Abstract
Growing evidence demonstrates that long noncoding RNAs (lncRNAs) are involved in the progression of various cancers including glioma. Nuclear enriched abundant transcript 1 (NEAT1), an essential lncRNA for the formation of nuclear body paraspeckles, was not fully explored in glioma. We aimed to determine the expression, roles, and functional mechanisms of NEAT1 in the progression of glioma. By real-time PCR, we suggested that NEAT1 was upregulated in glioma tissues than noncancerous brain tissues. Knockdown of NEAT1 reduced glioma cell proliferation, invasion, and migration. RNA immunoprecipitation assay combined with luciferase reporter assay confirmed miR-449b-5p-specific binding to NEAT1. Furthermore, we verified that c-Met was a directly target of miR-449b-5p. Rescue assays demonstrated NEAT1 functions a molecular sponge for miR-449b-5p and leads to the upregulation of c-Met. This regulation menchaism promotes glioma pathogenesis and may provide a potential target for the prognosis and treatment of glioma.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioma is the most common malignant tumor in central nervous system [1, 2]. Despite recent advances in treatment of surgery, radiotherapy, and chemotherapy, the prognosis of malignant glioma is still extremely poor [3, 4]. Therefore, the new potential biomarker for early diagnosis and novel therapeutic target needs to be explored. Although dozens of studies have indicated the complex gene interaction and molecular modulation network in the development of glioma, the research on revealing the oncogenes and tumor suppressors is still on the road [5, 6].
LncRNAs, which did not encode any proteins, are more than 200 nucleotides in length and have been shown to play key roles in imprinting control, cell differentiation, immune responses, human diseases, tumorigenesis, and other biological processes [7–9]. Expression profile revealed that many of lncRNAs were significantly altered in glioma tissue that implies that it may serve as oncogene or tumor suppressor in glioma development [10]. Indeed, several studies reported that lncRNAs related to glioma development and progression by regulating cell growth and metastasis [11, 12]. However, there are little studies about the role of nuclear enriched abundant transcript 1 (NEAT1) in glioma.
NEAT1 is one of the most highly regulated lncRNAs in the recent pangenomic datasets [13]. Twenty years ago, Guru et al. identified that it is transcribed from the familial tumor syndrome multiple endocrine neoplasia (MEN) type 1 locus on chromosome 11 and lacks any introns [14]. The alternation of NEAT1 expression has been reported in human malignancies, including leukemia and ovarian carcinoma, and plays important roles in tumorigenesis [15, 16]. To the present, lncRNAs have been demonstrated to function as competing endogenous RNAs (ceRNA) by competitively binding common microRNAs (miRNAs) [17–19]. So, lncRNAs can function as sponges for onco-miRNAs or tumor suppressor-miRNAs, and then could regulate tumor cell growth and metastasis [20, 21]. Therefore, we speculate that NEAT1 also has roles as miRNAs sponges to modulate the functions of miRNAs.
In the current study, we found that NEAT1 is upregulated in glioma tissues. And, we found that the upregulation of NEAT1 promotes the proliferation, invasion, and migration of glioma cells though regulate miR-449b-5p/c-Met axis.
Materials and methods
Clinical specimens
The 15 clinican tumor tissues were collected from surgical resections of brain glioma in the Department of Neurology, Shengjing Hospital, China Medical University, Liaoning Province, China. The adjacent normal tissues, which are defined as normal in the results, were obtained 2 cm away from the glioma tissue. These tissues were divided into two groups: grades I–II glioma group (n = 10) and grades III–IV glioma group (n = 5) according to the 2007 WHO classification of tumors in the central nervous system. All of the patients received surgery, and without preoperative chemotherapy or radiation therapy. The specimens were snap frozen in liquid nitrogen and stored at −80 °C until used in this study.
Cell lines
The glioma cell lines (U87, U373, and U251) were purchased from ATCC (Manassas, VA). All of the glioma cells were maintained in DMEM medium and supplemented with 10 % fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin.
Cell transfection
All of the constructs used to overexpression or knockdown of NEAT1 were purchased from RiboBio (RiboBio, China). The glioma cells were transfected by Opti-MEM I and Lipofectamine2000 reagents (Invitrogen, CA,USA) at approximately 50–70 % confluence according to the manufacturer’s instructions after 24 h of culture. Stable cell lines were created by selection with Geneticin (G418; Invitrogen, CA, USA).
