Abstract
Background
MicroRNAs (miRNAs) have long been established to remain stable in circulation, and dysregulated miRNAs in serum of tumor patients could potentially serve as novel biomarkers.
Aims
To determine whether certain serum miRNAs could represent potential diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma (PDAC).
Methods
About 35 patients diagnosed with PDAC at different stages between August 2007 and January 2009 were enrolled in this study. Sera from 15 chronic pancreatitis (CP) patients and 15 healthy individuals were treated as controls. Quantitative real-time polymerase chain reaction assays specific to mature miRNAs were used to quantify the relative levels of those PDAC-associated serum miRNAs.
Results
Of the seven miRNAs detected, three were identified as differentially expressed in PDAC and control groups. miR-21 was able to distinguish PDAC patients from CP (p = 0.033) and healthy subjects (p = 0.001), whereas miR-155 and miR-196a were able to differentiate sera with sick pancreas (PDAC/CP) from normal pancreas (p = 0.0002 and 0.010, respectively). Serum miR-196a expression levels in unresectable PDAC (stages III and IV) patients were significantly higher than those in resectable (stages I and II) patients (p = 0.001). Furthermore, serum miR-196a expression level was found to have a potential value in predicting median survival time of PDAC patients (high-level miR-196a, 6.1 months, (95% CI, 4.49–7.72) versus low-level miR-196a, 12.00 months, (95% CI, 5.92–18.08), p = 0.007).
Conclusions
Serum miR-196a could be a potential noninvasive marker for PDAC prognosis and selection of laparotomy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pancreatic cancer is one of the most devastating and rapidly fatal cancers, the incidence of which is almost equal to case-fatality. Surgical resection offers the most ideal treatment selection with prolonged survival for patients with resectable disease [1, 2]. However, the overwhelming majority of patients with newly diagnosed pancreatic cancer arrive with unresectable stage III and stage IV diseases [3, 4]. Good biomarkers, especially those that are non-invasive, are urgently needed in the field of diagnosing pancreatic cancer before they become unresectable. Due to the relatively easy accessibility of serum or plasma, circulating biomarkers remain one of the most promising means of diagnosis.
microRNAs (miRNAs) are endogenous, small (18–25 nt), non-coding RNAs that repress the expression of mRNAs by either cleavage or translational repression through perfectly or imperfectly binding to the 3′ untranslated region of target mRNAs. miRNAs were first discovered in 1993, and not until 8 years later were they found to be involved in multiple important biological processes, including development, differentiation, cancer, etc. The most recent release of the miRBase Registry (http://microrna.sanger.ac.uk/) (14.0, released on September 2009) lists more than 700 different miRNAs identified in humans [5]. These miRNAs may regulate expression of more than one-third of human genes [6].
In 2008, Lawrie et al. first established the existence of miRNA in circulation, which highlighted the potential of miRNAs as non-invasive diagnostic markers and their promising applications in therapy and prognostication against cancerous diseases [7]. This notion was further validated by similar studies and has extended beyond the clinical oncology domain. For instance, among 95 miRNAs screened, miR-92 emerged to be a potential plasma marker for colorectal cancer [8]. For PDAC, miR-21 [9], miR-155 [10], and miR-196a [11] have proved to be promising markers by analysis of cancerous tissues. However, whether the associated alteration in serum miRNA levels could be detected in PDAC patients remains to be determined.
The current study investigated whether circulating miRNAs could be detected in serum of patients with PDAC, and whether expression levels of specific miRNAs differed between PDAC patients and healthy individuals. Knowing that PDAC often occurs in a background of chronic pancreatitis (CP), we also use CP serum as a second control. Since “tumor is a wound that never heals” [12, 13] and highly expressed miRNAs in specific tissues may leak into circulation as biomarkers for tissue injuries [14–17], we hypothesized that those highly expressed miRNAs in PDAC tissues might be detectable in circulation as biomarkers. On the basis of this notion, we investigated in serum the expressions of miR-21, 155, 196a, 181a, 181b, 221, and 222, all of which were validated to be highly expressed in tissues of PDAC, to see if they could serve as good biomarkers for PDAC detection. If certain miRNAs levels were different between PDAC, CP, and normal, further analysis would be done to explore their relationship with tumor staging and survival.
