Abstract
The progression of colorectal cancer is commonly characterized by accumulation of genetic or epigenetic abnormalities, altering regulation of gene expression as well as normal protein structures and functions. Nonetheless, there are some questions that remain to be elucidated, such as the origin of cancer cells and populations of cells initiating and propagating tumor development. Currently, there are two rival theories describing the process of carcinogenesis. One is the stochastic model, arguing that any cell is capable of initiating and triggering the development of cancer. Meanwhile, the cancer stem cell model hypothesizes that only a small fraction of stem cells possesses cancer-promoting properties. Typically, colorectal cancer stem cells (CSCs) share the same molecular signaling profiles with normal stem cells or embryonic stem cells, such as Wnt, Notch, TGF-β, and Hedgehog. Nevertheless, CSCs differ from normal stem cells and the bulk of tumor cells in their tumorigenic potential and susceptibility to chemotherapeutic drugs. This may be a possible explanation of the high percentage of cancer recurrence in patients who underwent chemotherapeutic treatment and surgery. This review article focuses on the colorectal cancer stem cell biomarkers and the role of upregulated signaling pathways implicated in the initiation and progression of colorectal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third leading type of cancer with 1,361,000 newly recorded cases in 2012, accounting for more than 694,000 deaths worldwide [1]. It is one of the main causes of cancer-related deaths in the Western countries with the overall 5-year survival rates of 11 % for patients with metastatic disease [1].
Currently, the mechanisms of tumorigenesis and the precise origin of CRC cells still remain mostly unclear. Today tumors are considered not only simply as “bags packed with homogeneous malignant cells” but rather like hierarchically organized systems of different cell types including cancer, stromal, hematopoietic, endothelial, and infiltrating cells. These cell types can contribute to the tumor heterogeneity, influencing metabolic, physiological, and morphological changes, ultimately enhancing tumor growth, development, the probability of treatment failure, and tumor recurrence.
There are two major models describing the process of colorectal tumorigenesis, including the stochastic and cancer stem cell models. The “stochastic model” argues that tumors are biologically heterogeneous and not organized into systems with all cells within the tumor having an equal potential to tumor growth and metastasis [2]. Any functional discrepancies in the model are explained by stochastic factors, whether intrinsic (e.g., fluctuations of transcription factors or proteins) or extrinsic (e.g., environmental signals, immune responses) factors, altering normal cell behavior. The cellular heterogeneity in this model is explained by the hierarchy of the cell class emanating at the apex, distinct from the rest of the hierarchy and possessing different self-renewal capacities.
The cancer stem cell (CSC) model, proposed by Cohnheim in 1875 [3], is based on the idea that some cancer cells retain properties of embryonic cells. This theory suggests that different internal or external agents are able to cause DNA damage, and cells encompassing the damaged DNA could give rise to morphologically distinct tumor cell populations. Such populations share distinct biochemical, physiological, and genetic profiles on different stages of cancer progression. The modern perception of the theory arises from extensive studies of leukemia stem cells [4, 5], where it was found that cancer cells produced by tumor stem cells are similar to differentiated cells generated by progenitor cells [6]. Later, prospective CSCs were isolated for breast [7], brain [8], colon [9], head and neck [10], pancreatic [11], melanoma [12], hepatic [13], lung [14], prostate [15], and ovarian [16] tumors. In all cases, generated xenograft models provided evidences that CSC can be a common feature across different cancer types. In addition, the model claims that the cells within the tumor may vary by the cancer initiating and propagating potential, reflecting hierarchical organization similarities between tumorigenic and non-tumorigenic tissues.
Besides morphological, physiological, and metabolic differences of CSCs from normal stem cells, they also differ in their susceptibility to some chemotherapeutic drugs [17]. In fact, the high percentage of cancer recurrence in patients underwent chemotherapeutic treatment and surgery might be explained by the presence of CSC. Nonetheless, significant progress has been achieved in the identification of potential colorectal cancer stem cell biomarkers identifying the stage of the disease development, patient risk groups, and the chances of recurrence and dissemination, alongside with those who likely do not respond to certain types of treatment. Therefore, this review focuses on the current state of colorectal cancer stem cell biomarkers (CRCSC) and the pathways involved in the development of colorectal cancer from the perspective of the cancer stem cell model. This may be important due to the lack of understanding of specific biomarkers of CRCSC.
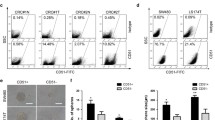
Table 1 summarizes the features of some cancer stem cell biomarkers, including their aliases, main functions, and clinical value.