RNA extraction and qRT-PCR
Total RNA was extracted from the cell lines by using TRIzol reagent. Then, RNA was synthesized into cDNA using M-MLV in a 10-μl volume. Real-time polymerase chain reaction (real-time PCR) was conducted using an SYBR Green mix in a 10-μl reaction volume on an ABI Prism 7900 Sequence Detector System (ABI, Foster City, CA). The expression of each miRNA was normalized to GAPDH and calculated using the 2−ΔΔCt method. Primer used in this study was shown as follows, GAPDH: forward 5′-TGCACCACCAACTGCTTAGC-3′, reverse 5′-GGCATGCACTGTGGTCATGAG-3′, miR-449b-5p: forward 5′-GGGAGGCAGTGTATTGTTA-3′, reverse 5′-CAGTGCGTGTCGTGGAGT-3′, NEAT1: forward 5-GUCUGUGUGGAAGGAGGAATT-3′, reverse 5′-UUCCUCCUUCCACACAGACTT-3′.
Cell proliferation assay
Twenty-hours after transfection, cells were seeded in 96-well plates at 8000 cells per well. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was used to determine cell viability at the point of 0, 12, 24, and 48 h after seeding. The absorbance at 570 nm was measured using a Quant Universal Microplate Spectropho-tometer (BioTek, Winooski, VT).
Apoptosis detection
Apoptosis was assessed using Annexin V-FITC/PI double staining kit (Beyotime, Jiangsu, China). Cells were harvested and stained with Annexin V-FITC and PI according to the manufacturer’s instructions. Cell samples were analyzed on flow cytometry (FACScan, BD Biosciences).
Cell migration and invasion assay, luciferase reporter assay, and pull-down assay
Cell migration and invasion assay, luciferase reporter assay, and pull-down assay were described previously [22, 23].
In vivo tumor xenograft studies
The protocol of constructing the stable cell lines was described as before [24]. Then, the stable si-NEAT1 overexpression U87 cells or control cells were inoculated with 4 × 106 cells per site bilaterally on the axillary fossae of female athymic nude mice aged 6–8 weeks. Tumor size was monitored by measuring the length and width with calipers, and volumes were calculated with the formula: (L × W 2) × 0.5, where L is the length and W is the width of each tumor. The mice used in this experiment were maintained under specific pathogen-free conditions and handled in accordance with NIH Animal Care and Use Committee regulations.
Real-time PCR-based miRNA expression profiling
Total miRNA was reverse-transcribed using Megaplex RT Primers (Applied Biosystems). The produced cDNAs were preamplified using Megaplex PreAmp Primers (Applied Biosystems), and the preamplified products were applied to a TaqMan Human MicroRNA Array Panel. Real-time PCR was performed using a 7900HT Fast Real-Time System (Applied Biosystems). The real-time PCR data obtained using the TaqMan MicroRNA Panel was analyzed with Applied Biosystems software (SDS ver. 2.3 and RQ manager ver. 1.2). For quantification, the relative CT method (−ΔΔCT method) was applied. U6 small nuclear B noncoding RNA (RNU6B) was used as the endogenous control. Each sample was run in triplicate to ensure quantitative accuracy.
Statistical analysis
All the data were presented as the mean ± SD. The significance of differences was carried out by two paired Student’s t test, and GraphPad 5.0 software was used for statistical analysis. P < 0.05 was considered statistically significant.
Results
NEAT1 was upregulated in glioma tissues and cell lines
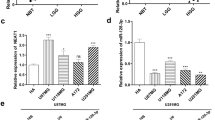
In order to identify the role of lncRNA NEAT1 in glioma, we performed real-time PCR to measure the expression of lncRNA NEAT1 in 15 glioma tissues, four normal brain tissues, and three glioma cell lines. Compared with normal brain tissues, glioma tissues and cell lines showed higher expression levels of lncRNA NEAT1 (P < 0.05, Fig. 1a). Furthermore, NEAT1 were increased with the rising pathological grades of gliomas (P < 0.05, Fig. 1b).
