Abstract
Squamous cell carcinoma of the lung (SCCL) is the most common and aggressive lung tumor with poor clinical outcome. Identification and development of potential genes in prognostic process could be beneficial for clinical management. Sequencing data of 300 SCCL samples at level 3 were downloaded from The Cancer Genome Atlas (TCGA) data portal. Single-factor survival analysis was performed by the Kaplan-Meier method. Functional annotation was conducted on the high-frequency genes filtered out by 1000 times of the least absolute shrinkage and selectionator operator regression analysis. Meanwhile, multi-factor survival analysis was conducted and ROC curve were produced. Risk coefficient and expression level of each gene were used in the division of high-risk and low-risk genes. The number of high-risk genes of each sample was obtained, and the survival condition of different samples was analyzed. Finally, the number of optimal high-risk genes was obtained. Seven thousand nine hundred ninety-eight differential expressed mRNAs were obtained, and 2041 potential prognostic genes were screened out. Twenty one of the 22 high-frequency genes were showed to have significant impact on prognostic process. Single-factor analysis was performed on the 22 models, and eight efficient models were obtained, and seven among them were proven to be significant. By random testing, ≥5 genes and ≥6 genes were proven to be most stable and ≥6 genes were finally recognized as the beneficial indicator to distinguish lung squamous cell carcinoma. Twenty-two potential genes differentially expressed in lung squamous cell carcinoma were identified as potential prognostic indicator in clinical outcome, and the novel model in this study could be applied in other cancer types.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer remains the leading cause of cancer-related death in many countries despite extensive preclinical and clinical research [1]. Lung cancer is one the major causes that influence the trends of overall cancer incidence [2]. It is characterized by late stage of presentation coupled with intrinsic resistance to cytotoxic chemotherapy [3]. Non-small cell lung cancer (NSCLC, accounting for 85 % of all lung cancers) and small cell lung cancer (SCLC, accounting for 15 %) are the two major forms of lung cancer [4]. NSCLC can be divided into three major histological subtypes: squamous cell carcinoma (SCC), adenocarcinoma (AC), and large cell lung cancer (LCC). Smoking causes all types of lung cancer but is most strongly linked with SCLC and squamous cell carcinoma, while adenocarcinoma is the most frequent type in patients who have never smoked [5–8].
The incidence of SCC in males is higher than females. Treatment of patients with SCC remains a vexing problem, and long-term survival beyond 5 years is extremely rare [9]. Despite various treatments for SCC patients, including surgery, radiotherapy, chemotherapy, or a comprehensive therapy approach, the survival rates for patients with SCC had not increased much [10]. Therefore, we aim to improve treatment and prevention of the disease by greater knowledge of the molecular origins and progression of lung cancer.
The Cancer Genome Atlas (TCGA) pilot is a feasible and powerful tool. The project can expand knowledge of the molecular basis of various cancers, and it aims to assess the value of large-scale multi-dimensional analysis of many molecular characteristics in human cancer, providing data rapidly to the research community [11]. Besides, the interim integrative analysis of DNA copy number, gene expression, and DNA methylation aberrations, along with network view of the pathways altered in the development of cancer, can be much helpful in clinical management. In this study, TCGA Research Network was established to generate the comprehensive catalog of genomic abnormalities driving tumorigenesis.
In the clinical setting, the evaluation of messenger RNA (mRNA) expression levels of selected potential genes may enable clinicians to tailor chemotherapy according to each individual’s gene profile and to produce a substantial improvement in the therapeutic outcome in terms of overall survival, time to progression, and response to therapy. But currently, not an effective model has been constructed to distinguish prognostic conditions of SCCL patients. The exploration of new markers in clinical management will hopefully improve survival and quality of life for patients with advanced SCCL.
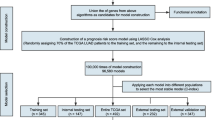
The main purpose of this study is to identify potential prognostic gene sets that are closely associated with tumor progression and survivals for SCCL patients in decreasing the dimensions by the least absolute shrinkage and selectionator operator (LASSO) regression model. Another goal is to construct a model that can distinguish the prognostic conditions of SCCL patients effectively. Here, we report that 22 potential genes could function as prognostic and predictive markers for survival of SCCL patients, and ≥6 gene model was constructed for the first time as indicator for SCC patients and can form the basis for multi-institutional randomized adjuvant trials for “high-risk” patients.