General criteria and methods of CRCSC identification
In order to study the biology of CRCSC, firstly, they have to be identified and, secondly, their nature has to be confirmed. The identification of CRCSC to this day has been relying mostly on molecular markers (described below in details) used in fluorescence-activated cell sorting (FACS), fluorescent microscopy, and other types of experiments. The original experiments clearly demonstrated the existence of the cells with the characteristics of CSC in colorectal cancer and were done by two independent groups of researchers led by John Dick [18, 19] and Ruggero De Maria [20–22].
It must be noted that CRCSC, as well as CSC from other organs and tissues, have to comply to certain criteria in order to be considered a CSC. The major criteria for CSC would be a possession of specific properties such as self-renewal, multilineage differentiation, as well as a tumorigenic potential. Self-renewal refers to the capability of the cell to produce one undifferentiated cell with the stem cell characteristics and another cell with the potential to give rise to differentiated lineage during cell division. The multilineage differentiation potential refers to the CSC ability to differentiate and give rise to several tissue cell types. For example, the intestinal crypt contains several cell types that include enterocytes, goblet cells, Paneth cells, and enteroendocrine cells. Therefore, the CRCSC should be able to produce cell types resembling the abovementioned cell types upon in vitro differentiation. The self-renewal and multilineage differentiation are two stemness parameters of the SC, while the tumorigenic potential is the parameter of the cancer cell, which in combination produces a CSC. Tumorigenic potential is another important feature of CRCSC, which is usually assessed by the ability of the cell to invade the tissue and produce a tumor upon transplantation. The assessment of the tumorigenic potential has been routinely done by injection of the studied cells to the severe combined immune-deficient mice. The experiments used to assess these parameters are described in details elsewhere, and they are out of scope of this review.
Identification of colorectal cancer stem cell biomarkers
CD24 is a mucin-like cell surface adhesion glycoprotein attached to the external side of the plasma membrane by a phosphatidylinositol anchor [23]. CD24 plays a role in cell proliferation and differentiation, expressed by a number of immune system cell types, including B lymphocytes [24], where it positively regulates proliferation and maturation of activated T cells [25] and granulocytes [26]. It has been reported that the protein can also be implicated in the development of a number of cancer types as breast, ovarian, lung, gastric, and colorectal [23]. Recently, it has been revealed that CD24 is expressed in nearly 50–68 % of CRCSC [27]. The expression of CD24 positively correlates with various stages of adenocarcinoma development, their degree of differentiation, and lymph node metastasis [28]. However, no significant correlation between a shortened period of patient survival and the presence of the marker was detected [29]. Although there are some evidences of CD24 as a tumor biomarker, the role of the protein as a CRCSC marker is less convincing and has not been fully proven.
CD26 (DPP4 or ADCP2) is a 110-kDA cell surface glycoprotein receptor expressed on the surface of various cell populations, including activated T, B, NK cells, macrophages, epithelial cells, and endothelial cells [30]. CD26 is composed of the transmembrane region, cytoplasmic domain, and extracellular stretch with dipeptidyl peptidase activities [31]. Originally identified for CD4+ memory T cell population, the marker can also serve a role in the activation of T cells through the collagen receptor molecule and possibly have a role in thymic ontogeny [32]. Other possible substrate targets of the CD26 molecule include RANTES, a crucial regulator of monocyte chemotaxis and interferon-inducible chemokines [33]. Recent works have also indicated a potential role of CD26 in the mobilization and migration of hematopoietic stem cells (HSC) [34]. A number of studies have found that the biomarker expression may be altered in a range of malignancies such as hepatocellular carcinoma, melanoma cells, lung adenocarcinoma, ovarian carcinoma, hematologic malignancies, and prostate carcinoma [30]. However, only one study has indicated the potential role of CD26 in the pathophysiology of CRCSC [35].
CD29 (β1-integrin) is a member of the large integrin superfamily. The protein comprises a large extracellular domain, transmembrane stretch, and a small intracellular domain. Functions of CD29 include activation of cell proliferation, growth, survival, and migration processes [36]. In normal intestinal mucosal cells, the marker has been found in the lower third part of crypts, which include progenitor and stem cells [37]. The combination of CD24/CD29 markers, which are found only in a small subpopulation of primary CR cancers, have been proposed as one of the tumor-initiating factors promoting SC proliferation, anchorage-independent growth, migration, and metastasis probably through regulation of the tumor microenvironment [38]. However, during late stages of CRC progression, CD29 expression is usually decreased, which has been associated with worse overall disease outcomes [39], and after calibrating the effect of the disease stage, was higher than in normal colon tissues [39]. Nevertheless, there are still some ambiguities about the correlation between CD29 expression and the size of stem cell populations, and its role in the prediction of malignancy outcomes [40].
