Abstract
MicroRNAs are a large group of non-coding RNAs that have emerged as regulators of various biological processes, especially carcinogenesis and cancer progression. Recent evidence has shown that microRNA-196a (miR-196a) is upregulated in most types of tumors and involved in multiple biological processes via translational inhibition and mRNA cleavage, such as cell proliferation, migration, and invasion, mostly functioning as an oncogene. Dysregulation of miR-196a promotes oncogenesis and tumor progression. In this review, we summarize the upstream regulators, target genes, signaling pathways, and single nucleotide polymorphisms of miR-196a, which collectively affect cell proliferation, migration, and invasion. In addition, we review the clinical outcomes and significance of miR-196a. miR-196a may serve as a novel biomarker or target for diagnosis, prognosis, and therapy in several human cancers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Non-coding RNAs are of crucial functional importance in diseases. They include the following members: microRNAs (miRNAs), PIWI-interacting RNAs, small nucleolar RNAs, long non-coding RNAs, and other types of non-coding RNAs [1]. miRNAs are small, non-protein-coding RNAs which are 19 to 25 nt in length. miRNAs function in many biological processes via translational inhibition and messenger RNA (mRNA) cleavage. Target genes of miRNA could be oncogenes or tumor suppressors. One target gene might be targeted by some miRNAs while one miRNA could have multiple target genes. Thus, the regulatory networks of miRNA are complex. Additionally, dysregulation of miRNAs is associated with cancer initiation, progression, diagnosis, therapy, and prognosis.
Previous miRNA profiling research demonstrated overexpression of miR-196a in many types of human malignancies [2–5]. Recently, the role of miR-196 in carcinogenesis and tumor progression has begun to emerge and more studies have suggested that miR-196a plays a vital role in most known processes of cancer, including proliferation, invasion, metastasis, and apoptosis [6–9]. Moreover, miR-196a is of great clinical significance as its performance in the prognosis of tumor patients, indicating that it has demonstrated diagnostic, prognostic, and therapeutic potential in human malignancies [10–12].
It is widely acknowledged that genetic variations, such as single nucleotide polymorphisms (SNPs), may be associated with cancer susceptibility, diagnosis, and prognosis. For example, the association between polymorphism rs11614913 in miR-196a-2 and breast cancer risk has been demonstrated [13]. Many studies have established that miR-196a is dysregulated in human malignancies, including lung cancer, gastric cancer, colorectal cancer, glioblastoma, esophageal cancer, breast cancer, and hepatocellular carcinoma [8, 14–19], showing that miR-196a plays a vital role in carcinogenesis. These findings suggest that miR-196a might be a novel and non-invasive prognostic factor for malignancies [20].
Biogenesis
miRNAs are small, non-protein-coding endogenous RNAs which are 19 to 25 nt in length and play a role in post-transcriptional regulation of gene expression via translational inhibition and mRNA cleavage [21]. The production of miRNAs occurs first in the nucleus. Primary RNAs (pri-miRNAs) with lengths of over 1000 nt are transcribed by RNA polymerase II. pri-miRNAs are then cleaved by RNase III endonuclease Drosha and its cofactor DGCR8 into long precursor RNAs (pre-miRNAs) of 60–70 nt. pre-miRNAs are transferred to the cytoplasm and then processed into mature miRNAs by Dicer, another RNase III endonuclease [22]. Double-stranded mature miRNA can be unwound into the single-stranded duplex of miRNA. Argonaute (Ago2) protein, the core component of RNA-induced silencing complex (RISC) [23, 24], binds mature miRNA, guiding RISC binding to the 3′-untranslated region (3′-UTR) of its target mRNAs by base pairing, thereby leading to degradation of target mRNAs or repression of protein translation.
Yekta et al. first demonstrated that miR-196 was transcribed from homeobox (HOX) clusters. In Homo sapiens, miR-196a-1 is located on chromosome 17 at 17q21.32 (Gene ID: 406972), downstream of HOXB8 and HOXB9 genes and upstream of HOXB10 gene. miR-196a-2 is located on chromosome 12 at 12q13.13 (Gene ID: 406973) between HOXC10 and HOXC9, in a highly conserved region. HOTAIR is located upstream of miR-196a-2. Interestingly, a nearly perfect complementary target site of miR-196a was predicted in HOXB8 mRNA. miR-196a with imperfect complementary sites, like HOX genes, functioned in translational repression [25].
Upstream regulation of miR-196a
Our research group showed that DNA demethylation may promote miR-196a expression. Through bioinformatic analysis, a canonical CpG island was found in the promoter region of miR-196a-1 but not miR-196a-2. After treatment of cells with DNA demethylation agents, the expression levels of miR-196a were markedly upregulated compared with controls, while the methylation frequency was decreased. Epigenetic modification of CpG islands could activate the transcription viability of miR-196a, leading to the upregulation of miR-196a [7].
MYC is another upstream regulator of miR-196a. Previous studies showed that miR-196a is a target gene of MYC, which functions as a transcriptional regulator of miR-196a [26]. ChIP assays showed the promoter of miR-196a was directly bound by MYC and miR-196a transcription was inhibited by MYC through its recruiting suppressors to putative MYC-binding sites. Together, these studies demonstrate that miR-196a expression is regulated by MYC, which binds to putative binding sites in the promoter of miR-196a and suppresses miR-196a expression [27, 28].
miR-196a affects tumor proliferation
Aberrant cell proliferation is one of the key features of cancer cells. Changes in cell cycle progression play a vital role in the regulation of cell proliferation. In mammalian cells, the G1/S phase is regulated by a series of precise expressions of cell cycle proteins, which activate the transcription of specific genes corresponding to various cyclin-dependent kinases (CDKs) and promote cell cycle progression [29–31]. Cell cycle progression is also negatively regulated by CDK inhibitors, and expressions of these inhibitors are often reduced in tumor cells, thus contributing to the dysregulated proliferation of malignant tumor cells [32].
