Abstract
LRRC3B has emerged as a tumor suppressor in several human cancers. However, its expression pattern and biological roles in human non-small-cell lung cancer (NSCLC) have not been explored. In the present study, we investigated clinical significance of LRRC3B in 101 NSCLC specimens. We found that LRRC3B expression was downregulated in NSCLC tissues compared with normal bronchial epithelium and that its downregulation significantly correlated with tumor–node–metastasis (TNM) stage (p < 0.0001), nodal metastasis (p < 0.0001), and poor patient prognosis (p = 0.0016, log-rank test). We also checked LRRC3B levels in several lung cancer cell lines and found that its expression was downregulated in four of nine lung cancer cell lines compared with normal human bronchial epithelial (NHBE) cell line. We further explored the biological role of LRRC3B. LRRC3B plasmid transfection in H460 and A549 cell lines inhibited proliferation, colony formation ability, and invading ability. Furthermore, we identified that LRRC3B could inhibit cell cycle progression with downregulation of cyclin D1 and decreased MMP9 expression. In addition, LRRC3B depletion in HBE cells promoted proliferation and invasion. In conclusion, our data suggested that LRRC3B may serve as an important tumor suppressor in NSCLC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of death among malignant tumors worldwide [1–3]. Although targeted therapies have been established, mutations such as EGFR and KRAS causing activation of these gene products are identified only in a limited number of patients. Many complex genetic, epigenetic, and microenvironmental factors play important roles in growth and invasion of tumor cells [4–7]. Thus, identification of these oncogenes/tumor suppressors and elucidation of their mechanism are important missons for development of a new treatment.
Leucine-rich repeat (LRR)-containing 3B (LRRC3B) is a putative LRR-containing transmembrane protein. LRRs are protein motifs characterized by repetition of hydrophobic residues, especially leucine, and that are separated by conserved distance [8–10]. LRR-containing proteins participate in many important cellular processes such as immunity, cell adhesion, signal transduction, and apoptosis [11]. Several studies using human cancer microarray have shown that LRRC3B is downregulated in breast and colorectal cancers, suggesting involvement of LRRC3B in carcinogenesis [12]. It was reported that LRRC3B messenger RNA (mRNA) was downregulated in gastric cancer tissues and that introduction of LRRC3B in gastric cancer cells inhibited colony formation and tumorigenesis in nude mice [13]. However, there was no report concerning protein expression pattern and clinical significance of LRRC3B in human non-small-cell lung cancer. Its biological roles in lung cancer cells also remain unexplored.
Here, we investigated LRRC3B protein expression in non-small-cell lung cancers (NSCLCs) and analyzed its correlation with clinicopathological factors. We also examined the function of LRRC3B as a tumor suppressor in lung cancer cell lines and provide evidence that LRRC3B inhibits lung cancer cell proliferation and invasion.
Materials and methods
Tissue samples
The study was approved by the ethical committee of Shengjing hospital. A total of 101 cases of NSCLC were retrieved from the Pathology Archive of the First Affiliated Hospital of China Medical University from 2008 to 2011. The histological diagnosis was evaluated by two independent pathologists according to the WHO classification. Clinical data about the patients were obtained from patient records.
Immunohistochemistry
Tumor specimens were fixed with neutral formalin and embedded in paraffin, and 4-μm-thick sections were prepared. The sections were deparaffinized in xylene, rehydrated in graded alcohol series, and boiled in 0.01 M citrate buffer for 2 min in an autoclave. Endogenous peroxidase activity was blocked using hydrogen peroxide, which was followed by incubation with normal goat serum to reduce non-specific binding. The primary antibody for LRRC3B (1:100, Sigma, USA) was incubated overnight at 4 °C. Sections were stained in parallel with non-immune IgG to provide a negative control. Antibody binding was then detected by the EliVision plus kit (Maixin, Fuzhou, China) and DAB plus kit (Maixin, Fuzhou, China). Sections were counterstained with hematoxylin, dehydrated, and mounted.
Two independent investigators examined all tumor slides randomly. Five views were examined per slide, and 100 cells were observed per view at ×400 magnification. Immunostaining of LRRC3B was scored following a semi-quantitative scale by evaluating in representative tumor areas the intensity and percentage of cells. Cytoplasmic staining was considered as positive. The intensity of LRRC3B staining was also scored as 0 (no staining), 1 (weak), and 2 (strong). Percentage scores were assigned as 1 (1–25 %), 2 (26–50 %), 3 (51–75 %), and 4 (76–100 %). The scores of each tumor sample were multiplied to give a final score of 0 to 8, and the total expression of LRRC3B was determined as low expression, score ≤4, and high expression (+), score >4.