Patients and Methods
Patients and Serum Samples
In total, 65 serum samples were used in this study, including 35 from PDAC patients, 15 from CP patients, and 15 from normal persons. Histopathologic diagnosis of PDAC were determined by the WHO criteria and confirmed by post-pancreatectomy pathology. All serum samples were collected 1 day before surgery and then properly stored. Diagnosis of CP was determined according to the Asia-Pacific consensus for chronic pancreatitis [18]. The demographic data and patient information are shown in Table 1. Samples of serum from the individuals enrolled in our study were stored at −80°C within 3 h after collection. Post-resection survival time of PDAC patients was calculated from the initiation date of surgery until the date of death or the last follow-up visit.
Informed consent was obtained from participants for the use of their blood samples in this study. This project was approved by the Ethics Committee of Shanghai Changhai Hospital (Shanghai, China). All patients were considered sporadic cases on the basis that no family histories of PDAC or chronic pancreatitis were reported. Tumors were staged according to the sixth edition of the AJCC tumor-node-metastasis (TNM) staging system. No patient had received chemotherapy or radiotherapy before blood sampling.
Serum Collection
Blood samples for miRNA detection were collected form individuals of three different groups, and were allowed to sit at room temperature for a minimum of 30 min and a maximum of 2 h. Separation of the clot were accomplished by centrifugation at 1,200 g at 4°C for 20 min. Each serum sample (500 μl at least) was moved into a 1.5-ml Eppendorf tube, leaving enough serum in the original tube such that the lowest point of the meniscus did not touch the clot. Then the samples were stored at −80°C waiting for further extraction for total RNA isolation.
RNA Extraction
All serum samples were thawed on ice and 200 μl of each sample was transferred to a tube containing 750 μl TRI Reagent BD (Molecular Research Center, Inc., Cincinnati, USA) and 20 μl 5 mol/l acetic acid. Five microliters of synthetic C. elegans miRNAs cel-miR-39 (50 pmol/l, synthetic RNA oligonucleotides synthesized by Qiagen) was added to each denatured sample as the spiked-in control [17, 19]. RNA was isolated using the TRI Reagent BD following the manufacturer’s protocol. Each obtained RNA pellet was resuspended in 40 μl nuclease-free water and stored at −80°C.
Serum miRNA Quantification by Real-Time Quantitative RT-PCR
A TaqMan miRNA real-time RT-PCR kit (Applied Biosystems) was used to detect and quantify the mature miRNA existing in total RNA extracted from sera. Briefly, 2 μl of serum-derived total RNA (from 10 μl of serum) was reverse transcribed by TaqMan® miRNA RT Kit. Negative controls were included with every real-time RT-PCR assay, and no amplification of the signal was detected when nuclease-free water was added instead of RNA or cDNA sample. Data were analyzed with 7500 software v.2.0.1. (Applied Biosystems), with the automatic Ct setting for adapting baseline and threshold for Ct determination. RT-PCR assays were performed in triplicate on each cDNA sample. Data obtained were translated in log2 (relative level).
Statistical Analysis
With SPSS 13.0 software (SPSS Inc., IL, USA), relative quantifications of the studied miRNAs were compared between groups using Student's t test, ANOVA, or nonparametric methods. Receiver operating characteristic (ROC) curves were established for discriminating patients with resectable or unresectable PDAC. The median survival time of post-operative survival analysis was evaluated by the Kaplan–Meier method and log-rank analysis. A p value of less than 0.05 (two-sided) was defined as statistically significant.
Results
Patient Information
A total of 65 participants including 35 PDAC patients, 15 CP patients, and 15 healthy volunteers were recruited in current study (Table 1). There was no significant difference in age and sex distribution between three groups.