CD44 is a cell adhesion, transmembrane hyaluronic acid receptor glycoprotein and acts as a transducer of extracellular signals influencing downstream Wnt/Beta-catenin pathways [41]. CD44 is involved in homing and activation of lymphocytes, hematopoiesis, angiogenesis, cell adhesion, and migration mechanisms [42]. Recently, CD44 was found in a number of CSCs including breast, head and neck, non-small lung, and colon cells [43]. In vitro experiments showed higher clonogenicity and tumor colony forming potential of CD44+ cells compared to the knockdown cell lines [44]. Moreover, only CD44+, but not CD44−, cells retain morphological characteristics of tumor cells from which they were derived; meanwhile, deletion or overexpression of CD44 in APC Min/+mice reduced tumor initiation properties [45]. Thus, CD44 has been suggested as a potential co-CSC marker, since in xenograft, CD44+/CD166+ cells have a higher tumorigenicity potential compared to CD44−/CD166− cells possibly through the involvement in the activation of the tyrosine kinase receptor c-Met [46]. The combination of CD44 and CD54 has been proven to specifically identify rectal cancer cells [47]. Nonetheless, the role of CD44 regarding development of malignancies and metastases is still debatable. This explains that many cancer types typically express multiple CD44 splice variants [48]. The marker’s prognostic relevance regarding survival of patients, lymph node size, and tumor grade progression is inconsistent and controversial, with further studies warranted to understand the suitability of the CD44 molecule as a CRCSC marker.
Pentaspan-transmembrane cholesterol-interacting CD133 protein, also known as prominin-1, contains the extracellular N-terminus, intracellular C-terminus with two cysteine-rich loops, five tyrosine phosphorylation sites, and two stretches of N-terminus glycosylation sites [49]. CD133 was firstly discovered as a cell surface marker of several subsets of hematopoietic cells, intestine bottom crypt cells, and also bone marrow-derived endothelial progenitor cells [50]. Currently, CD133 has been described for a wide variety of cancer tissues as brain [51], lung [52], liver [53], prostate [54], and colon [55]. Several studies have also proved that the glycoprotein may act as a CRC stem cell marker [9, 56–58] demonstrating raised abilities of CD133+ enriched cell populations to engraft and initiate solid tumor formation in immunodeficient mice compared to unsorted CRC cell populations. This can be explained by hyperactivation of Wnt/Ras–Raf–Mek–Erk signaling pathways [59]. Further, expression of the SRp20 splicing factor, a newly identified target gene of the Wnt/β-catenin pathway [60], suggested that the expression of CD133 and CXCR4 in the tumor microenvironment acts through the CXCR4SDF-1 paracrine axis and usually correlates with poor overall survival rates of patients diagnosed with stage 2 and 3 colorectal cancers.
According to Mohammadi et al. [61], the share of CD133+ decreases through a non-dysplastic subset of sessile serrated adenoma–polyp–lesions (SSA/P/L) (premalignant CR lesions) to non-dysplastic serrated hyperplastic polyps (HP) and ultimately through the passage of CSC to dysplasia, adenomas, and then cancers, remaining constantly higher than in normal tissue cells [62]. This fact indicates that upregulation of CD133 expression more probably occurs during early stages of CRC progressions, contributing to the entire process of cancer expansion [63]. Other potential routes of CD133+ expression in CSC include epigenetical mechanisms proposed by Yi JM et al. [64], who found that hypermethylation of the CD133 gene promoter in a CpG island may lead to the upregulation of CD133.
A number of papers evidenced a high prognostic relevance of CD133 in the promotion of malignancies. Several studies stated that higher levels of CD133 expression usually correlate with worse prognostic outcomes [65, 66], and it may associate with the 5-FU-based chemotherapy resistance. Furthermore, CD133+ cell populations are more resistant to conventional radiation therapy, explaining increased chances of CRC relapse and radiotherapy-related risks [67]. However, the usefulness of CD133 is still debatable, as CD133− cell populations are also able to generate tumor development in immune-deficient mice [68]. This fact was explained by Feng et al. [69] suggesting that CD133+SW620 and CD133−SW620 colon cancer cells can switch between the two subpopulations in the presence of environmental stress and hypoxia-inducible factors (HIFs). Particularly, the P5 promoter of the CD133 gene is regulated by HIF-1α and HIF-2α through one of two E-twenty six (ETS) binding sites in the human embryonic colon and kidney cancer cells [70]. All these findings suggest a pivotal role of CD133 in cancer initiating and progression processes supporting its potential role as a prognostic biomarker in CRCSC.
