infection in Kashmir, India. Vet Rec. 2006;18:587–90.
Yus E, Guitián J, Díaz A, Sanjuán ML. Outbreak of malignant catarrhal fever in cattle in Spain. Vet Rec. 1999;145:466–7.
Acknowledgments
The authors would like to thank Indian Council of Agricultural Research, New Delhi for provision of funds, Director, NIVEDI and Director, NIHSAD, Bhopal for encouragement and necessary infrastructural facilities. We also thank Dr. K. P. Suresh, Biostatistician, NIVEDI for help in the sampling strategy. Thanks are also due to Department of Animal Husbandry and Veterinary Services, Govt of Karnataka for help during the survey. We also acknowledge Dr. C. Pasare, University of Texas-Southwestern Medical Center, USA for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Premkrishnan, G.N., Sood, R., Hemadri, D. et al. Cross-sectional study indicates nearly a quarter of sheep population in Karnataka state of India is infected with ovine herpesvirus 2. VirusDis. 26, 180–188 (2015). https://doi.org/10.1007/s13337-015-0274-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-015-0274-8