Materials and methods
Data source
The SCC microRNA (miRNA) expression profiles were downloaded from TCGA dataset. Three hundred samples with squamous lung carcinoma were included. The level 3 RNAseq data were extracted. The data platform was UNC__IlluminaHiSeq_RNASeqV2. mRNAs with no signal or whose signal was 0 were eliminated.
Data preprocessing
The standard miRNA expression profiles were extracted from the original downloaded data; mRNAs with no signal or whose signal was 0 were eliminated. To eliminate the batch effect, the generalized linear model (GLM) in Limma package of R project was used for standardization between samples.
Survival analysis
miRNA expression profiles related to squamous lung carcinoma survival were identified by the Kaplan-Meier survival analysis. Survival and prognostic conditions in each clinical stage were painted. The Cox proportional hazard regression risk ratios were used to determine influences of miRNA expressions as well as clinicopathological factors (age, gender, and recurrence) on patient survival by multiplying the ratios for all factors present [12]. SPSS (version 17.0; SPSS Inc.) was used to perform the survival analysis while the GraphPad Prism (version 5.04; GraphPad Software, Inc.) was used to generate the survival curve.
Differentially expressed gene screening
Genes whose expression value in each sample were 20 % higher than the 1.5-folds of median or lower than 1/1.5-folds of all the samples and whose variance was significantly larger than the median of all the genes in each sample (p < 0.05) were filtered out as the differentially expressed genes among cancer samples.
Potential prognostic gene screening
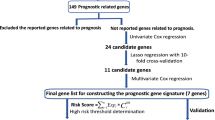
Single-factor survival analysis was performed on the differentially expressed genes to all the cancer samples by survival package [13] in R language. Two conditions were satisfied: p < 0.5 and s (variance) > 0.2. Genes meeting the above conditions were figured out as the prognostic genes to squamous cell cancer of the lung.
LASSO regression model
LASSO [14] was proposed by Tibshirani. It is an algorithm to obtain a refined model by constructing a penalty function and then define coefficient of some index as zero, thus simplifying index sets. AIC and BIC principles can help to achieve the reduction of dimensions by simplifying the variance sets of statistical model. LASSO model of penalized package [15] in R language was performed on the potential prognostic genes, and after 1000 times LASSO regression, the left genes were counted. Finally, the frequency of each gene was obtained, and genes with frequency higher than 100 were recognized as prognostic genes with high frequency.
Functional enrichment analysis
Functional enrichment analysis was performed on these high-frequency genes by Database for Annotation, Visualization, and Integrated Discovery (DAVID) [16]. Single-factor survival analysis and multi-factor survival analysis were conducted on these genes in order to obtain the roles they played on the prognostic process. Besides, ROC curve was generated (survival ROC package of R software [17]).
Construction of prognostic model
High-risk and low-risk genes were divided by the following rules [18]: (1) HR of single-factor survival analysis of genes was higher than 1 and gene expression level was on the top 20 % of all the samples. (2) HR of single-factor survival analysis of genes was lower than 1 and gene expression level was on the low 20 % of all the samples. (3) Genes which do not meet the above conditions were marked as low-risk expression genes. The model was constructed by counting the number of genes with high-risk expression, and the model that had the great impact on prognosis was screened. Samples were divided by the number of their corresponding genes with high-risk expression: ≥1 gene, ≥2 genes, ≥3 genes, etc. The samples in each category and the survival time to obtain the survival model of each category were counted. Survival analysis was performed on each model to get the prognostic condition, and then the model that significantly affected prognosis was to be found.
Model stability testing
The samples were selected randomly in the original sample sets, and the above steps were repeated in order to testify the stability of the model. The significance in survival analysis of each model was observed, and 1000 repeats were carried out in the attempt to find the most stable model.