Distinct miRNA Signatures of Sera from PDAC Patients, CP Patients, and Normal Individuals
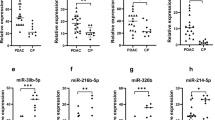
Expression levels of seven PDAC-associated miRNAs (miR-21, 155, 196a, 181a, 181b, 221, and 222) were compared in serum samples from patients with PDAC (n = 35), CP (n = 15), and healthy controls (n = 15). Although these miRNAs were previously identified to be relatively abundant in PDAC tissues, our results showed that serum miRNA signature was not fully consistent with that of solid tumors. We found that miR-21 could differentiate PDAC from CP and healthy individuals (p = 0.033 and 0.001, respectively), corresponding to an average fold change of 2.26 (PDAC vs. CP and normal), whereas miR-155 and miR-196a could differentiate sera with sick pancreas (PDAC and CP), from those from healthy controls (p = 0.0002 and 0.010), corresponding to an average fold change of 5.86 and 36.14, respectively. However, the expression levels of the other four miRNAs (miR-181a, 181b, 221, and 222) exhibited no significant differences between three studied groups (p = 0.156, 0.207, 0.698, and 0.254) (Fig. 1). To highlight the potential value of tumor-abundant miRNAs’ elevation in serum, we also tested the relative level of a non-tumor-associated miRNA (miR-16). Our results showed no significant difference of this non-tumor-associated miRNA among studied groups, suggesting that PDAC or CP would not cause general elevation of serum miRNAs.
Differential amounts of eight miRNAs in sera of PDAC, CP, and normal. miR-21 (a) was able to distinguish PDAC from CP and normal, miR-155 (b) and miR-196a (c) were able to distinguish normal from PDAC and CP. There is no general elevation in the expression (d) of either those tumor-associated miRNAs (miR-181a, 181b, 221, and 222) or those non-tumor-associated miRNAs (e.g., miR-16)
Relationship Between Serum miR-196a Expression and Resectability of PDAC
Next, to evaluate the clinical significance regarding three elevated miRNAs in serum, we subgrouped PDAC into different clinical stages according to the sixth edition of the current AJCC staging system (Table 2) [20]. As surgical resection offers the only chance of cure for PDAC patients, and whether the primary tumor can be removed represents the strongest prognostic factor for patients with this disease, differentiation of the patients with resectable PDAC from those unresectable becomes critical for surgeons’ selection of laparotomy. By classifying 35 PDAC patients into resectable group (stages I and II) and unresectable group (stages III and IV), it was found that serum miR-196a expression level in the unresectable group was significantly higher than that in the resectable group (p = 0.001) (Fig. 2a). ROC curve analyses revealed that the serum level of miR-196a was a useful biomarker for differentiating PDAC resectable from those unresectable PDAC, the ROC curve area being 86.4% (95% CI = 0.745–0.983) (Fig. 2b). At the cut-off value of −5.22 (log2 relative level), the sensitivity was 100% and the specificity was 75% (Fig. 2a). No significant correlation was observed between expression levels of the other six miRNAs (miR-21, 155, 181a, 181b, 221, 222) and resectable status in this series of cases (p = 0.159, 0.736, 0.375, 0.833, 0.520, 0.136, respectively) (Fig. 2c). Expression levels of miR-196a in sera are associated with the TNM staging of PDAC and thus provide objective clues for the selection of laparotomy on the part of surgeons.
Validation of the association between serum miR-196a level and PDAC’s resectability. a Scatter plot of miR-196a expression level according to resectability of PDAC. Serum levels of miR-196a could discriminate PDAC resectable from those unresectable (p = 0.001). The dashed line indicates a 100% sensitivity and a 75% specificity threshold with the cut-off of −5.22 (log2 relative level). b Serum miR-196a yielded a receiver operating characteristic (ROC) curve value of 86.4% (95% CI = 0.745–0.983). c Serum levels of other tumor-associated miRNAs (miR-21, 155, 181a, 181b, 221, 222) are not substantially different between two groups
Predictive Value of Serum miR-196a for PDAC Survival
Knowing that surgically removing primary tumor represents the strongest prognostic factor for patients with PDAC and high level serum miR-196a was identified to be associated with less probability of resectability, the impact of serum expression level of miR-196a on survival of PDAC patients was investigated. Of the 35 PDAC patients, 32 were still in the follow-up list by June 2009 and the other three were lost from the follow-up plan (Table 2). Kaplan–Meier survival curves were generated and compared by log-rank analysis. Based on the relative quantification data of miR-196a, −5.22 (log2 relative level) was taken as the cut-off value to classify all PDAC patients into high miR-196a and low miR-196a groups. Patients with high expression of miR-196a had a median survival of 6.1 months (95% CI, 4.49–7.72) versus 12.0 months (95% CI, 5.92–18.08) for those with low expression (p = 0.007). High expression of miR-196a, which was seen in 48.6% of tumors, resulted in a 1-year expected survival of zero versus 40.6% for low expression (Fig. 3). This result was consistent with the study published in JAMA, which indicated that high expression of miR-196a-2 in pancreatic tissue was found to predict poor survival [11].