We demonstrated that miR-196a directly targets the CDK inhibitor p27kip1 by interacting with its 3′-UTR [33]. p27kip1 is one of the important members of the CDK inhibitor Kip/Cip family, the first to be discovered in the G1 arrest induced by TGF-β. Expression of miR-196a is inversely correlated with the p27kip1 protein level in gastric cancer. Upregulation of p27kip1 in the nucleus could inhibit cell proliferation and the transition from G1 to S. Additionally, inhibition of proliferation could be rescued by overexpression of miR-196a [33].
Our research group also showed that HOXA5, another target of miR-196a that functions as a tumor suppressor, may be negatively regulated by miR-196a. HOXA5 was reported to participate in the growth of mouse lung [34]. Aberrant expression of HOXA5 has been correlated with human cancers and other diseases [35]. A previous study showed that HOXA5 exerts its oncogenic function by affecting p53 expression in breast cancer [36]. Knockdown of HOXA5 promotes non-small cell lung cancer (NSCLC) cell proliferation, suggesting that miR-196a regulates NSCLC cell proliferation via downregulation of HOXA5 [7].
NFKBIA, also known as IκBα, has been identified as an important modulator of apoptosis and invasion [37]. The activation of NF-κB requires phosphorylation and degradation of IκBα, which serves as a negative regulator of NF-κB. IκBα is associated with distal metastasis of oral squamous cell cancer [38]. In glioblastoma, miR-196a facilitates cell proliferation and suppresses cell apoptosis in cancer cells via repression of IκBα. Yang et al. validated the interplay between miR-196a and IκBα [39]. The 3′-UTR of NFKBIA was confirmed as a target of miR-196a in pancreatic cancer. The proliferation-promoting effect of miR-196a in pancreatic cancer is mediated by suppression of NFKBIA [8]. Another target of miR-196a in pancreatic cancer is ING5 or inhibitor of growth 5. miR-196a promotes proliferation and inhibits apoptosis by downregulating the expression of ING5 [6].
Forkhead box O proteins play vital roles in a multitude of biological processes including cell growth, proliferation, differentiation, and tumorigenesis [40]. The growth promotion effect of miR-196a may be partially attributed to downregulating FOXO1 and p27kip1 [9].
NTN4 is a functional target of miR-196a in cervical carcinogenesis and may participate in the oncogenic functions of miR-196a. miR-196a directly binds to the 3′-UTR of NTN4 to repress gene expression. The NTN4 mRNA level is reciprocally related to the expression level of miR-196a [41].
UBE2C, a novel target of miR-196a in breast cancer, is overexpressed and positively correlated with miR-196a in breast cancer, which is inconsistent with negative regulation mechanism. The authors postulate that miR-196a upregulates the expression of UBE2C and HER2 may be regulated by UBE2C, leading to the promotion of cell proliferation in breast cancer. Detailed mechanism that miR-196a positively regulates UBE2C expression needs further studies to be elucidated [18].
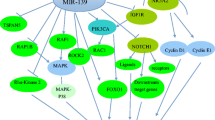
In esophageal cancer and breast cancer, miR-196a functions as a tumor promoter by targeting Annexin A1 [42, 43]. Other predicted target genes of miR-196a include ERG, KRT5, SPRR2C, S100A9, NRP2, TYMS, and LSP1. These target molecules of miR-196a have also been confirmed with luciferase assays, although their biological functions have not been completely elucidated. We have summarized the effects of miR-196a on proliferation in Table 1 and Fig. 1.
miR-196a regulates tumor cell migration and invasion
In addition to proliferation, migration and invasion are two other major biological features of tumors. Migration and invasion are also correlated with cancer diagnosis, progression, and prognosis. However, the underlying molecular mechanisms behind these processes are complicated and need further investigation. Several studies have suggested that miRNAs play a critical role in modulating both tumor migration and invasion [51].
miR-196a/miR-196b is overexpressed in gastric cancer tissues. The upregulation of miR-196a is associated with poor clinical prognosis and metastatic activity. Radixin is a direct target of miR-196a, as validated by bioinformatics algorithm and luciferase reporter assays. Radixin also acts as a tumor suppressor, and the restoration of radixin expression blocks migration and invasion induced by miR-196a. miR-196a promotes cell migration and invasion of gastric cancer in vitro and in vivo, which is partially attributed to silencing radixin [44].
In oral cancer, miR-196a exerts its function in migration and invasion by negatively regulating NME4 expression. NME4, also known as nm23-H4, belongs to the nm23 protein family and is a tumor suppressor [52]. The nm23 family participates in cell migration and adhesion [53]. NME1, another member of this family, functions as a metastatic inhibitor under the regulation of TP53 [54]. miR-196a regulates the expression level of NME4 and subsequently activates the phosphorylation of JNK. Accumulating evidence indicates miR-196a-NME4 signaling regulates JNK phosphorylation, resulting in downregulation of TIMP1 expression and upregulation of MMP1/9 expression, which is involved in tumor progression pathways, such as invasion, metastasis, and angiogenesis [55]. These findings suggest that the miR-196a-NME4-JNK-TIMP1-MMP signal pathway plays a critical role in modulating tumorigenesis in oral cancer [45].