CD166 or activated leukocyte cell adhesion molecule (ALCAM) is a transmembrane protein with five extracellular immunoglobulin-like domains [71]. CD166 is present on the surface of T and B lymphocytes [71], myeloid [72], and mesenchymal stem cells [73]. Apart from hematopoietic cells, the presence of CD166 has also been reported for lung [74], breast [75], ovarian [76], and prostate [77] cancers. In colonic epithelial cells, CD166 was observed at the base crypt and progenitor niche cells or CRCSC [78]. Further, when xenografted on immunodeficient CD44+/CD166+/EpCAM+ mice [38], the tumorigenicity potential of the cells increased compared to wild-type mice, suggesting some not well-characterized functions. In a group of 111 patients with CRC diagnosis [79], CD166 expression correlates with a shortened period of survival. Furthermore, Lugli et al. [46] have demonstrated positive correlation between the lymph node metastasis, tumor size, the number of colon polyps, and worse clinical prognosis [47]. In contrast to these findings, Horst et al. [80] have not found any statistically significant association between CD166 markers and CRC outcomes. These evidences suggest that upregulation of CD44 and CD166 must have a role in colon polyps to carcinoma transition events; however, the exact role remains yet to be defined.
Aldehyde dehydrogenase (ALDH1) was detected as a CSC marker for lung, prostate, breast, and pancreatic cancers and in a number of myelomas and leukemias [81]. High levels of ALDH1 were reported for a number of stem cell populations of various lineages, including colon progenitor cells [82]. Functions of ALDH1 include catalysis and irreversible oxidation of aldehydes to their corresponding carboxylic acids [83]. Several groups recorded reduced survival time for ovarian cancer [83]. An increased expression of ALDH1 is associated with increasing stage of cancer development and for normal tissues for poorly differentiated tumors and for metastatic colon patients [40]. Recently, ALDH1 was also detected in malignant colonic stem cells [84, 85]. During progression from normal epithelium cells to adenomas in APC mice, expression of ALDH1 was detected further up towards the apex of the crypt [84].
Bmi1 has also been reported as a stem cell biomarker, playing a crucial role in the maintenance and self-renewal processes of stem cells in several tissue types [86–88], where Bmi acts as an epigenetic chromatin modifier of multiple target genes [89, 90]. As a component of the polycomb repressive complex (PRC1), Bmi1 is implicated in the stimulation E3 ubiquitin-protein ligase activity of RNF2/RING2 [91]. Usually, in the gastrointestinal tract, Bmi1-expressing cells are abundantly expressed in the distal part of the small intestine [92]. Through a lineage tracing mechanism and in situ hybridization, it has been shown that Bmi1-expressing cells could be found at +4 and +5 positions, possibly playing an essential role in the crypt architecture [93]. Moreover, the selective removal of Bmi1-expressing cells carried out by diphtheria toxin (DT) treatment results in the disruption of the normal crypt composition. The logical question that arises is whether Bmi1-expressing cells take part in compensation of the loss of Lgr5 positive cells. As it was hypothesized, the number of Bmi1-expressing cells increased three times than in controls upon removal of Lgr5-expressing cells [92]. Bmi-1 has also been reported playing a role in the behavior and functioning of several cancer stem cell types including lung [94], mammary [95], prostate [96], glioma [97], and medulloblastomas [98]. However, although the oncoprotein has been correlated with the development, expansion, and metastasis of colorectal cancer, usually resulting in poor clinical outcomes and survival rates [99–102], the biomarker has rarely been reported playing a role in CRCSC [102, 103]. For example, Kreso et al. [103] detected the expression of Bmi1 in the LS174T cell line, a well-characterized human colon adenocarcinoma line. Injection of Bmi1 knockdown cells into interleukin-2 receptor γc–deficient mice (NSG) and non-obese diabetic–severe combined immune-deficient mice (NOD–SCID) resulted in reduction of tumor initiation properties and sphere formation potential compared to control cells, possibly through the CDKN2A locus, encoding p16INK4a and p14ARF [104]. Functioning of CRCSC and CRC progression seems to be dependent on the functioning of the canonical Bmi1 signaling pathway [103].