The expression of NEAT1 is downregulated in glioma. a Quantitative analysis of the expression levels of NEAT1 normalized against those of GAPDH using qRT-PCR. There are four adjacent normal tissues, 15 glioma tissues and glioma cell lines U373, U87, and U251. b Quantitative analysis of the expression levels of NEAT1 in adjacent normal tissues, low-grade glioma (I-II), and high-grade glioma (III-IV). *P < 0.05 compared with the normal group
Knockdown of NEAT1 inhibited cell proliferation, migration, and invasion and promoted apoptosis of glioma cells
To assess the potential functional role of NEAT, NEAT1-siRNA plasmids were constructed, and the efficiency was measured in glioma cell lines (60 % decreases in U87 cells, 70 % decreases in U251 cells, Fig. 2a). As shown in Fig. 2b, knockdown of NEAT1 resulted in a significantly decreased proliferation of U87 and U251 glioma cells compared to the respective si-NC group (P < 0.05). Transwell assays were used to detect the effects of si-NEAT1 on the invasiveness and migratory ability of glioma cells. As shown in Fig. 2c, knockdown of NEAT1 reduced the invasiveness by roughly 75 % in U87 cells and by roughly 65 % in U251 cells. Corresponding effects were also observed in a parallel migration assay, which indicated a significant reduction of migration in U87 and U251 cells compared with the respective si-NC group (P < 0.05). Moreover, Fig. 2d shows that knockdown of NEAT1 significantly promoted the apoptosis in both U87 and U251 cells compared with the si-NC group (P < 0.05).
Knockdown of NEAT1 inhibited cell proliferation, migration, and invasion and promoted apoptosis of glioma cells. a NEAT1 expression levels were evaluated using qRT-PCR in si-NEAT1-transfected U251 and U87 cells. b MTT assay was performed to determine the proliferation of U251 and U87 cell lines stably knockdown of NEAT1. c Transwell assay was performed in U251 and U87 cells stably knockdown of NEAT1 to investigate changes in cell migration and invasiveness. d The apoptotic percentages of U251 and U87 cells stably knockdown of NEAT1 were detected by flow cytometry. The LR region suggested the early apoptotic cells. Data were presented as mean ± SD from three independent experiments. *P < 0.05. Scale bar represents 20 μm
Screen and verified miR-449b-5p could directly target NEAT1
A growing body of literature has confirmed that lncRNA may function as a competing endogenous RNA (ceRNA) or a molecular sponge in modulating the concentration and biological functions of miRNA. To determine the miRNAs that are interaction with NEAT1, we used the bioinformatics, starBase2.0 for miRNAs prediction (shown in Table 1). We also performed real-time PCR-based miRNA profiling to explore the differentially expressed miRNAs between U87/NEAT1 and U87 negative control cells. Combining the bioinformatics and miRNA profiling analysis, we got miR-449b-5p, miR-335-3p, miR-449a-5p, miR-329-3p, miR-342-3p, and miR-107 as the candidate miRNAs (Fig. 3a). Among these six miRNAs, miR-449b-5p (MIMAT0003327) shared the most reduction in miRNA profiling assay; so, we selected it for further study. The binding site of miR-449b-5p to NEAT1 was shown (Fig. 3b). Figure 3c indicates that the knockdown of NEAT1 significantly increased the expression level of miR-449b-5p (P < 0.05). Meanwhile, overexpression of NEAT1 decreased miR-449b-5p expression in both glioma cell lines (Fig. 3d). To further investigate whether NEAT1 was a functional target of miR-449b-5p, dual-luciferase reporter assay was performed. The results showed that compared with the scramble group, miR-449b-5p could reduce pmirGLO-NEAT1-WT (wild-type) luciferase activity but could not affect the mutated pmirGLO-NEAT1-Mut luciferase activity (P < 0.05, Fig. 3e). Furthermore, we used pulldown assay to test whether NEAT1 could pull down miR-449b-5p, using a biotin-labeled specific NEAT1 probe. MiR-449b-5p was precipitated as analyzed by qRT-PCR (Fig. 3f). These data demonstrated that miR-449b-5p could directly bind to NEAT1 at the miRNA recognition site.