In pancreatic cancer, miR-196a facilitates cell migration via targeting NFKBIA. NFKBIA functions as a suppressor of migration and invasion that may be partially attributed to inhibition of epithelial-mesenchymal transition (EMT), which is considered to be essential for cancer invasion and migration [56, 57]. miR-196a also represses the expression of ING5 to promote invasion [6]. Zhang et al. reported that miR-196a enhanced cervical cancer cell migration abilities through decreasing the expression of NTN4. Research in our lab has shown that HOXA5, a direct target of miR-196a, not only has influence on tumor proliferation but also facilitates migration and invasion. Annexin A1, a target of miR-196a, is reported to be involved in tumor migration and invasion in esophageal cancer, breast cancer, and head and neck cancer [42, 43].
miR-196a can also function as a tumor suppressor through regulating corresponding target genes. In breast cancer, HOXC8 plays an oncogenic role through its contribution to cell migration and invasion. HOXC8 upregulates and downregulates genes such as cadherin-11, embigin, transgelin, and calponin [58], which are associated with cell migration. Furthermore, the ratio of miR-196a to HOXC8 mRNA could reflect the cell line migration status. miR-196a exerts its anti-metastatic role by inhibition of HOXC8 expression [48]. In melanoma, HOXC8 is an indispensable part of melanomagenesis. Three migration-associated genes, cadherin-11, calponin-1, and osteopontin, are increased or decreased under the regulation of HOXC8. HOXC8 is also negatively regulated by miR-196a. Both in breast cancer and melanoma, the 3′-UTR of HOXC8 is targeted by miR-196a.
Another signal cascade was detected in malignant melanoma. HOXB7 activates bFGF to promote ETS-1 expression. BMP4, important in melanoma progression, is regulated by ETS-1 and functions in migration of melanoma. However, miR-196a binds to the 3′-UTR of HOXB7 to repress its expression, and in fact, the expression level of miR-196a is decreased in melanoma cells while the level of HOXB7 is increased. This suggests that miR-196a suppresses the oncogenic function of HOXB7 through miR-196a-negative regulation of HOXB7 [47, 49].
Furthermore, in esophageal cancer, RAP1A was overexpressed because the SNP rs6573 change (from A to C) is involved in the miR-196a-binding site. RAP1A facilitates cancer cell migration and invasion via MMP2. miR-196a functions in suppression of migration and invasion by inhibiting the expression of RAP1A [46]. Table 1 summarizes how miR-196a affects tumor invasion and migration through different pathways. We have summarized the effects of miR-196a on migration and invasion in Table 1 and Fig. 1.
SNP in miR-196a
An SNP is a variation of a single nucleotide occurring at a specific position in the genome. SNPs are more likely to take place in non-coding regions than in coding regions. SNPs could alter the expression of corresponding miRNAs, potentially contributing to cancer risk. The expression level of miR-196a could be influenced by genetic polymorphisms. rs11614913 CC or rs11614913 CT was significantly associated with high levels of miR-196a [59, 60]. The rs11614913 SNP is located in the 3′ strand of mature miR-196a-2. Some studies showed that expression of miR-196a is upregulated by the rs11614913 C allele [59–61]. The variant allele of miR-196a-2 (rs11614913, C to T substitution) was markedly related to a reduced risk for breast cancer, lung cancer, and hepatocellular carcinoma [62–64]. SNPs of miR-196a are also closely correlated with decreased susceptibility in colorectal cancer [65]. However, the association between rs11614913 and gastric cancer susceptibility is still controversial. Wang et al. indicated that the variant homozygote CC was linked to reduced risk of gastric cancer in a Chinese population [66]. Xu et al. found no significant correlation between rs11614913 and gastric cancer risk, and correlations only existed in specific subgroups [67]. Further studies should be conducted to validate whether rs11614913 is correlated with gastric cancer.
The SNP rs6573 A to C substitution affects the binding of miR-196a to the 3′-UTR of RAP1A, which is one of the target genes of miR-196a. SNP rs6573 also inhibited RAP1A expression induced by miR-196a [46]. These data suggest that abnormal expression of miR-196a in many types of cancers may contribute to the presence of miR-196-a-2 SNPs. Genetic variants in its mature region could serve as a promising biomarker, which may be helpful to evaluate cancer susceptibility.
miR-196a in cancer diagnosis and prognosis
Biomarkers are traceable substances that are introduced into the body to examine the status of diseases, evaluate pathogenic processes, and assess pharmacologic responses. There are several reasons supporting the selection of miRNAs as potential biomarkers. First, the expression of miRNAs is dysregulated in human cancer and tissue-specific. Second, miRNAs show high stability in blood, in urine, and even in formalin-fixed tissues. At the same time, miRNAs can also be detected in samples obtained less invasively.
Aberrant expression of miR-196a has been documented in different types of cancers (Table 2). miR-196 is upregulated in a multitude of cancers including head and neck cancer, glioblastoma, oral cancer, breast cancer, pancreatic cancer, cervical cancer, gastric cancer, lung cancer, esophageal cancer, and acute myeloid leukemia. Darda and colleagues detected a number of differentially expressed miRNAs between normal cells and cancer cells using the Affymetrix miRNA array, and miR-196a was the only miRNA highly expressed in head and neck squamous cell carcinoma (HNSCC) [68]. Hou et al. reported that miR-196a was significantly increased in cervical cell lines and cancer tissues in comparison with normal cervical squamous cells and normal cervical tissue [9]. We previously discovered that the expression level of miR-196a was markedly increased in gastric tissues and cell lines compared with normal counterparts [33]. We also found that miR-196a expression was markedly upregulated in gastric cancerous tissues by 22.9-fold compared with corresponding normal tissues [7]. These results imply that miR-196 may function as an oncogene in the development and progression of cancers.