The epithelial cell adhesion molecule (EpCAM) is a transmembrane glycoprotein mediating epithelial specific intracellular adhesion, cell migration, proliferation, differentiation, and signaling [105, 106]. Expression of EpCAM was observed on epithelium-derived cell populations and a number of human cancer tissues including colon, normal colon progenitor cells, and cancer initiating cells from colon, pancreas, breast, and prostate carcinomas [9, 57, 107]. Initially, EpCAM was discovered using FACs focusing on tumorigenic propensities of EpCAMhigh/CD44+ and EpCAMlow/CD44− [44]. Not surprisingly, EpCAMhigh/CD44+ were able to initiate progression of a tumor, whereas EpCAMlow/CD44− failed to produce any colonies. Subsequently, when xenografted on NOD/SCID mice, the active EpCAM produced distinct cancer heterogeneity, usually observed in original tumors [44]. Similarly, Dylla et al. [108] proposed that CRCSC chemoresistance may be attributed to the presence of EpCAM+/CD44+ cell surface markers. The combination of EpCAMhigh/CD44+/CD166+ cell lines appears to have a higher proliferation and cell colony formation potential, as well as a higher resistance to drugs with less spontaneous rates of apoptosis compared to EpCAM− populations [109]. EpCAM expression has also been found to be reversely correlated with the primary tumor grade and with higher expression rates observed for earlier events of cancer progression [46]. It has also been noted that decreased EpCAM expression was associated with the tumor invasion potential of a metastatic cell, infiltration of the invasive tumor margin, and the presence of lymph node metastasis. Similarly, Langan et al. [39] found that decreased EpCAM expression rates correlate with the lymph node stage (N0 vs. N1 vs. N2). These facts suggest EpCAM involvement in cellular signaling processes necessitates further investigation of the protein for better understanding of its clinical, prognostic, and therapeutic value.
Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) is a member of GPCR class A orphan receptor proteins. The protein consists of a seven transmembrane and a leucine-rich extracellular stretch. Lgr5 has firstly been discovered in colon crypt base columnar progenitor cells harboring Wnt/β-catenin pathway enhancing mutations [110]. After that, the overexpression of Lgr5 was reported for esophageal carcinomas, ovarian carcinomas, and hepatocellular carcinomas and a number of stem cell lines including CRCSC [111]. The loss of Lgr5 expression may be a necessary prerequisite for upregulation of genes responsible for epithelial mesenchymal transition (EMT) [112]. Moreover, elevated expression of Lgr5 correlates with low expression of miR-200c and high expression of vimentin [112], one of the EMT characteristics of tumors, resulting in increased cancer invasiveness and lymph node metastasis. In adenocarcinomas, expression of the protein is localized on peripheral regions, which may lead to the loss of Lgr5+ cell polarity and promote cell migrations and tumor–host interface (carcinoma in situ) [113]. In a recent investigation [114], it has been shown that patients with simultaneous expression of stem cell genes including Lgr5 may lead to 10-fold elevated risks of colon cancer emergence compared to those with low levels. Further, Takahashi et al. [113] reported that overexpression of Lgr5 correlates with higher levels of lymph node metastasis and decreased rates of patient survival. These findings defined a crucial role of Lgr5 in CR carcinogenesis and may be a suitable marker of CSC.
Musashi-1 (Msi-1) is a RNA-binding protein that firstly has been discovered for neuronal stem cells, where it competes with elF4G translation initiating factor [115] of two messenger RNAs (mRNAs): p21/Waf1. Further, Battelli et al. [116] investigated the HEK293T cell line and mNumb, an agonist of the Notch receptor [117]. These mRNAs are implicated in cell proliferation and tumorigenesis [118]. Several authors have also linked Msi-1 to stem and tumor cell biology, identifying Msi-1 as an intestinal stem cell putative biomarker discovered on the LT97 adenoma-like cell line and in primary colon tumors [119]. Further, knockdown of Msi-1 in HCT116 adenocarcinoma xenografts resulted in impaired tumor-initiating activities and ultimately tumor growth [118, 119]. These data suggest that Musashi-1 might play an important role in CSC carcinogenesis and tumor progression. In CRCSC, Msi-1 is involved in the control of “stemness” regulating Wnt/Notch pathways [120, 121]. However, apart from the presence of Msi-1 on CRCSC as a stem cell progenitor, the role of the protein was extensively studied only for the nervous system [122] and no preliminary data about the influence of Msi-1 can be concluded. Taken together, these facts indicate the importance of Msi-1 in the development of CRCSC, stipulating further investigations.
Colorectal cancer stem cell signaling pathways
Wnt signaling pathway
The Wnt (Wingless) signaling pathway is an evolutionary conservative pathway found in all metazoan animals [123]. This pathway regulates an impressive array of intracellular processes as cell proliferation and differentiation [124], cell polarity and motility [125], stem cell fate determination, and tissue renewal, particularly in intestinal crypt [126]. In contrast, dysregulation of the cascade has been reported to play a role in many epithelial malfunctions, including colon tumorigenesis through the canonical (Wnt/β-catenin dependent) or the non-canonical (Wnt/β-catenin independent) pathways [127]. The relation of the non-canonical pathway in CSC theory is not well characterized. It is considered, however, that it can antagonize β-catenin-dependent hyperactivation of the Wnt pathway, suggesting some anti-oncogenic activities [128].