While we were preparing these results for publication, another research group reported that a panel of four miRNAs (miR-21, 210, 155, and 196a) in combination had a reasonable power to differentiate PDAC from healthy controls [21], which was somewhat similar to our results. Given the totally different prognoses that two diseases have and common features that they hold, notably that PDAC often occurs in a background of CP, we think it is worthy to use CP serum samples as the other control besides healthy individuals. Though Wang et al.’s work suggested that a panel of four miRNAs, including miR-155 and miR-196a, could be used to diagnose PDAC with good sensitivity and specificity, our results showed that the expression levels of miR-155 and 196a in CP sera were also significantly elevated compared to normal controls, which may lower their diagnostic efficacy for PDAC. Furthermore, although no significant difference in serum miR-21 and 155 in cancer samples of different stages was observed in our study, which was consistent with the finding of Wang et al., we found that miR-196a was able to differentiate resectable PDAC from unresectable PDAC, implying that miR-196a may be a potential marker for laparotomy selection. Further follow-up study consolidated serum miR-196a’s prognostic value for PDAC. Sampling bias may be the key factor leading to the distinction between two articles. As our sample size is small, a large and well-defined cohort is required in independent studies for further confirmation.
Discussion
Although there is a long history of investigation of circulating DNA as biomarkers in many fields, studies of circulating RNA are still in its infancy. Since tremendous RNases in circulation were widely thought to be a challenge to RNAs [22], we used to speculate that miRNAs could not keep intact in plasma/serum [23]. RNAs detected in cell-free blood were considered as degraded fragments of large-molecular-weight RNAs. However, this opinion has been changed since Lawrie et al. [7] demonstrated that miR-21, the finding circulating miRNA, existed in sera from diffuse large B-cell lymphoma patients and high expression levels of it were found to be associated with improved relapse-free survival times. Nowadays, researches on circulating miRNAs have emerged in many fields, including prostate cancer [19], acute leukemia [24], fetal medicine [25], stroke [26], colorectal cancer [8], drug-induced liver injury [16], etc. Though the enigmatic mechanism of their existence in serum/plasma remains to be elucidated, the stability of miRNAs has been established in almost all the literature concerning this field. Our study further validated the existence of miRNAs in circulation and firstly showed their potential utility as diagnostic or prognostic biomarkers for PDAC.
In this study, we identified differential expressions of three PDAC-associated miRNAs: miR-21, miR-155, and miR-196a. miR-21 was able to differentiate PDAC patients from those with benign pancreas (CP/normal), whereas miR-155 and miR-196a could differentiate sick pancreas (PDAC/CP) from normal pancreas. Too much attention has been previously attached to the protein-based biomarkers, while those newly developed up-regulated miRNAs in sera, which are often associated with down-regulated proteins, used to be neglected. Hence we deduced that a cluster of up-regulated miRNAs combined with protein-based biomarkers will notably improve the diagnosis of cancers, perhaps those masses “suspicious for malignancy”. A preliminary diagnostic strategy for these patients might be set up, with the combination of the above three serum miRNAs.
miR-21 is a small multi-faceted miRNA that has been proved to be up-regulated in numerous cancers, including glioma, breast cancer, ovarian cancer, colorectal cancer, gastric cancer, pancreatic cancer, etc. Thus it was used as the best hit in a number of medium-scale and high-scale profiling experiments designed for the detection of miRNAs dysregulated in cancers. A list of direct miR-21 targets have been identified recently, including PDCD4, RECK, NFIB, tropomyosin 1, etc. It should be noted that PDCD4 [27] and RECK [28] are two tumor suppressors that have been proven to be involved in the tumorigenesis of PDAC, which may partly explain the role that the miR-21 elevation plays. Nevertheless, miR-21 has also been identified to be consistently induced in response to hypoxia, a pathophysiological setting accompanying cancer, and elevated expression of miR-21 was reported in sera of other tumors, both of which lower the specificity of this potential biomarker. As stressed by Dillhoff M et al. [9], miR-21 was overexpressed in pancreatic cancer tissue and could be a potential predictor of survival, while the serum results from the current study only demonstrated its overexpression but did not find its prognostic value. Further tissue-serum paired study is encouraged to determine the diagnostic value of miR-21.