NEAT1 is a direct target of miR-449b-5p. a Screen of the candidate miRNAs that interacted with NEAT1 by real-time PCR-based miRNA expression profiling and starBase (v2.0). Coanalysis of the downregulated miRNAs in stable overexpression NEAT1 U87 cells compared to the control U87 cells and the miRNA list that potentitally target NEAT1 predicted by starBase (v2.0), shown in Table 1, we got six candidate. b Sequence alignment of miR-449b-5p with the putative binding sites within the wild-type regions of NEAT1. c, d Detection of miR-449b-5p using qRT-PCR in the si-NEAT1 or NEAT1 overexpression glioma cell lines compared with control group. e The luciferase report assay demonstrated that overexpression of miR-449b-5p could reduce the intensity of fluorescence in U87 and U251 cells transfected with the NEAT1-WT vector, while had no effect on the NEAT1 mutant vector. f Detection of miR-21 using qRT-PCR in the sample pulled down by biotinylated NEAT1 probe. Three independent assays were performed, and the data were represented as the mean ± SD. *P < 0.05
Repression of miR-449b-5p could reverse the si-NEAT1-induced glioma cell tumorigenesis inhibition
To study whether NEAT1 functions through miR-449b-5p, we synthesize miR-449b-5p knockdown mimics (anti-miR-449b-5p), which can inhibit endogenous miR-449-b-5p, to perform a rescue experiment. Real-time PCR analysis showed that overexpression of anti-miR-449b-5p mimics can decrease miR-449b-5p level (Fig. 4a). Furthermore, MTT, invasion, and migration assays were employed in U87 and U251 cells cotransfected with si-NEAT1 and anti-miR-449b-5p mimics or their control oligomers and vectors. The results show that blocking the expression of miR-449b-5p could rescue si-NEAT1-induced glioma cell viability, invasion, and migration inhibition (Fig. 4b–e). Meanwhile, blockage of miR-449-5p also decreases the apoptotic cell numbers which was increased by si-NEAT1 (Fig. 4f, g). These results showed that NEAT1 act its tumor oncogene roles though miR-449-5p in glioma cells.
Blockage of miR-449b-5p largely reversed si-NEAT1-induced inhibitory effects on glioma cells. a Real-time PCR detects the efficiency of the anti-miR-449b-5p in blockage of miR-449b-5p expression in glioma cell lines. b, c Blockage of miR-449b-5p partly reversed siNEAT1-induced inhibition of proliferation in U251 and U87 cells determined by MTT assay. OD570 nm was measured at 0, 24, and 48 h after transfection. d, e Blockage of miR-449b-5p largely reversed siNEAT1-induced suppression of migration and invasion in U251 and U87 cells detected by Transwell assay. f, g Blockage of miR-449b-5p partly reversed siNEAT1-induced apoptosis in U251 and U87 cells detected by flow cytometry. Representative date was shown. Data were presented as mean ± SD from three independent experiments. *P < 0.05
miR-449b-5p directly target oncogene c-Met
Having demonstrated that NEAT1 could negatively regulate miRNA-449b-5p expression, we then examined the functional aspect. To access this goal, we screen Targetscan to predict the potential targets of miR-449b-5p. As a result, the top 100 potential targets were identified. Among these genes, c-Met, a well-known oncogene, is upregulated in a large number of malignancies, which led us to believe that c-Met may be a direct target of miR-449b-5p in glioma. Two putative binding sites of the microRNA in the 3′-UTR of c-Met are described in Fig. 5a, and the complementary seed sequences were mutated which should not bind miR-449b-5p. Then, we conducted luciferase reporter assays to confirm that miR-449b-5p could target c-Met. As shown in Fig. 5b, the luciferase activity generated by the reporter vector with c-Met-3′-UTR WT1 decreased by 63.5 % after cotransfection with miR-449b-5p when compared with the control group. However, the activity generated by the reporter vector with c-Met-3′-UTR mut-1 was no longer changed than that of the control group. Furthermore, the second binding sites in the 3′-UTR of c-Met aslo affect the activity generated by the reporter vectors when compared with the control groups (Fig. 5c). These results indicated that miR-449b-5p might suppress the expression of c-Met by strongly and directly binding to the two putative sites in its 3′-UTR. The following real-time PCR and Western blot assays showed that the relative mRNA and protein level of c-Met in U87 and U251 were significantly decreased after transfection with miR-449b-5p, while blockage of miR-449b-5p could increase c-Met expression level in glioma cells (Fig. 5d–f).