Meanwhile, in the canonical pathway, the activity of the Wnt cascade is regulated by the amount of unbound β-catenin in cytoplasm [129]. Generally, the level of cytoplasmic β-catenin is maintained through the ubiquitin-mediated proteosomal degradation of the multiprotein complex containing β-catenin, axin, adenomatous polyposis coli (APC), and glycogen synthase kinase-3β (GSK-3β) [130]. However, upon binding of Wnt to the low density lipoprotein receptor protein (LRP)/Frizzled complex, its downstream targets are phosphorylated, thereby abrogating GSK-3β activities, which results in enhanced rates of non-phosphorylated β-catenin accumulation in the cytoplasm [129]. Non-phosphorylated β-catenin translocates to the nucleus, where it binds to the T cell transcription factor (TCF) and lymphoid enhancer-binding factors (LEF), simultaneously recruiting transcriptional coactivators as p300, cAMP response element-binding protein (CREB), and other components of the machinery [131, 132]. This leads to the expression of genes responsible for cell fate, proliferation, stem cell maintenance, and embryogenesis including Axin2, c-MYC, and ASCL2 [130].
In normal intestine cells, expression of Wnt is detected in the bottom crypt cells, which is essential for the maintenance of stem cell compartmentalization and ultimately organization and patterning of the intestinal tract [133]. For example, the blockade of Wnt cascades by overexpression of Dickkopf-1 (DKK1), a functional antagonist of Wnt, or deletion of TCF4 and its target proteins as EphB2 and B3 receptors, which are also responsible for positioning of stem and differentiated cells, results in the abnormal colon tissue structure [134]. Furthermore, disruption of other targets of the Wnt cascade as ASCL2 [130], LGR5 [135], and CD44 [136], which are confined by the crypt bottom, results in hyperplasia and the loss of the stem cell compartments [137]. Similarly, EpCAM signaling mechanisms seem to be closely interconnected with Wnt signaling networks [138]. For example, proteolysis of EpCAM by preselin-2 and TACE molecules results in the release of EpICD, an intracellular domain of EpCAM. After translocation of EpICD into the nucleus, it associates and regulates transcription of Wnt target genes as β-catenin, FHL2, and Lef1 [139]. Some studies indicate that CD133 also can promote β-catenin signaling functions and therefore inhibition of cancer cell differentiation [140]. Activities of the Wnt pathway have been shown reciprocally regulated by Bmi signaling [141]. For instance, it has been shown that Bmi1 can suppress the DKK1 molecule, therefore leading to upregulation of the Wnt signaling pathway and transcription of target genes as c-Myc, Lgr5, CD44, and Bmi1 [141]. Further, hyperactivation of DKK1 can modulate ALDH-1 activities having some impacts on cancer cell survival mechanisms, possibly through the upregulation of JNK, RhoA, and MAPK mechanisms [142].
Interestingly, mutations activating and stabilizing β-catenin or inactivating the APC gene result in the permanent activation of the Wnt transcriptional program even in the absence of any extracellular signal. Further, silencing of β-catenin using siRNA, usually leads to decreased colonosphere formation, an important feature in the process of tumorigenesis and serving as surrogates of tumors [41]. In contrast, hyperactivation of TCF/LEF and c-Myc increases the formation potential of colonospheres [134]. The described events are considered to be the major cause of malignant transformation present virtually in all CRC patients.
NOTCH signaling pathway
The NOTCH signaling pathway is a highly conservative system presented almost in all multicellular organisms. It has been revealed that the NOTCH signaling pathway is fundamental for determination of cell fate, hematopoiesis, embryogenesis, and colorectal epithelium maturation, as well as for the maintenance of the balance between cellular proliferation and differentiation, migration, apoptosis, adjacent cell to cell communication, and the development of immune cells [143]. In mammalian cells, the NOTCH signaling cascade is comprised of two structurally distinct families: Delta-like ligands (DLL1, 3, and 4) and four transmembrane Notch homologue receptors (1, 2, 3, 4) interacting with two different families of ligands: Delta-like ligands (DLLs) and Jagged ligands [144], which results in the activation of a number of downstream target genes such Deltex, Hes-1, and asp21 [145, 146]. Activation of the NOTCH pathway incepts upon binding of the NOTCH ligands to NOTCH receptors located on affecter cells [147]. Once the ligand binds to a corresponding receptor, the NOTCH receptor undergoes conformational changes exposing the previously protected site to the proteolytic cleavage guided by γ-secretase and metalloprotease, releasing the Notch intracellular domain (NICD) from the NOTCH complex [148]. The released cytoplasmic NICD fragment undergoes nuclear translocation, where it forms a complex with MAML-1, p300/CBP, Myc, p21, and core-binding factor-1 (CBF-1), modulating activities of hairy enhancer of split Hes-1, 5, 6, 7, and Math1 [149].