miR-155 is a typical multifunctional miRNA and the first miRNA showed to increase in cancer. Elevated expression of miR-155 has been verified in a cluster of neoplastic diseases, e.g., 10–14-fold up-regulated in PDAC as compared to that in normal pancreatic tissue [10]. The distinguished ability of miR-155 to differentiate sick pancreas (PDAC and CP) from those healthy controls also suggests its diagnostic utility as a first-line serum biomarker for early PDAC.
In addition, this study demonstrated for the first time that the expression level of miR-196a in serum might significantly impact postoperative survival. The expression level of miR-196a in the unresectable (stages III and IV) PDAC serum was significantly higher than that in the resectable group (stages I and II) (p = 0.001), which may provide new objective clues for selection of laparotomy. Notably, our result is quite consistent with the data of Bloomston et al. [11], who found that the expression level of miR-196a in PDAC tissue was able to distinguish between long- and short-term survivors. It would be of great value if further studies could elucidate the correlation of these two levels from different sources. We hypothesize that miR-196a may be packaged inside exosomes that are secreted from PDAC cells enriched of this survival-related miRNA. Exosomes are a type of intraluminal vesicles derived from multivesicular bodies that can be released into the extracellular milieu by exocytic fusion with the plasma membrane [29]. Though exosomes were long considered to shed unwanted proteins from cells undergoing terminal differentiation [30], several recent independent studies showed that secreted exosomes contained biofunctional compositions, including proteins, mRNAs, and miRNAs. They could be delivered to other cells and function [31]. Although the mechanism by which miR-196a promotes invasion and metastasis has not been elucidated in PDAC, its involvement in tumorigenesis and progression has been reported in other cancers [32–34]. Several studies have even evaluated its diagnostic potential in clinical settings [35, 36]. Whether targeting miR-196a by antisense oligonucleotides complementary to it (named AMO or antagomirs) to abolish its function may eventually improve the prognosis of PDAC needs to be confirmed in future studies.
Though the results are promising, several limitations exist in current study. Firstly, the sample size is small. Therefore additional investigations with larger cohorts of healthy people and patients are needed to extensively evaluate the miRNAs as practical biomarkers for PDAC. qPCR by relative quantification approach becomes less accurate if being used to measure those low-level miRNAs that fall out of the linear of the assay. More accurate approaches are encouraged to further validate the results of current study. A paired tissue study has not been done but we think it would not affect the goal of serum biomarker screening. A fall down level of these miRNAs after operation by dynamic evaluation would strengthen their role in PDAC. Finally, miRNAs other than those reported in this article might also be identified as serum PDAC markers in future reports.
As a conclusion, our results indicate that sera from patients with PDAC have a unique miRNA expression pattern compared to normal as well as chronic pancreatitis. This study suggests that the amount of miRNAs in serum have potential as diagnostic and prognostic biomarkers for cancer, including PDAC. miR-196a could prove to be a potential noninvasive molecular marker for PDAC prognosis and selection of laparotomy. As circulating microRNAs “are not the molecular remnants, of once-living tumor cells, but rather of utmost functional importance” [37], we will further study the mechanism by which miRNAs affect PDAC’s biology and explore the possible therapeutic effects involved in it.
Abbreviations
- miRNA:
-
microRNA
- PDAC:
-
Pancreatic ductal adenocarcinoma
- CP:
-
Chronic pancreatitis
- ROC:
-
Receiver operating characteristic
- TNM:
-
Tumor-node-metastasis
References
Gallagher SF, Zervos EE, Murr MM. Distal pancreatectomy. In: Von Hoff DD, Evans DB, Hruban RH, eds. Pancreatic Cancer. Boston, MA: Jones and Bartlett Publishing Company; 2005:299–312.
Yen TW, Abdalla EK, Pisters PW, Evans DB. Pancreaticoduodenectomy. In: Von Hoff DD, Evans DB, Hruban RH, eds. Pancreatic Cancer. Boston, MA: Jones and Bartlett Publishing Company; 2005:265–286.