Cell culture and transfection
Human NSCLC cell lines H460 and A549 and normal bronchial cell line HBE were purchased from ATCC. The cells were cultured in RPMI-1640 (Gibco, Invitrogen, NY, USA) supplemented with 10 % fetal bovine serum at 37 °C in 5 % CO2. pCMV6-LRRC3B plasmid and the control empty vector pCMV6 were purchased from Origene (Origene, Rockville, MD, USA). Cells were transfected using Attractene reagent (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
siGENOME SMARTpool siRNA for LRRC3B and non-targeting siRNA were obtained from Dharmacon (ThermoFisher, USA). siRNA transfection was performed using DharmaFECT 1 reagent.
Real-time RT-PCR
Real-time PCR was performed using SYBR Green mastermix kit form ABI (Applied Biosystems). Using ABI 7500 Fast Real-Time PCR System, the thermo cycle was set as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. The relative expressions of target genes were calculated using 2−ΔΔCt method. Experiments were repeated in triplicate.
The primer sequences were listed as follows: LRRC3B for, 5′-TCCAATCATGAGACAGCCCAC-3′, LRRC3B rev, 5′-TCTGCCAGCATGTTCATCCAA-3′; and β-actin for, 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′, β-actin rev, 5′-CACCTTCTACAATGAGCTGCGTGTG-3′.
Western blotting
Total protein was extracted using Pierce lysis buffer (ThermoFisher, USA). Proteins were quantified using Bradford method. About 50 μg of protein was loaded and separated with SDS-PAGE. Then, proteins were transferred to PVDF membrane (Millipore, Bedford, MA, USA), which was blocked using 5 % non-fat milk. Membranes were incubated overnight at 4 °C with primary antibodies against LRRC3B (1:800, Sigma, USA), cyclin D1, and MMP9 (1:1000, Cell Signaling, USA). After washing, the membrane was incubated with a horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotechnology) at 37 °C for 2 h. Protein bands were visualized with the ECL and captured using DNR BioImaging Systems (DNR, Jerusalem, Israel).
MTT proliferation assay
After transfection, cells were seeded in 96-well plates at about 2000 cells per well. Then, 20 μl of 5 mg/ml MTT solution was added to each well and incubated for 4 h at 37 °C. The medium was removed from each well and the resulting MTT formazan was solubilized in 150 μl of DMSO. Each solution was measured spectrophotometrically at 490 nm. Each experiment was performed in triplicate.
Colony formation assay
Cells (about 1000) were seeded in 6-cm cell culture dishes and incubated for 2 weeks. Then, colonies were stained using Giemsa. The number of colonies with more than 50 cells was counted.
Matrigel invasion assay
Cell invasion assay was performed using a 24-well Transwell chamber with a pore size of 8 μm (Costar, Cambridge, MA). The inserts were coated with 20 μl Matrigel (1:3 dilution, BD Bioscience, San Jose, CA, USA). Forty-eight hours after the transfection, cells were trypsinized and 3 × 105 cells in 100 μl of serum-free medium were transferred to the upper Matrigel chamber and incubated for 16 h. Medium supplemented with 10 % FBS was added to the lower chamber as the chemoattractant. After incubation, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4 % paraformaldehyde and stained with hematoxylin. The number of invaded cells was counted in 10 randomly selected high-power fields under the microscope. This experiment was performed in triplicate.
Statistical analysis
The statistical package SPSS (SPSS, Chicago, IL, USA) was used for all analyses. The χ 2 test was used to examine the correlation between LRRC3B status and clinical parameters. Student’s t test was used to compare cell experiment data. A p value of <0.05 was considered significant.
Results
Expression pattern and clinical significance of LRRC3B in NSCLC
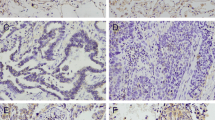
We investigated LRRC3B expression in 101 NSCLC tissue specimens and 20 normal lung tissue specimens by immunohistochemistry. In normal lung tissues, positive cytoplasmic staining was observed in normal bronchial epithelial cells and alveolar cells (Fig. 1a). As for lung cancer tissues, 62 cases showed strong cytoplasmic LRRC3B staining and 39 cases (38.6 %) showed downregulated LRRC3B staining compared with normal tissue (Fig. 1c, d). LRRC3B protein was mainly accumulated in the cytoplasmic compartment of tumor cells. We analyzed the relationship between LRRC3B downregulation and the clinicopathologic factors. As shown in Table 1, LRRC3B downregulation correlated with advanced tumor–node–metastasis (TNM) stage (p < 0.0001) and positive nodal metastasis (p < 0.0001). No difference in the LRRC3B was found in relation to the age, gender, differentiation, and tumor size.