In normal intestinal cells, all components of the NOTCH signaling cascade are expressed in intestinal bottom crypts of various stages of development and differentiation [150], where activities of NOTCH pathways are essential for the regulation of progenitor cell and goblet cell differentiation [151]. Nevertheless, several studies showed that the aberrant hyperactivation of Notch signaling components is significantly higher in cancer cells compared to normal colonic mucosa cells. Aberrantly expressed NOTCH pathways are observed in the pathogenesis of a number of cancer types as pancreatic cancer [152], prostate cancer [153], Ewing sarcoma [154], cervical cancer [155], and colon cancer [156]. In a study conducted by Reedijk and colleagues [156], it has been shown that the expression of Jagged ligands, Notch1, and HES1 in cancer cells was comparable to or slightly higher than that in normal intestinal crypt cells. Another study conducted by Meng and colleagues [157] revealed a correlation between the expression of hes1, Notch1, and nicd genes with colon cancer progression. Moreover, overexpression of these genes is hypothesized to be involved in chemoresistance of CRCSC.
Overexpression of the Notch signaling cascade has been reported to be associated with poor malignancy prognosis and responses for medical treatment in solid tumors as breast tumors [156]. It has been shown that Notch signaling-induced ALDH1A1 deacetylation is associated with the aggressive metastatic cancer phenotype [158]. Notch activities can also be regulated by MSI-1 signaling, resulting in the promotion of cell survival and enhancement of cell proliferation [159]. Similarly, the blockade of the Notch pathway depletes CD133 positive cells inhibiting the growth and propagation of tumor cells [160]. However, it appears to be that upregulation of Notch is mainly associated with the development of primary CRC, rather than metastatic CRC, indicating that hyperactivation of NOTCH takes a place in early stages of CRC development and downregulated in advanced CRCs [161]. For example, it was demonstrated that elevated levels of Jagged1, Jagged2, DLL1, DLL3, DLL4, and Notch receptors are present in 75 % of all CRC tissues [149]. The Notch/STAT3/p63/Jagged signaling-induced hyperactivation can also lead to the upregulation of mTOR/PI3K/AKT, EGF, MAML-1, cyclin D1, c-Myc pathways, Bcl-2, Bcl-XL, and IAP family members, as well as a nuclear NFkB factor, constitutively active in human CRC tissues [143]. All these factors contribute to uncontrolled cell proliferation, altered goblet cell differentiation, mucin formation, and tumor cell chemoresistance to chemotherapeutic agents. Interestingly, not all components of the Notch cascade are implicated in the development of CRC. It is found, for example, that the expression of Notch1 and Hes-1, but not Notch2, Jagged1, and DLL3, is associated with the grade of tumor progression and increases from normal colon cells to adenomatous polyps and metastatic colon cancers [149]. Notwithstanding this fact, Notch signaling does not always function as an oncogenic factor and can act as a tumor suppressor depending on the cellular context. For instance, KLF4 C2H2 zinc-finger-containing transcription factor in haploinsufficient mice showed increased susceptibility of colon cancer development [162] assuming anticancer properties of some components of the Notch cascade.
TGF-β signaling pathway
TGF-β is a superfamily which is comprised of more than 30 proteins, including activins, bone morphogenetic proteins (BMP), and inhibins [163]. The TGF-β signaling cascade, its components, and downstream target genes have been reported involved in cell proliferation, motility and migration, differentiation, cell adhesion, extracellular matrix composition, and apoptosis [164]. It is now clear that TGF-β is an inhibitor of the cell cycle and is expressed in normal epithelial, nodal, and immune cells, playing an essential role in tissue and organ homeostasis [165]. On the cellular level, the pathway is one of the most commonly altered cell signaling cascades found in various cancer types [166]. In addition, hyperactivation of the pathway promotes angiogenesis and immunosuppression, which may be important in the context of carcinogenesis, tumor invasion, and metastasis [167].
The cascade initiates upon binding of TGF-β1 ligands to three types of TGF-β receptor isoforms (TGFB1, TGFB2, and TGFB3), inducing the assembly and autophosphorylation of the receptor–ligand heteromeric complex [168]. This leads to the recruitment and phosphorylation of receptor-regulated SMADs (Smad1, Smad2, Smad3, Smad5, and Smad8) [168]. Phosphorylated SMAD forms homo-oligomeric and hetero-oligomeric complexes with the SMAD co-mediators (Smad4 and Smad10) through β2SP and SARA adaptor molecules [169]. The complex is then translocated into the nucleus where it regulates transcriptional activities of the multiple target genes [168]. Alternatively, the TGF–βR complexes can be subjected to degradation upon binding of inhibitory SMADs (Smad6 and Smad7) and SMAD ubiquitinated regulatory factor (SMURF) [170].