The validation of c-Met was a direct target of miR-449b-5p. a The alignment of miR-449b-5p binding sites in the c-Met-3’UTR (wild-type and mutant) was shown. b The luciferase report assay demonstrated that overexpression of miR-449b-5p could reduce the luciferase intensity in U87 cells transfected with the c-Met-WT vector, while had no effect on the c-Met mutant 1 vector or c-Met mutant 2 vector. c, d The effect of miR-449b-5p on c-Met mRNA levels was analyzed by real-time PCR in U87 and U251 cells. e, f The effect of miR-449b-5p on c-Met protein levels was analyzed by Western blot in U87 and U251 cells. All the experiments were repeated three times, and all data are representative of three independent experiments (*P < 0.05)
NEAT1’s oncogenic activity is in part through negative regulation of miRNA-449b-5p and then modulating c-Met in glioma cells
Having confirmed that NEAT1 could regulate miR-449b-5p expression and miR-449b-4p directly target c-Met, whether NEAT1’s oncogenic activity was though miR-449b-5p/c-Met axis remains unclear. Figure 6a, b, d, and e shows that upregulated c-Met in U251 and U87 glioma cells by transfecting with pcDNA3/c-Met largely reversed the inhibitory effect of si-NEAT1 or miR-449b-5p on glioma cell proliferation, migration, and invasion. Moreover, overexpression of c-Met largely suppressed the cell apoptosis promoted by si-NEAT1 or miR-449b-5p (Fig. 6c). These results strongly suggested that NEAT1’s oncogenic activity is at least in part through miRNA-449b-5p/c-Met axis in glioma cells.
Ectopic expression of c-Met without 3′UTR restores the effects of siNEAT1 or miR-449b-5p on cell proliferation, migration, and invasion in glioma cells. a–e pcMV6/c-Met counteracts the inhibition of growth, migration, and invasion of U87 and U251 cells caused by siNEAT1 or miR-449b-5p compared with that observed in control cells. The cell growth capacity was detected using MTT assays, and the cell invasion and migration capacity were determined using the transwell assays. The cell apoptosis was detected by flow cytometry. Data were presented as mean ± SD from three independent experiments. *P < 0.05
Kncokdown of NEAT1 inhibits glioma cell growth in vivo
The stable si-NEAT1 knockdown U87 cell line as well as a control cell line was established to confirm that NEAT1’s oncogenic activity is in part through the regulation of miRNA-449b-5p axis in vivo. Cells of either line were injected into the axillary fossa of SCID mice, and the tumor growth activity was measured. As shown in Fig. 7a, the average volume of tumors derived from the si-NEAT1 group was only 30 % of that in the control group (n = 7 animals per group, P < 0.05). Furthermore, we measured the expression level of miR-449b-5p in two groups of tumors, the results revealed that the si-NEAT1 group has a higher expression of miR-449b-5p (Fig. 7b) and lower expression of c-Met (Fig. 7c) compared with the control group. The relative representative IHC image is shown in Fig. 7d. These results were consistent with the effects of NEAT1 knockdown in vitro and strongly suggested that NEAT1 regulate glioma cell proliferation, invasion, and migration through miR-449b-5p/c-Met axis (Fig. 7e).
NEAT1’s oncogenic activity is in part through miRNA-449b-5p/c-Met axis in vivo. a Tumor growth curves of subcutaneous implantation models of U87 cell are shown. The tumor derived from stable overexpression of siNEAT1-U87 cells was smaller than that of control group (n = 7 in each group). b Real-time PCR results demonstrated that the expression of miR-449b-5p was higher in the siNEAT1-U87-induced tumors compared to the control group. c Immunohistochemical staining of c-Met demonstrated that the expression of c-Met was lower in the siNEAT1-U87 induced tumors compared to the control group. d Representative IHC image is shown. Scale bar represents 20 μm. e Schematic representation of the proposed molecular mechanism of long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-3p/c-Met axis
Discussion
Studies indicated that LncRNAs play an important role in multiple process in cells, from guidance of the chromatin-modifying complexes to acting as “molecular sponges” for the regulation of microRNAs [9]. For instance, the function of many lncRNAs have been revealed, such as lncRNA GAS5 [25, 26]. Another example was long noncoding RNA CCAT1, which has been indicated as miR-218-5p sponges in promoting gallbladder cancer development [27]. Recently, Hirose et al. investigated the role of NEAT1 in transcriptional regulation through the sequestering of SFPQ from the RNA-specific adenosine deaminase B2 (ADARB2) gene in response to proteasome inhibition [28]. And, NEAT1 also has been found to be correlating with poor survival in patients with breast cancer [29]. Chakravarty D et al. provide evidence that NEAT1 drives oncogenic growth by altering the epigenetic landscape of target gene promoters to favor transcription in prostate cancer [30]. However, the detailed mechanism of NEAT1 in glioma is not clear. In the present study, we explored the role of NEAT1 in glioma cells and found that it participated in the “competitive endogenous RNAs (ceRNA)” regulatory network and act as endogenous miRNA sponges to bind to miRNAs and regulate their function. Finally, we investigated that NEAT1 regulates miR-499b-5p expression and suggested that NEAT1 could promoter glioma tumorigenesis through miR-449b-5p/c-Met axis.