Expression of LRRC3B in non-small-cell lung cancers. a Positive cytoplasmic staining of LRRC3B in normal bronchial epithelium and normal alveolar cells. b Positive staining of LRRC3B in a case of lung adenocarcinoma. c Negative LRRC3B staining in a case of lung adenocarcinoma. d Negative staining of LRRC3B in lung squamous cell carcinoma. (Magnification is ×200.) e Survival analyses of patients with LRRC3B expression and those without. The overall survival was significantly lower in patients with LRRC3B-negative NSCLCs than in patients with LRRC3B-positive NSCLCs
Furthermore, Kaplan–Meier survival analysis showed a significantly lower overall survival in patients with negative LRRC3B compared with those with positive expression (p = 0.0016, log rank test; Fig. 1e). In addition, univariate analysis showed that TNM stage and LRRC3B status were both significant prognostic factors (TNM stage: hazard ratio, 2.110, p = 0.0001; LRRC3B status: hazard ratio, 0.433, p = 0.0025), and multivariate analysis using a Cox regression model indicated that TNM stage was an independent, unfavorable prognostic factor (TNM stage: hazard ratio, 1.946, p = 0.0010; Table 2).
LRRC3B is downregulated in lung cancer cell lines and inhibits cell proliferation and invasion
Relative expression level of LRRC3B was analyzed by western blot and real-time RT-PCR in a panel of lung cancer cell lines. In accordance with tissue samples, the LRRC3B protein expression was remarkably decreased in NSCLC cell lines, especially in H460, H358, HCC827, and A549, compared with normal human bronchial epithelial (NHBE) cell line (Fig. 2a). A549 and H460 cell lines were selected for LRRC3B transfection. We upregulated LRRC3B expression using LRRC3B plasmid, and transfection efficiency was confirmed by western blot analysis (Fig. 2b). LRRC3B upregulation in A549 and H460 cells greatly inhibited the proliferation rate (control vs siRNA at day 5; A549, 0.783 ± 0.032 vs 0.568 ± 0.028, p < 0.05; H460, 1.397 ± 0.051 vs 1.076 ± 0.056, p < 0.05) and the potential of colony formation (control vs siRNA; A549, 426 ± 37.3 vs 135 ± 26.8, p < 0.05; H460, 257 ± 35.5 vs 118 ± 20.3, p < 0.05; Fig. 3a, b). To characterize the effect of LRRC3B on cell invasion, Matrigel invasion assay was performed in A549 and H460 cells. As shown in Fig. 3c, a significant reduced invading ability (A549, 59.7 %; H460, 65.1 %) was observed in cells with LRRC3B transfection compared with empty controls.
LRRC3B expression in lung cancer cell lines and its transfection efficiency. a Endogenous expression of LRRC3B was examined in NHBE and lung cancer cell lines by western blot and real-time RT-PCR. A549, H358, H460, and HCC827 cell lines have a significant downregulated LRRC3B expression. b Western blot analysis showed that pCMV6-LRRC3B plasmid markedly increases its levels in H460 and A549 cells compared with control
LRRC3B restoration inhibits cell proliferation and invasion. a MTT assay in H460 and A549 cells transfected with LRRC3B plasmid. Time-dependent decrease in cell proliferation after LRRC3B transfection compared with control. b Colony formation assay was performed in cells transfected with LRRC3B plasmid and cells transfected with control. A marked decrease in colony formation is seen in the groups with LRRC3B restoration. c Matrigel invasion assay showed that LRRC3B transfection decreased cell invasion in A549 and H460 cell lines. *p < 0.05
LRRC3B inhibits cell cycle and regulates cyclin D1 and MMP9 expression
The aforementioned results indicate that LRRC3B leads to decreased cellular proliferation and invasion. We further checked the role of LRRC3B on cell cycle progression. As shown in Fig. 4a, LRRC3B upregulation inhibited G1-S transition in H460 and A549 cell lines. To underline the possible mechanisms, we examined a panel of growth- and invasion-related proteins. As shown in Fig. 4b, LRRC3B transfection significantly inhibited cell cycle protein cyclin D1 and invasion-related protein MMP9.