Specificity analysis of subtype disease
All samples were divided into subtype according to the clinical experience so as to testify the sample specificity of patient samples. The subtype with more than 100 samples was selected out and was verified by the obtained model.
Results
Data processing
Gene expressions (20,254) from 300 samples were obtained after data processing from TCGA dataset. Data with undetected mRNA or no signal were eliminated. Meanwhile, the corresponding survival information of the 300 samples was obtained.
Survival analysis of different clinical stages
miRNA expression profiles related to survival were identified using the Kaplan-Meier survival analysis, and statistical significances of overall survival (OS) and progression-free survival (PFS) were determined using the log-rank test. Survival analysis was performed on SPSS (version 17.0; SPSS Inc.), and the survival curve was generated by GraphPad Prism (version 5.04; GraphPad Software, Inc.) (Fig. 1). As can be seen from the figure, various methods used to classify the disease in clinical cannot differentiate the risk degree of the disease, indicating the model is necessary to estimate the risk after prognosis accurately.
Differentially expressed gene screening
Seven thousand nine hundred ninety-eight differentially expressed genes were identified out by the forth step of the method, and all the selected genes fulfilled the two conditions mentioned above. Two thousand forty-one genes that had the potential roles on prognostic process were filtered out by single-factor analysis (Table 1).
Prognostic gene screening
LASSO was used to identify gene-gene interaction in genome-wide association studies. In this study, 22 prognostic genes, that is, BCAR3, PCDHGB4, PLIN2, SCD5, STC2, TGM2, APLN, GNB3, ZNF813, COBL, SDK2, NGFR, FKBP10, NR1I3, TNFSF11, BSPRY, C12orf53, GALNT14, NHLRC1, KLF12, and TREM1, were with high frequency after 1000 times regression by LASSO method (Table 2). And, the frequency of each gene was obtained. Twenty-one genes among them had frequency more than 100, and they were shown to have significant prognostic roles by single-factor analysis.
Survival analysis
The DAVID classification system, a powerful bioinformatic tool for classifying genes according to their function, was used to identify gene families that may play significant roles in specific pathways, biological processes, and molecular functions. In this study, we used it to classify the differentially expressed sequences (obtained after Bonferroni test application) in the comparisons between conditions. DAVID analysis was performed on the 22 high-frequency genes, and the result was shown in Table 3. Ten genes were highly enriched on four molecular functions: GO:0005509—calcium ion binding, GO:0046872—metal ion binding, GO:0043169—cation binding, and GO:0043167—ion binding (Table 3).
Multi-factor survival analysis of high-frequency genes
Multi-factor survival analysis results showed that Wald test p = 8.902e-11, revealing that the overall multi-factor survival analysis on the 22 genes was significant. The ROC curve was generated (Fig. 2). The average AUC were all above 0.05, indicating their effective roles on differentiating the disease from the normal samples on prognostic process.
Construction of efficient models
Genes with high-risk expression and corresponding with each sample were listed in Table 4. Eight efficient models were obtained, and seven among them had great impact on prognostic process. Single-factor survival analysis of the eight models was shown in Fig. 3. As can be seen, the fifth and the sixth had the largest number of samples, and the survival curve can differentiate the high-risk samples from the low-risk ones.
Stability testing of high-risk models
Random sample selecting was used to screen out the most stable prognostic model. Eventually, 1000 random significance distributions by survival analysis of each model were revealed in the box plot (Fig. 4). It showed that 5 genes and 6 genes were the most stable in the random selection.
Specificity testing of the subtype of SLLC
Different division was done by the different subtype of the samples so as to verify the specificity of the subtype of the disease (Table 5). There were fewer samples in most division, and subtype with sample number more than 100 was selected to be performed on the nine to ten steps. Single-factor survival analysis of the model corresponding with each subtype was obtained (Table 6). Five genes and 6 genes were shown to have the most favorable division effect in different subtype of the disease.