Combined detection of miR-148a and miR-196a in serum enabled the identification of gastric cancer, with a sensitivity of 80 %, specificity of 96.97 %, and an area under the curve value of 0.924 [2]. Likewise, high sensitivity and specificity in the prediction of malignancy has been demonstrated by collective detection of miR-196a and miR-196b. Thus, miR-196a may be beneficial in the early diagnosis of oral cancer [70].
However, different from upregulation of miR-196a in most cancers, miR-196a expression was decreased in melanoma, breast cancer, and acute lymphoblastic leukemia compared with non-tumor tissues [47, 50] or cell lines [27, 49]. These data indicate that miR-196a may be a potential diagnostic biomarker in human cancers.
The molecular mechanisms leading to the poor prognosis and high recurrence rate of human cancer have not been soundly elucidated, although several molecular mechanisms are known. Hence, a better understanding of therapy involved in disease prognosis would offer more optimal treatments for various cancers. The upregulation of miR-196a is associated with poor prognosis in lung cancer, pancreatic cancer, cervical cancer, and gastric cancer, which is consistent with its functions in cell proliferation, invasion, and metastasis in these tumors.
High expression of miR-196a has been observed in NSCLC samples and cell lines, and the level is increased compared with corresponding non-tumor tissues. Moreover, the miR-196a expression level was much higher in pathological stages III–IV than in pathological stages I–II, indicating that miR-196a expression was undoubtedly connected with the malignant degree of NSCLC [7]. miR-196a is differentially expressed in esophageal adenocarcinoma (EAC) and Barrett’s esophagus (BE) compared with normal squamous mucosa, and its dysregulation is closely associated with the progression from low-grade dysplasia to high-grade dysplasia, suggesting that miR-196a could be used as an indicator to identify BE-EAC carcinogenesis [4, 17, 71]. Inconsistent with this observation, miR-196a is significantly downregulated in melanoma compared with healthy melanocytes and the expression level of miR-196a plays a role in early detection of melanoma progression [49]. In colorectal cancer (CRC), the level of miR-196a is associated with the oncogenic phenotype and the miR-196a-2 C>T polymorphism may contribute to CRC susceptibility [72, 73]. Similarity, overexpression of miR-196a and miR-196b induce the immunophenotype in acute myeloid leukemia and acute T-lymphoblastic leukemia, which indicated that miR-196a may be useful in the prognosis of leukemia [50]. Recent evidence also shows that dysregulation of the levels of miR-196a and miR-196b in plasma samples may serve as novel prognostic markers in oral cancer. Chi-squared analysis indicated that just pathological T stage may contribute to a high miR-196 level; in other words, larger tumor size was associated with high expression of miR-196s [70]. Darda et al. identified miR-196a and HOXB9 shared the same primary transcript and presented high expression in HNSCC than in normal oral mucosa [68]. Furthermore, miR-196a may serve as a diagnostic biomarker in laryngeal cancer [74]. In breast cancer, the expression level of miR-196a was significantly correlated with clinicopathological characteristics of patients and demonstrated that miR-196a is favorable for the prognosis of breast cancer [18]. With respect to the prediction of miR-196a-related prognosis, Sun et al. identified that the dysregulation of miR-196a in gastric cancer tissues was correlated with tumor size and advanced pathologic stage. Gastric cancers with tumor size over 5.5 cm and clinical stage IV had higher expression of miR-196a and may lead to poor prognosis [33].
Many studies have shown that miR-196a as a direct prognosis factor is related to clinical outcome of patients with malignant cancers. In comparing the expression levels of the miR-196 family members in glioblastoma cell lines, miR-196a was more differentially expressed compared with miR-196b. Increasing miR-196 levels were closely correlated with poor overall survival (OS) rates of glioblastoma patients (P = 0.021; hazard ratio, 2.81) [16]. Dou et al. found that the rs11614913 CC polymorphism of miR-196a was related to lower risk of glioma in a Chinese population, with an odds ratio of 0.74 [75]. A previous study indicated that the miR-196a and FOXO1 expression levels along with tumor stage were potential prognostic biomarkers for OS in cervical cancer [9]. Overexpression of miR-196a was highly associated with poor OS and disease-free survival in 92 cervical cancer patients [9]. In addition, increasing evidence has showed that serum miR-196a, as a reliable biomarker, was associated with FIGO stage (P = 0.004), grade (P = 0.011), tumor size (P = 0.031), and lymph node metastasis (P = 0.018) in cervical cancer. The 5-year OS rate for low miR-196a expression patients with CC was 73.13 %, which was higher than that of the high miR-196a expression patient group (39.47 %) [76]. Furthermore, ovarian cancer patients with high levels of miR-196a had significantly poor OS and recurrence-free survival time compared with patients with lower levels of miR-196a, suggesting that miR-196a could be used as a clinical prognostic factor in ovarian cancer patients [77]. miR-196a-2 rs11614913 in human renal cell carcinoma was associated with patient survival time. Individuals with the C variant allele (CC and TC genotypes) compared with the TT wild-type genotype, especially in stage I/II, had a higher 5-year survival rate [12]. Liu et al. demonstrated miR-16 and miR-196a as early discriminating markers for PCa are more effective than serum marker CA19-9, especially in early tumor screening [78]. miR-196a, as a potential biomarker, was also significantly upregulated in pancreatic ductal adenocarcinoma (PDAC) and intestinal-type intraductal papillary mucinous neoplasms and predicted a worse survival outcome [79–82]. Data from 35 PDAC patients showed that the median survival time (6.1 months) was low in the high-level miR-196a group compared with the low-level miR-196a group (12.00 months) [82]. The relapse-free survival and OS rates of patients with high-level miR-196a diagnosed with gastrointestinal stromal tumors were markedly lower than those of patients with low-level miR-196a in both univariate and multivariate analyses [83].