LRRC3B transfection inhibits cell cycle progression, with cyclin D1 and MMP9 donwregulation. a Cell cycle analysis showed that LRRC3B transfection decreased cell percentage in S phase and increased the cell percentage in G1 phase. b Western blot analysis showed that LRRC3B restoration could decrease the protein expression of cyclin D1 and MMP9
LRRC3B depletion promotes proliferation and invasion
We used siRNA to knock down LRRC3B expression in normal bronchial cell line HBE cells and examined its biological effect. As shown in Supplementary Fig. S1a–d, LRRC3B depletion increased HBE growth rate, colony formation ability, and invading ability. In addition, expression of MMP9 and cyclin D1 was slightly upregulated in LRRC3B-depleted cells (Supplementary Fig. S1e).
Discussion
Epigenetic inactivation of LRRC3B, which is caused by methylation and/or deletions, has been investigated in a number of cancers. Reduced LRRC3B mRNA expression and hypermethylated promoter region of LRRC3B were found in gastric cancer, colorectal cancer, and clear cell renal cell carcinoma [13–15]. Upregulation of LRRC3B by transfection inhibited colony formation of gastric cancer cells [13]. However, there was no study concerning protein expression of LRRC3B in cancer tissues and its correlation with clinicopathological parameters. In addition, biological roles of LRRC3B in human non-small-cell lung cancer remain elusive. In this study, we demonstrated that LRRC3B protein expression was significantly decreased in 38.6 % of NSCLC tissues, which was significantly correlated with lymph node metastasis and TNM stage, suggesting LRRC3B as a putative tumor suppressor in NSCLC. Importantly, we showed that loss of LRRC3B was correlated with poor patient prognosis. To data, this is the first report concerning protein expression pattern and clinical significance of LRRC3B in human cancers. We also examined LRRC3B expression in several lung cancer cell lines. LRRC3B expression was significant lower in four of nine cancer cell lines compared with NHBE, which was consistent with immunohistochemical findings showing LRRC3B as a tumor suppressor.
Using LRRC3B plasmid, we demonstrated that its restoration in NSCLC cell lines with low endogenous expression significantly inhibited proliferation and cell cycle progression. Previous reports showed evidence supporting LRRC3B as a tumor suppressor. It has been reported that restoring the expression of LRRC3B was sufficient to suppress gastric cancer proliferation and tumor growth in mice [13]. In addition, LRRC3B could also inhibit colony formation in renal cell carcinoma line [16]. Our result was in accord with these studies, suggesting the role of LRRC3B as a tumor suppressor in lung cancer cells. To data, the effect of LRRC3B on cell cycle and cell invasion has not been investigated. The mechanism of LRRC3B as a tumor suppressor also has not been elucidated. Thus, we checked cell cycle progression and related proteins and found that LRRC3B upregulation inhibited S phase percentage and cyclin D1 expression. Cyclin D1 plays a critical role during cell cycle progression at G1-S checkpoint. Many studies have demonstrated that cyclin D1 was overexpressed in non-small-cell lung cancer and correlated with malignant progression and poor prognosis [17, 18]. These results suggested that LRRC3B could regulate cell proliferation through modulation of cyclin D1 status. The role of LRRC3B on cell invasion has not been reported previously. In the present study, we found that LRRC3B inhibited invading ability of lung cancer cell lines. LRRC3B restoration decreased MMP9 level, an important mediator of invasion in many types of cancers [19–22]. Thus, the role of LRRC3B on invasion inhibition may be due to its role on MMP9 downregulation.
In conclusion, this study demonstrated downregulation of LRRC3B protein expression in NSCLC and its correlation with TNM stage, nodal status, and poor prognosis. Our results demonstrated that LRRC3B restoration in lung cancer cells could inhibit proliferation and invasion, possibly through regulation of cyclin D1 and MMP9. Given these findings, LRRC3B serves as an important tumor suppressor in non-small-cell lung cancer.
References
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57(1):43–66.
Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1(1):49–52.
Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, et al. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–8.
Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ, et al. CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol.
Dong QZ, Zhao Y, Liu Y, Wang Y, Zhang PX, Jiang GY, et al. Overexpression of SCC-S2 correlates with lymph node metastasis and poor prognosis in patients with non-small-cell lung cancer. Cancer Sci. 101(6):1562–9.