Discussion
SCC is one major subtype of lung cancer, but there were few biomarkers to aid patient management. Currently, despite advances in treatment modalities, the prognosis of SCC patients is very poor. Recent studies suggested that microRNA biomarkers could be useful for stratifying lung cancer subtypes [19], but microRNA signatures varied between different populations [20]. In this study, we identified 22 differentially expressed genes from the most significantly altered genes by using data from TCGA dataset. And we found that the 22 genes had the potential to serve as prognostic genes in clinical management. What is more, the result of single-factor survival analysis showed that 21 genes among them had significant impact on prognostic process. For example, breast cancer anti-estrogen resistance protein 3 (BCAR3) was once reported to be a candidate marker in classifying epithelial-like and mesenchymal-like phenotypes observed in NSCLCs [21] and homologous to the cell division cycle protein CDC48 [22], thus increasing the reliability of its potential role in classifying prognostic conditions of SCCL patients. Another selected gene, PCDHGB4, was also reported to be associated with lung cancer since PCDH hypermethylation was proven to be a frequent event found in all Wilms’ tumor subtypes [23]. The expression of PCDHGB4 may be involved in methylation process since hypermethylation was found to be concordant with reduced PCDH expression in tumors [24]. Other genes, PLIN2, may be involved in the development and maintenance of adipose tissue while pathways related with SCD5 were fatty acid metabolism. And the promoter methylation of transglutaminase 2 (TGM2) was identified as good responders of cisplatin in NSCLC. Therefore, we suspected that the occurrence of SCC may be in association with the metabolism of adipose. Combined with the result of enrichment analysis, the four most enriched GO terms were calcium ion binding (GO:0005509), metal ion binding (GO:0046872), cation binding (GO:0043169), and ion binding (GO:0043167), and we proposed that there may be close association between DNA methylation and ion binding ability, thus resulting in the occurrence of SCC, which needs further research to support our idea.
Since different clinical classification methods cannot make an accurate distinction between high-risk and low-risk ones, the necessity to make a prognostic model became more urgent. Walter’s study confirmed that NSCLC can be divided into two phenotypically distinct subtypes of tumor [21]. As for the squamous cell carcinoma of the lung, in this study, ≥6 gene model was constructed to distinguish prognostic condition of patient cases, providing reference for clinical therapy. The model we constructed in this study can help to predict recurrence and death in a large population of patients with SCC. The current model of 300 cancer samples from patients can be used to stratify high-risk future populations for adjuvant therapy. Nowadays, due to the development of molecular and gene profiles, molecular stratification for patients’ outcome is increasingly emphasized [25, 26], which leads to the extensive investigation and exploration of molecular markers. Therefore, the construction of the model can be used to predict recurrence of individual patients with SCC significantly, and it was consistent across all early stages of NSCLC. In this study, samples were selected randomly to ensure the reliability, and through 1000 times random LASSO regression analysis, the most stable prognostic model was found out. Herein, the model we constructed in this article was more convincing and feasible for further potential application in clinical practice. The model can also be used to identify a subgroup of patients who were at high risk for recurrence; thus, we can determine who might be best treated by adjuvant chemotherapy. In addition, the functional enrichment analysis on the 22 high-frequency genes showed that four molecular functions, namely GO:0005509—calcium ion binding, GO:0046872—metal ion binding, GO:0043169—cation binding, and GO:0043167—ion binding, were highly enriched. We speculated that the ion binding may be associated with the methylation of DNA since there were early reports about the cytotoxic effect of metal ions and their complexes on DNA interactions [27, 28], which still need further research.
Conclusion
In conclusion, we identified 22 potential genes, BCAR3, PCDHGB4, PLIN2, SCD5, STC2, TGM2, APLN, GNB3, ZNF813, COBL, SDK2, NGFR, FKBP10, NR1I3, TNFSF11, BSPRY, C12orf53, GALNT14, NHLRC1, KLF12, and TREM1, which may function as prognostic indicator of squamous lung cell carcinoma, and the ≥6 gene model constructed based on these high-risk genes can help in the early recurrences and death in localized SCC. Thus, patients with high risk for recurrence and death can receive timely adjuvant therapy. As for the association between four highly enriched GO terms and the prognostic process of SCCL, there still needs further research to prove our hypothesis.