In conclusion, miR-196a can serve as a candidate prognostic indicator in human malignancies. Further research is needed to provide better understanding of the oncogenic networks mediated by miR-196a in human cancer.
miR-196a in drug resistance
Multiple studies have shown that expressions of miRNAs are significantly correlated with the development and progression of several cancers. Thus, discovery of the critical roles of miR-196a regulation of gene expression may provide insights into the prevention and treatment of malignant cancers. Endocrine therapy is the current therapy for estrogen receptor (ER)-positive breast cancer. However, resistance to tamoxifen often occurs in breast cancer patients and seriously affects hormone treatment efficacy. Overexpression of HER2, one of the multiple ER-target genes in tamoxifen-resistant breast cancer cells, is caused by estrogen receptor-alpha (ERalpha) and HOXB7, which functions as a cofactor. miR-196a and MYC restored sensitivity of breast cancer cells to tamoxifen through targeting HOXB7 both in vitro and in vivo [27, 28]. Treatment of ovarian carcinoma cell lines with miR-196a inhibitors caused cell death, indicating that miR-196a replacement therapy may reverse OS and recurrent-free survival of patients [77]. Chemotherapy resistance is currently a serious issue in many cancers. Li et al. reported miR-196a as a cisplatin-resistant factor in A549/DDP NSCLC cells and demonstrated that miR-196a regulates multiple important drug-resistant proteins, including MDR1, MRP1, ERCC1, survivin, and Bcl-2. NSCLC cell proliferation and cell apoptosis are regulated by miR-196a [84]. In addition, the expressions of miR-196a and other miRNAs were upregulated in the HCPT-resistant gastric cancer cells [85]. Suh et al. indicated that miR-196a may be a target which was significantly associated with the clinical response to radiotherapy in HNSCC. miR-196a also induces resistance to these radiotherapeutic agents by targeting ANXA1 [43]. Furthermore, miR-196a-2 polymorphism as a susceptibility factor was related to cirrhosis-related hepatocellular carcinoma and may be associated with hepatocellular carcinoma recurrence after liver transplantation [61, 86, 87]. Based on this evidence, we conclude that the miR-196a gene could be used as a pivotal therapeutic target for gene therapy in patients with different carcinomas, opposing a new strategy that may increase the sensitivity of cancer cells to antitumor drugs based on miR-196a, opening a new era in cancer therapy.
Conclusion
These data pave the road for further characterization of the function of miR-196a in human malignancies and propose that miR-196a plays an important role in early diagnosis, prognosis, and development of personalized therapies in human cancer. Several target genes of miR-196a have highlighted the potential for small molecules to regulate miR-196a expression and affect biological processes such as proliferation, migration, and invasion. In addition, SNPs in miR-196a could contribute to cancer risk and may be associated with cancer prognosis and therapy. More studies should be conducted to further elucidate the complex regulatory mechanisms, and more work should be performed to apply miRNA-based diagnosis and therapy to clinical practice.
References
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature Reviews Genetics. 2009;10:155–9.
Zheng G, Xiong Y, Xu W, Wang Y, Chen F, Wang Z, Yan Z. A two-microRNA signature as a potential biomarker for early gastric cancer. Oncol Lett. 2014;7:679–84.
Hong TH, Park IY. MicroRNA expression profiling of diagnostic needle aspirates from surgical pancreatic cancer specimens. Annals of surgical treatment and research. 2014;87:290–7.
Slaby O, Srovnal J, Radova L, Gregar J, Juracek J, Luzna P, Svoboda M, Hajduch M, Ehrmann J. Dynamic changes in microRNA expression profiles reflect progression of Barrett’s esophagus to esophageal adenocarcinoma. Carcinogenesis. 2015;36:521–7.
Ge J, Chen Z, Li R, Lu T, Xiao G. Upregulation of microRNA-196a and microRNA-196b cooperatively correlate with aggressive progression and unfavorable prognosis in patients with colorectal cancer. Cancer Cell Int. 2014;14:128.
Liu M, Du Y, Gao J, Liu J, Kong X, Gong Y, Li Z, Wu H, Chen H. Aberrant expression miR-196a is associated with abnormal apoptosis, invasion, and proliferation of pancreatic cancer cells. Pancreas. 2013;42:1169–81.
Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, De W, Wang ZX. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012a;12:348.
Huang F, Tang J, Zhuang X, Zhuang Y, Cheng W, Chen W, Yao H, Zhang S. MiR-196a promotes pancreatic cancer progression by targeting nuclear factor kappa-B-inhibitor alpha. PLoS One. 2014;9:e87897.
Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;110:1260–8.
Karsy M, Arslan E, Moy F. Current progress on understanding microRNAs in glioblastoma multiforme. Genes & cancer. 2012;3:3–15.
Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas. 2011;40:1165–71.
Du M, Lu D, Wang Q, Chu H, Tong N, Pan X, Qin C, Yin C, Wang M, Zhang Z. Genetic variations in microRNAs and the risk and survival of renal cell cancer. Carcinogenesis. 2014a;35:1629–35.