Fidler IJ, Kripke ML. Genomic analysis of primary tumors does not address the prevalence of metastatic cells in the population. Nat Genet. 2003;34(1):23. author reply 25.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–6.
Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374(6518):183–6.
Kajava AV, Kobe B. Assessment of the ability to model proteins with leucine-rich repeats in light of the latest structural information. Protein Sci. 2002;11(5):1082–90.
Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11(6):725–32.
Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277(3):519–27.
Bianchini M, Levy E, Zucchini C, Pinski V, Macagno C, De Sanctis P, et al. Comparative study of gene expression by cDNA microarray in human colorectal cancer tissues and normal mucosa. Int J Oncol. 2006;29(1):83–94.
Kim M, Kim JH, Jang HR, Kim HM, Lee CW, Noh SM, et al. LRRC3B, encoding a leucine-rich repeat-containing protein, is a putative tumor suppressor gene in gastric cancer. Cancer Res. 2008;68(17):7147–55.
Tian XQ, Zhang Y, Sun D, Zhao S, Xiong H, Fang J. Epigenetic silencing of LRRC3B in colorectal cancer. Scand J Gastroenterol. 2009;44(1):79–84.
Kondratov AG, Stoliar LA, Kvasha SM, Gordiyuk VV, Zgonnyk YM, Gerashchenko AV, et al. Methylation pattern of the putative tumor-suppressor gene LRRC3B promoter in clear cell renal cell carcinomas. Mol Med Rep. 2012;5(2):509–12.
Haraldson K, Kashuba VI, Dmitriev AA, Senchenko VN, Kudryavtseva AV, Pavlova TV, et al. LRRC3B gene is frequently epigenetically inactivated in several epithelial malignancies and inhibits cell growth and replication. Biochimie. 2012;94(5):1151–7.
Liu J, Liao Q, Zhang Y, Sun S, Zhong C, Liu X. Cyclin D1 G870A polymorphism and lung cancer risk: a meta-analysis. Tumour Biol. 33(5):1467–76.
Liu Y, Wang L, Lin XY, Wang J, Yu JH, Miao Y, et al. The transcription factor DEC1 (BHLHE40/STRA13/SHARP-2) is negatively associated with TNM stage in non-small-cell lung cancer and inhibits the proliferation through cyclin D1 in A549 and BE1 cells. Tumour Biol. 34(3):1641–50.
Dong QZ, Wang Y, Tang ZP, Fu L, Li QC, Wang ED, et al. Derlin-1 is overexpressed in non-small cell lung cancer and promotes cancer cell invasion via EGFR-ERK-mediated up-regulation of MMP-2 and MMP-9. Am J Pathol. 2013;182(3):954–64.
Jian H, Zhao Y, Liu B, Lu S. SEMA4b inhibits MMP9 to prevent metastasis of non-small cell lung cancer. Tumour Biol. 2014;35(11):11051–6.
Song H, Tian Z, Qin Y, Yao G, Fu S, Geng J. Astrocyte elevated gene-1 activates MMP9 to increase invasiveness of colorectal cancer. Tumour Biol. 2014;35(7):6679–85.
Feng X, Miao G, Han Y, Xu Y. CARMA3 is overexpressed in human glioma and promotes cell invasion through MMP9 regulation in A172 cell line. Tumour Biol. 2014;35(1):149–54.
Acknowledgments
The study was supported by Outstanding Scientific Fund of Shengjing Hospital (No. 201205).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
LRRC3B depletion promotes growth, invasion, cyclin D1 and MMP9. A. Western blot analysis showed that siRNA markedly decreases its levels in HBE cells compared with control. B. MTT assay showed that LRRC3B depletion increased cell growth rate. C. Colony formation assay showed that LRRC3B depletion increased colony number. D. Matrigel invasion assay showed that LRRC3B depletion facilitated HBE cell invasion. E. Western blot analysis showed that LRRC3B depletion slightly upregulated cyclin D1 and MMP9. (GIF 62 kb)
Rights and permissions
About this article
Cite this article
Kan, L., Li, H., Zhang, Y. et al. LRRC3B is downregulated in non-small-cell lung cancer and inhibits cancer cell proliferation and invasion. Tumor Biol. 37, 1113–1120 (2016). https://doi.org/10.1007/s13277-015-3833-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3833-0