Bilimoria KY, Bentrem DJ, Ko CY, et al. Validation of the 6th edition AJCC Pancreatic Cancer Staging System: report from the National Cancer Database. Cancer. 2007;110:738–744.
Sener SF, Fremgen A, Menck HR, Winchester DP. Pancreatic cancer: a report of treatment and survival trends for 100, 313 patients diagnosed from 1985–1995, using the National Cancer Database. J Am Coll Surg. 1999;189:1–7.
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. Mirbase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158.
Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20.
Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large β-cell lymphoma. Br J Haematol. 2008;141:672–675.
Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–1381.
Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008;12:2171–2176.
Gironella M, Seux M, Xie MJ, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–16175.
Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908.
Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor generation and wound healing. N Engl J Med. 1986;315:1650–1659.
Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563.
Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–1949.
Laterza OF, Lim L, Garrett-Engele PW, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem. 2009;55:1977–1983.
Wang K, Zhang S, Marzolf B, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407.
Wang GK, Zhu JQ, Zhang JT, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666.
Tandon RK, Sato N, Garg PK. Chronic pancreatitis: Asia-Pacific consensus report. J Gastroenterol Hepatol. 2002;17:508–518.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518.
Katz MH, Hwang R, Fleming JB, Evans DB. Tumor-node-metastasis staging of pancreatic adenocarcinoma. CA Cancer J Clin. 2008;58:111–125.
Wang J, Chen J, Chang P, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2:807–813.
Tsui NB, Ng EK, Lo YM. Stability of endogenous and added RNA in blood specimens, serum, and plasma. Clin Chem. 2002;48:1647–1653.
Reddi KK, Holland JF. Elevated serum ribonuclease in patients with pancreatic cancer. Proc Natl Acad Sci USA. 1976;73:2308–2310.
Tanaka M, Oikawa K, Takanashi M, et al. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS One. 2009;4:e5532.
Gilad S, Meiri E, Yogev Y, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148.
Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966.
Ma G, Guo KJ, Zhang H, et al. Expression of programmed cell death 4 and its clinicopathological significance in human pancreatic cancer. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2005;27:597–600.
Masui T, Doi R, Koshiba T, et al. RECK expression in pancreatic cancer: its correlation with lower invasiveness and better prognosis. Clin Cancer Res. 2003;9:1779–1784.
van Niel G, Porto-Carreiro I, Simoes S, Raposo G. Exosomes: a common pathway for a specialized function. J Biochem. 2006;140:13–21.
Johnstone RM. The Jeanne Manery-Fisher memorial lecture 1991. Maturation of reticulocytes: formation of exosomes as a mechanism for shedding membrane proteins. Biochem Cell Biol. 1992;70:179–190.
Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808–818.
Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916.
Hu Z, Chen J, Tian T, et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest. 2008;118:2600–2608.
Luthra R, Singh RR, Luthra MG, et al. MicroRNA-196a targets annexin A1: a microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene. 2008;27:6667–6678.
Maru DM, Singh RR, Hannah C, et al. MicroRNA-196a is a potential marker of progression during Barrett’s metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus. Am J Pathol. 2009;174:1940–1948.
Szafranska AE, Doleshal M, Edmunds HS, et al. Analysis of microRNAs in pancreatic fine-needle aspirates can classify benign and malignant tissues. Clin Chem. 2008;54:1716–1724.
Jackson DB. Serum-based microRNAs: are we blinded by potential? Proc Natl Acad Sci USA. 2009;106:E5.
Acknowledgments
All authors would like to thank Xiaofei Ye, Ph.D. from the Statistics Department of SMMU for data collection and analysis.
Funding support
The project was supported by the China Key Technology R&D Program (Grant No. 2006BAI02A12) and National Nature Science Foundation of China (30971344).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiangyu Kong, Yiqi Du, and Guokun Wang are the co-first authors, and have contributed equally to this work.
An erratum to this article can be found at http://dx.doi.org/10.1007/s10620-010-1410-3
Rights and permissions
About this article
Cite this article
Kong, X., Du, Y., Wang, G. et al. Detection of Differentially Expressed microRNAs in Serum of Pancreatic Ductal Adenocarcinoma Patients: miR-196a Could Be a Potential Marker for Poor Prognosis. Dig Dis Sci 56, 602–609 (2011). https://doi.org/10.1007/s10620-010-1285-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-010-1285-3