Hoffman AE, Zheng T, Yi C, Leaderer D, Weidhaas J, Slack F, Zhang Y, Paranjape T, Zhu Y. MicroRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res. 2009;69:5970–7.
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, Huang KH, Lin WC. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes, chromosomes & cancer. 2012;51:394–401.
Ahmed FE, Ahmed NC, Vos PW, Bonnerup C, Atkins JN, Casey M, Nuovo GJ, Naziri W, Wiley JE, Mota H, Allison RR. Diagnostic microRNA markers to screen for sporadic human colon cancer in stool: I. Proof of principle. Cancer genomics & proteomics. 2013;10:93–113.
Guan Y, Mizoguchi M, Yoshimoto K, Hata N, Shono T, Suzuki SO, Araki Y, Kuga D, Nakamizo A, Amano T, Ma X, Hayashi K, Sasaki T. MiRNA-196 is upregulated in glioblastoma but not in anaplastic astrocytoma and has prognostic significance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:4289–97.
Maru DM, Singh RR, Hannah C, Albarracin CT, Li YX, Abraham R, Romans AM, Yao H, Luthra MG, Anandasabapathy S, Swisher SG, Hofstetter WL, Rashid A, Luthra R. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–8.
Han Q, Zhou C, Liu F, Xu G, Zheng R, Zhang X. MicroRNA-196a post-transcriptionally upregulates the UBE2C proto-oncogene and promotes cell proliferation in breast cancer. Oncol Rep. 2015;34:877–83.
Hao YX, Wang JP, Zhao LF. Associations between three common microRNA polymorphisms and hepatocellular carcinoma risk in Chinese. Asian Pacific journal of cancer prevention: APJCP. 2014;14:6601–4.
Zhang C, Yao C, Li H, Wang G, He X. Combined elevation of microRNA-196a and microRNA-196b in sera predicts unfavorable prognosis in patients with osteosarcomas. Int J Mol Sci. 2014;15:6544–55.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III drosha initiates microRNA processing. Nature. 2003;425:415–9.
Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell cycle (Georgetown, Tex). 2010;9:4455–60.
Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human risc couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–40.
Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science (New York, NY). 2004;304:594–6.
Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, Fu Y, Weng Z, Kuznetsov VA, Sung WK, Ruan Y, Dang CV, Wei CL. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc Natl Acad Sci U S A. 2006;103:17834–9.
Jin K, Park S, Teo WW, Korangath P, Cho SS, Yoshida T, Gyorffy B, Goswami CP, Nakshatri H, Cruz LA, Zhou W, Ji H, Su Y, Ekram M, Wu Z, Zhu T, Polyak K, Sukumar S. HOXb7 is an ERalpha cofactor in the activation of HER2 and multiple er target genes leading to endocrine resistance. Cancer discovery. 2015;5:944–59.
Jin K, Sukumar S. A pivotal role for HOXB7 protein in endocrine resistant breast cancer. Oncoscience. 2015;2:917–9.
Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–60.
Pagliuca FW, Collins MO, Choudhary JS. Coordinating cell cycle progression via cyclin specificity. Cell cycle (Georgetown, Tex). 2011;10:4195–6.
Banyai G, Baidi F, Coudreuse D, Szilagyi Z. Cdk1 activity acts as a quantitative platform for coordinating cell cycle progression with periodic transcription. Nat Commun. 2016;7:11161.
Matus DQ, Lohmer LL, Kelley LC, Schindler AJ, Kohrman AQ, Barkoulas M, Zhang W, Chi Q, Sherwood DR. Invasive cell fate requires G1 cell-cycle arrest and histone deacetylase-mediated changes in gene expression. Dev Cell. 2015;35:162–74.
Sun M, Liu XH, Li JH, Yang JS, Zhang EB, Yin DD, Liu ZL, Zhou J, Ding Y, Li SQ, Wang ZX, Cao XF, De W. MiR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol Cancer Ther. 2012;11:842–52.
Foronda D, de Navas LF, Garaulet DL, Sanchez-Herrero E. Function and specificity of Hox genes. The International journal of developmental biology. 2009;53:1404–19.
Bhatlekar S, Fields JZ, Boman BM. Hox genes and their role in the development of human cancers. Journal of molecular medicine (Berlin, Germany). 2014;92:811–23.
Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–8.
Bredel M, Scholtens DM, Yadav AK, Alvarez AA, Renfrow JJ, Chandler JP, IL Y, Carro MS, Dai F, Tagge MJ, Ferrarese R, Bredel C, Phillips HS, Lukac PJ, Robe PA, Weyerbrock A, Vogel H, Dubner S, Mobley B, He X, Scheck AC, Sikic BI, Aldape KD, Chakravarti A, GRT H. NFKBIA deletion in glioblastomas. N Engl J Med. 2011;364:627–37.
Lin CW, Hsieh YS, Hsin CH, CW S, Lin CH, Wei LH, Yang SF, Chien MH. Effects of NFKB1 and NFKBIA gene polymorphisms on susceptibility to environmental factors and the clinicopathologic development of oral cancer. PLoS One. 2012;7:e35078.
Yang G, Han D, Chen X, Zhang D, Wang L, Shi C, Zhang W, Li C, Chen X, Liu H, Zhang D, Kang J, Peng F, Liu Z, Qi J, Gao X, Ai J, Shi C, Zhao S. MiR-196a exerts its oncogenic effect in glioblastoma multiforme by inhibition of IkappaBalpha both in vitro and in vivo. Neuro-Oncology. 2014;16:652–61.