References
Levi F, Maisonneuve P, Filiberti R, La Vecchia C, Boyle P. Cancer incidence and mortality in Europe. Soz Präventivmed. 1989;34:S3–83.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29.
Haura EB, Zheng Z, Song L, Cantor A, Bepler G. Activated epidermal growth factor receptor–stat-3 signaling promotes tumor survival in vivo in non-small cell lung cancer. Clin Cancer Res. 2005;11:8288–94.
Sanchez‐Palencia A, Gomez‐Morales M, Gomez‐Capilla JA, Pedraza V, Boyero L, Rosell R, et al. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129:355–64.
Vavalà T, Levra MG, Novello S. Lung cancer in never smokers: a different disease. Curr Respir Care Rep. 2014;3:26–34.
Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–43.
Zudaire I, Lozano MD, Vazquez M, Pajares MJ, Agorreta J, Pio R, Zulueta JJ, Yankelevitz D, Henschke C, Montuenga LM. Molecular characterization of small peripheral lung tumors based on the analysis of fine needle aspirates. 2008.
Wistuba II, Berry J, Behrens C, Maitra A, Shivapurkar N, Milchgrub S, et al. Molecular changes in the bronchial epithelium of patients with small cell lung cancer. Clin Cancer Res. 2000;6:2604–10.
Sekine I, Saijo N. Novel combination chemotherapy in the treatment of non-small cell lung cancer. Expert Opin Pharmacother. 2000;1:1131–61.
Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–30.
McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8.
Harpole DH, Herndon JE, Wolfe WG, Iglehart JD, Marks JR. A prognostic model of recurrence and death in stage I non-small cell lung cancer utilizing presentation, histopathology, and oncoprotein expression. Cancer Res. 1995;55:51–6.
O’Quigley J, Moreau T. Cox’s regression model: computing a goodness of fit statistic. Comput Methods Prog Biomed. 1986;22:253–6.
Tibshirani R. The lasso method for variable selection in the cox model. Stat Med. 1997;16:385–95.
Goeman JJ. L1 penalized estimation in the cox proportional hazards model. Biom J. 2010;52:70–84.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57.
Heagerty PJ, Lumley T, Pepe MS. Time-dependent roc curves for censored survival data and a diagnostic marker. Biometrics 2000: 337–344.
Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci. 2013;110:7413–7.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98.
Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83.
Walter K, Holcomb T, Januario T, Du P, Evangelista M, Kartha N, et al. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clin Cancer Res. 2012;18:2360–73.
Near RI, Zhang Y, Makkinje A, Borre PV, Lerner A. And‐34/bcar3 differs from other NSP homologs in induction of anti‐estrogen resistance, cyclin D1 promoter activation and altered breast cancer cell morphology. J Cell Physiol. 2007;212:655–65.
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang C, et al. A 5-microRNA signature for lung squamous cell carcinoma diagnosis and hsa-mir-31 for prognosis. Clin Cancer Res. 2011;17:6802–11.
Nevins JR, Harpole D, Potti A, West M, Dressman H. Prediction of lung cancer tumor recurrence. Google Patents, 2007.
Hirsch FR, Varella-Garcia M, Bunn PA, Di Maria MV, Veve R, Bremnes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol. 2003;21:3798–807.
Volm M, Koomägi R, Mattern J. Prognostic value of vascular endothelial growth factor and its receptor flt‐1 in squamous cell lung cancer. Int J Cancer. 1997;74:64–8.
McKee ML, Kerwin SM. Synthesis, metal ion binding, and biological evaluation of new anticancer 2-(2′-hydroxyphenyl) benzoxazole analogs of uk-1. Bioorg Med Chem. 2008;16:1775–83.
Williams RJ. Metal ions in biological systems. Biol Rev. 1953;28:381–412.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Duan, K., Li, L., Tan, Xd. et al. A novel model for identification of prognostic indicator for clinical outcome of squamous cell lung carcinoma. Tumor Biol. 37, 15873–15881 (2016). https://doi.org/10.1007/s13277-016-5407-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5407-1