Studies provide evidence that the alternation of miRNA in carcinoma and noncarcinoma cells with great molecular and clinical implications [31]. In addition, other studies explained that the miRNAs contributed to cell proliferation, apoptosis, metastasis, and development in glioma. Galina Gabriely et al. found that human glioma growth is controlled by microRNA-10b [32]. Sung-Suk Suh et al. reveal the microRNAs/TP53 feedback circuitry in glioblastoma multiforme [33]. To the present, so much research indicated which miRNA drives glioma. In this study, we focus on search which miRNA was drive by NEAT1 in glioma. At last, we capture miR-449b-5p by bioinformatics analysis, pull-down assay, and micro-array assays. To our knowledge, there was little study about miR-449b-5p, especially its function in cancers. However, its clustered miRNA (hsa-mir-449b-chr5: 55170646–55170742 [−]; hsa-mir-449a-chr5: 55170532–55170622 [−]), miR-449a, has been reported in several types of cancers. Jeon et al. found that miR-449a/b was downregulated in 21 pairs of lung cancer tissues and induced growth arrest by targeting HDAC-1 [34]. Ren et al. indicated that miR-449a caused cell cycle arrest and cell senescence in A549 and 95D cells [35]. In our study, we found that miR-449b act as a target of NEAT1; then, we validated that c-Met was a direct target of miR-449b-5p. These results suggested that NEAT1 promotes glioma pathogenesis, at least, partly through regulating miR-449b-5p/c-Met axis.
To our knowledge, this may be the first study to show the role and function of NEAT1 in glioma. And, we may first discover the NEAT1/miR-449b-5p/c-Met axis in glioma. Therefore, understanding the key role of “lncRNA-miRNA” module in glioma will lead to the identification of new therapeutic targets for treating glioma. At last, we will perform further investigation for this regulation mechanism in other carcinomas.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29.
Hegi ME, Janzer RC, Lambiv WL, Gorlia T, Kouwenhoven MC, Hartmann C, et al. European Organisation for R, Treatment of Cancer Brain T, Radiation Oncology G, National Cancer Institute of Canada Clinical Trials G: Presence of an oligodendroglioma-like component in newly diagnosed glioblastoma identifies a pathogenetically heterogeneous subgroup and lacks prognostic value: Central pathology review of the eortc_26981/ncic_ce.3 trial. Acta Neuropathol. 2012;123:841–52.
Wang Y, Jiang T. Understanding high grade glioma: Molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139–46.
Ohkawa Y, Momota H, Kato A, Hashimoto N, Tsuda Y, Kotani N, Honke K, Suzumura A, Furukawa K, Ohmi Y, Natsume A, Wakabayashi T, Furukawa K: Ganglioside gd3 enhances invasiveness of gliomas by forming a complex with platelet-derived growth factor receptor alpha and yes. The Journal of biological chemistry 2015.
Lin JJ, Zhao TZ, Cai WK, Yang YX, Sun C, Zhang Z, Xu YQ, Chang T, Li ZY: Inhibition of histamine receptor 3 suppresses glioblastoma tumor growth, invasion, and epithelial-to-mesenchymal transition. Oncotarget 2015.
Maruyama R, Suzuki H. Long noncoding rna involvement in cancer. BMB Rep. 2012;45:604–11.
Esteller M. Non-coding rnas in human disease. Nat Rev Genet. 2011;12:861–74.
Mercer TR, Dinger ME, Mattick JS. Long non-coding rnas: Insights into functions. Nat Rev Genet. 2009;10:155–9.
Han L, Zhang K, Shi Z, Zhang J, Zhu J, Zhu S, Zhang A, Jia Z, Wang G, Yu S, Pu P, Dong L, Kang C: Lncrna pro fi le of glioblastoma reveals the potential role of lncrnas in contributing to glioblastoma pathogenesis. International journal of oncology 2012;40:2004–2012.
Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, et al. A pituitary-derived meg3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88:5119–26.
Zhang JX, Han L, Bao ZS, Wang YY, Chen LY, Yan W, et al. Chinese Glioma Cooperative G: Hotair, a cell cycle-associated long noncoding rna and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro-Oncology. 2013;15:1595–603.
Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, et al. Extensive regulation of the non-coding transcriptome by hypoxia: Role of hif in releasing paused rnapol2. EMBO Rep. 2014;15:70–6.
Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, et al. A transcript map for the 2.8-mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res. 1997;7:725–35.
Kim YS, Hwan JD, Bae S, Bae DH, Shick WA. Identification of differentially expressed genes using an annealing control primer system in stage iii serous ovarian carcinoma. BMC Cancer. 2010;10:576.
Zeng C, Xu Y, Xu L, Yu X, Cheng J, Yang L, et al. Inhibition of long non-coding rna neat1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer. 2014;14:693.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A cerna hypothesis: The rosetta stone of a hidden rna language? Cell. 2011;146:353–8.
Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, et al. Coding-independent regulation of the tumor suppressor pten by competing endogenous mrnas. Cell. 2011;147:344–57.
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous mirna sponge lincrna-ror regulates oct4, nanog, and sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80.
Karreth FA, Tay Y, Perna D, Ala U, Tan SM, Rust AG, et al. In vivo identification of tumor- suppressive pten cernas in an oncogenic braf-induced mouse model of melanoma. Cell. 2011;147:382–95.
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microrna-mediated network of rna-rna interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81.
Wang Y, Wang Y, Li J, Zhang Y, Yin H, Han B: Crnde, a long-noncoding rna, promotes glioma cell growth and invasion through mtor signaling. Cancer letters 2015.
Wang P, Liu YH, Yao YL, Li Z, Li ZQ, Ma J, et al. Long non-coding rna casc2 suppresses malignancy in human gliomas by mir-21. Cell Signal. 2015;27:275–82.
Wan HY, Guo LM, Liu T, Liu M, Li X, Tang H. Regulation of the transcription factor nf-kappab1 by microrna-9 in human gastric adenocarcinoma. Mol Cancer. 2010;9:16.
Tani H, Torimura M, Akimitsu N. The rna degradation pathway regulates the function of gas5 a non-coding rna in mammalian cells. PLoS One. 2013;8, e55684.
Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP: Noncoding rna gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Science signaling 2010;3:ra8.
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY, Gong W, et al. Long non-coding rna ccat1 promotes gallbladder cancer development via negative modulation of mirna-218-5p. Cell Death Dis. 2015;6, e1583.
Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, et al. Neat1 long noncoding rna regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–83.
Choudhry H, Albukhari A, Morotti M, Hider S, Moralli D, Smythies J, Schodel J, Green CM, Camps C, Buffa F, Ratcliffe P, Ragoussis J, Harris AL, Mole DR: Tumor hypoxia induces nuclear paraspeckle formation through hif-2alpha dependent transcriptional activation of neat1 leading to cancer cell survival. Oncogene 2014.
Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, et al. The oestrogen receptor alpha-regulated lncrna neat1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383.
Zhong X, Coukos G, Zhang L. Mirnas in human cancer. Methods Mol Biol. 2012;822:295–306.
Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, et al. Human glioma growth is controlled by microrna-10b. Cancer Res. 2011;71:3563–72.
Suh SS, Yoo JY, Nuovo GJ, Jeon YJ, Kim S, Lee TJ, et al. Micrornas/tp53 feedback circuitry in glioblastoma multiforme. Proc Natl Acad Sci U S A. 2012;109:5316–21.
Jeon HS, Lee SY, Lee EJ, Yun SC, Cha EJ, Choi E, et al. Combining microrna-449a/b with a hdac inhibitor has a synergistic effect on growth arrest in lung cancer. Lung Cancer. 2012;76:171–6.
Ren XS, Yin MH, Zhang X, Wang Z, Feng SP, Wang GX, et al. Tumor-suppressive microrna-449a induces growth arrest and senescence by targeting e2f3 in human lung cancer cells. Cancer Lett. 2014;344:195–203.
Conflicts of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhen, L., Yun-hui, L., Hong-yu, D. et al. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumor Biol. 37, 673–683 (2016). https://doi.org/10.1007/s13277-015-3843-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3843-y