Kousteni S. FoxO1: a molecule for all seasons. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2011;26:912–7.
Zhang J, Zheng F, Yu G, Yin Y, Lu Q. MiR-196a targets netrin 4 and regulates cell proliferation and migration of cervical cancer cells. Biochem Biophys Res Commun. 2013;440:582–8.
Luthra R, Singh RR, Luthra MG, Li YX, Hannah C, Romans AM, Barkoh BA, Chen SS, Ensor J, Maru DM, Broaddus RR, Rashid A, Albarracin CT. MicroRNA-196a targets annexin a1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–78.
Suh YE, Raulf N, Gaken J, Lawler K, Urbano TG, Bullenkamp J, Gobeil S, Huot J, Odell E, Tavassoli M. MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. International journal of cancer Journal international du cancer. 2015;137:1021–34.
Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY, Lin KH. MicroRNA-196a/-196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett. 2014;351:222–31.
Lu YC, Chang JT, Liao CT, Kang CJ, Huang SF, Chen IH, Huang CC, Huang YC, Chen WH, Tsai CY, Wang HM, Yen TC, You GR, Chiang CH, Cheng AJ. OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Mol Cancer. 2014;13:218.
Wang K, Li J, Guo H, Xu X, Xiong G, Guan X, Liu B, Li J, Chen X, Yang K, Bai Y. MiR-196a binding-site snp regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33:2147–54.
Mueller DW, Bosserhoff AK. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. International journal of cancer Journal international du cancer. 2011;129:1064–74.
Li Y, Zhang M, Chen H, Dong Z, Ganapathy V, Thangaraju M, Huang S. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res. 2010a;70:7894–904.
Braig S, Mueller DW, Rothhammer T, Bosserhoff AK. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cellular and molecular life sciences: CMLS. 2010;67:3535–48.
Coskun E, von der Heide EK, Schlee C, Kuhnl A, Gokbuget N, Hoelzer D, Hofmann WK, Thiel E, Baldus CD. The role of microRNA-196a and microRNA-196b as ERG regulators in acute myeloid leukemia and acute t-lymphoblastic leukemia. Leuk Res. 2011;35:208–13.
Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2013;34:455–62.
Boissan M, Lacombe ML. Nm23/NDP kinases in hepatocellular carcinoma. J Bioenerg Biomembr. 2006;38:169–75.
Qu L, Liang L, Su J, Yang Z. Inhibitory effect of upregulated DR-nm23 expression on invasion and metastasis in colorectal cancer. European journal of cancer prevention: the official journal of the European Cancer Prevention Organisation (ECP). 2013;22:512–22.
Chen SL, Wu YS, Shieh HY, Yen CC, Shen JJ, Lin KH. P53 is a regulator of the metastasis suppressor gene Nm23-H1. Mol Carcinog. 2003;36:204–14.
Groblewska M, Siewko M, Mroczko B, Szmitkowski M. The role of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in the development of esophageal cancer. Folia histochemica et cytobiologica / Polish Academy of Sciences, Polish Histochemical and Cytochemical Society. 2012;50:12–9.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90.
Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, Baba H, Mori M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293–9.
Lei H, Wang H, Juan AH, Ruddle FH. The identification of Hoxc8 target genes. Proc Natl Acad Sci U S A. 2005;102:2420–4.
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L, Zeng Y, Miao R, Jin G, Ma H, Chen Y, Shen H. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–8.
Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, Sun AM, Liu Y. A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a chinese population. Arch Med Res. 2011;42:144–8.
Li XD, Li ZG, Song XX, Liu CF. A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in chinese patients with cirrhosis. Pathology. 2010b;42:669–73.
Lee SJ, Seo JW, Chae YS, Kim JG, Kang BW, Kim WW, Jung JH, Park HY, Jeong JY, Park JY. Genetic polymorphism of miR-196a as a prognostic biomarker for early breast cancer. Anticancer Res. 2014;34:2943–9.
Hong MJ, Choi YY, Jang JA, Jung HJ, Lee SY, Lee WK, Yoo SS, Lee J, Cha SI, Kim CH, Lee E, Jeon HS, Son JW, Park JY. Association between genetic variants in pre-microRNAs and survival of early-stage NSCLC. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2013;8:703–10.
Xu Y, Li L, Xiang X, Wang H, Cai W, Xie J, Han Y, Bao S, Xie Q. Three common functional polymorphisms in microRNA encoding genes in the susceptibility to hepatocellular carcinoma: a systematic review and meta-analysis. Gene. 2013;527:584–93.
Du W, Ma XL, Zhao C, Liu T, YL D, Kong WQ, Wei BL, JY Y, Li YY, Huang JW, Li ZK, Liu L. Associations of single nucleotide polymorphisms in miR-146a, miR-196a, miR-149 and miR-499 with colorectal cancer susceptibility. Asian Pacific journal of cancer prevention: APJCP. 2014b;15:1047–55.
Wang S, Tao G, Wu D, Zhu H, Gao Y, Tan Y, Wang M, Gong W, Zhou Y, Zhou J, Zhang Z. A functional polymorphism in miR196a2 is associated with risk and prognosis of gastric cancer. Mol Carcinog. 2013;52(Suppl 1):E87–95.
Xu Q, Liu JW, Yuan Y. Comprehensive assessment of the association between miRNA polymorphisms and gastric cancer risk. Mutation research Reviews in mutation research. 2015;763:148–60.
Darda L, Hakami F, Morgan R, Murdoch C, Lambert DW, Hunter KD. The role of HOXB9 and miR-196a in head and neck squamous cell carcinoma. PLoS One. 2015;10:e0122285.
Villegas-Ruiz V, Juarez-Mendez S, Perez-Gonzalez OA, Arreola H, Paniagua-Garcia L, Parra-Melquiadez M, Peralta-Rodriguez R, Lopez-Romero R, Monroy-Garcia A, Mantilla-Morales A, Gomez-Gutierrez G, Roman-Bassaure E, Salcedo M. Heterogeneity of microRNAs expression in cervical cancer cells: over-expression of miR-196a. International journal of clinical and experimental pathology. 2014;7:1389–401.
Lu YC, Chang JT, Huang YC, Huang CC, Chen WH, Lee LY, Huang BS, Chen YJ, Li HF, Cheng AJ. Combined determination of circulating miR-196a and miR-196b levels produces high sensitivity and specificity for early detection of oral cancer. Clin Biochem. 2015;48:115–21.
Kan T, Meltzer SJ. MicroRNAs in Barrett’s esophagus and esophageal adenocarcinoma. Curr Opin Pharmacol. 2009;9:727–32.
Shi L, Zhang C, Zhao D, Liu K, Li T, Tian H. MiR-196a-2 c>t polymorphism as a susceptibility factor for colorectal cancer. Int J Clin Exp Med. 2015;8:2600–6.
Schimanski CC, Frerichs K, Rahman F, Berger M, Lang H, Galle PR, Moehler M, Gockel I. High miR-196a levels promote the oncogenic phenotype of colorectal cancer cells. World J Gastroenterol. 2009;15:2089–96.
Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, Hirasawa A, Iwamaru A, Ishikura T, Hanaoka H, Okubo K, Onozaki T, Zama T. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One. 2013;8:e71480.
Dou T, Wu Q, Chen X, Ribas J, Ni X, Tang C, Huang F, Zhou L, Lu D. A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in chinese population. J Cancer Res Clin Oncol. 2010;136:1853–9.
Liu P, Xin F, Ma CF. Clinical significance of serum miR-196a in cervical intraepithelial neoplasia and cervical cancer. Genetics and molecular research: GMR. 2015;14:17995–8002.
Fan Y, Fan J, Huang L, Ye M, Huang Z, Wang Y, Li Q, Huang J. Increased expression of microRNA-196a predicts poor prognosis in human ovarian carcinoma. International journal of clinical and experimental pathology. 2015;8:4132–7.
Liu J, Gao J, Du Y, Li Z, Ren Y, Gu J, Wang X, Gong Y, Wang W, Kong X. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. International journal of cancer Journal international du cancer. 2012b;131:683–91.
Aso T, Ohtsuka T, Tamura K, Ideno N, Kono H, Nagayoshi Y, Ohuchida K, Ueda J, Takahata S, Shindo K, Aishima S, Oda Y, Mizumoto K, Tanaka M. Elevated expression level of microRNA-196a is predictive of intestinal-type intraductal papillary mucinous neoplasm of the pancreas. Pancreas. 2014;43:361–6.
Xue Y, Abou Tayoun AN, Abo KM, Pipas JM, Gordon SR, Gardner TB, Barth Jr RJ, Suriawinata AA, Tsongalis GJ. MicroRNAs as diagnostic markers for pancreatic ductal adenocarcinoma and its precursor, pancreatic intraepithelial neoplasm. Cancer genetics. 2013;206:217–21.
Szafranska-Schwarzbach AE, Adai AT, Lee LS, Conwell DL, Andruss BF. Development of a miRNA-based diagnostic assay for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn. 2011;11:249–57.
Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, Zhang Z, Zhu J, Jing Q, Qin Y, Li Z. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: miR-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–9.
Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y. Upregulation of miR-196a and hotair drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72:1126–36.
Li JH, Luo N, Zhong MZ, Xiao ZQ, Wang JX, Yao XY, Peng Y, Cao J: Inhibition of microRNA-196a might reverse cisplatin resistance of A549/DDP non-small-cell lung cancer cell line. Tumour biology the journal of the International Society for Oncodevelopmental Biology and Medicine 2015
Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L, Liu SX, Lin J, Xiao HS. Genome-wide analysis of microRNA and mRNA expression signatures in hydroxycamptothecin-resistant gastric cancer cells. Acta Pharmacol Sin. 2011;32:259–69.
Kim HY, Yoon JH, Lee HS, Cheong JY, Cho SW, Shin HD, Kim YJ. MicroRNA-196a-2 polymorphisms and hepatocellular carcinoma in patients with chronic hepatitis B. J Med Virol. 2014;86:446–53.
Xu X, Ling Q, Wang J, Xie H, Wei X, Lu D, Hu Q, Zhang X, Wu L, Zhou L, Zheng S. Donor miR-196a-2 polymorphism is associated with hepatocellular carcinoma recurrence after liver transplantation in a Han Chinese population. International journal of cancer Journal international du cancer. 2016;138:620–9.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No.81272601, 81472198), the Key Clinical Medicine Technology Foundation of Jiangsu Province (No.BL2014096), the Medical Key Talented Person Foundation of the Jiangsu Provincial Developing Health Project (No.RC2011080), the Innovation Team Project of the Second Affiliated Hospital of Nanjing Medical University, and the “333 high class Talented Man Project” (No.2011-III-2630).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zhen-Yao Chen and Xin Chen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Chen, ZY., Chen, X. & Wang, ZX. The role of microRNA-196a in tumorigenesis, tumor progression, and prognosis. Tumor Biol. 37, 15457–15466 (2016). https://doi.org/10.1007/s13277-016-5430-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5430-2