Constant proliferation and differentiation is one of the main properties of gastrointestinal epithelium cells. Therefore, mutations in stem cell populations most likely result in increased chances of tumorigenesis and exacerbate probabilities of additional mutation occurrence. Targeting these cell populations may provide a rationale for better strategies for treatment of patients with colon cancer. Some of the TGF targets are important cell checkpoint genes as p21 (CDKN1A), p27 (CDKN1B), and p15 (CDKN2B) [171]. Hyperactivation of these genes leads to cell cycle arrest [165], as well as Myc suppression [172]. However, once p21Cip1 is released from the Myc suppression, it transactivates by the SMAD–FOXO complex [172]. In normal intestinal epithelium cells, TGF-β signaling serves as a tumor growth suppressor initiating apoptosis and inhibiting cell proliferation. The mechanism of TGF-β-mediated apoptosis includes activation of the death-associated kinases (DAPk) and SHIP, inhibiting downstream targets of Akt signaling [173]. In addition, apart from the tumor suppression role, TGF-β signaling also plays a role in the conditioning of mucosa-resident cells [164]. Therefore, development of CRC requires suppression of the pathway in early stages of development. However, during late stages of advanced primary tumor development, TGF-β promotes colorectal cancer tumorigenesis and associates with increased chances of cancer recurrence, likelihood of tumor relapse, and poor chances of survival [164]. Recently, it has been demonstrated that CD166 (pos) cell lines may have a higher proliferative potential compared to CD166(neg) cell, probably through the loss of TGF-β suppressive activities [174]. Although it is still not clear how the inhibitory effects of the cascade are switched to cancer-promoting properties, is has been suggested that TGF-β is capable of activating SMAD-independent pathways, as mTOR/Akt/PI3K, JNK, and MAPK signaling cascades [173, 175, 176]. Moreover, the tbr2 gene seems to be prone to microsatellite instabilities, replication errors, and frameshift mutations, fluctuating from 40 to 80 % of all CRC cases [177, 178]. Interestingly, reconstitution of the tbr gene functions showed reduced tumorigenicity in in vivo experiments [179]. Similarly, mice with homozygous deletion of the smad and β2SP genes (β2SP±/Smad4±) demonstrated increased chances of aggressive colonic adenoma development compared with the wild-type controls [180–182].
Hedgehog pathway
The Hedgehog (Hh) signaling pathway is one of the key regulators of stem cell maintenance, polarity, migration, and differentiation during the embryonic development [183]. There are three homologues of Hh receptor: Desert Hedgehog (Dhh), Indian Hedgehog (Ihh), and Sonic Hedgehog (Shh) [184]. Currently, it has been demonstrated that deregulation of this pathway correlates with higher chances of tumor progression in a wide variety of tissues, including CRC [144]. Several studies have also indicated that hyperactivation of Hh may regulate cancer stem cell maintenance. The cascade starts from RASP-mediated acylation of the Hh N-terminus, which is then released through the transmembrane transporter Dispatched [185]. Association of the released Hh with Drosophila patched gene (Ptch1) stabilizes the receptor Smoothened (Smo), allowing it to internalize and to activate signaling cascades by impeding its localization to the primary cilium, instead of the plasma membrane resulting in the activation of the cubitus interruptus (Gli) gene family of zinc-finger transcription factors [184]. There are three homologues of Gli proteins: Hh target genes are activated by Gli1, Gli2 serves as an activator and repressor, and Gli3 represses transcription of the target gene [184]. It seems that Hh signaling is dependent on the balance of the Gli proteins [186]. However, in CRC stem cells and primary human colon carcinoma cells, the Hh-Gli pathway seems to be overactivated, which affects cell proliferation, tumor growth, survival, and metastases of cancerous cells [187]. Interestingly, Hh signaling is also involved in the maintenance of CD44+/CD24−/low phenotype in CSCs, contributing to the development of invasive cancer forms [188]. Inhibition of Hh cascades by cyclopamine has been reported to inhibit epithelial-to-mesenchymal transition (EMT) and metastasis [189], indicating the potential therapeutic value of Hh pathway for drug treatment.
Crosstalk between signaling pathways
Aberrant activation of individual stem cell pathways in CSCs rarely operates in isolation, which potentially can have some effects on how stem cells respond to extracellular cues. A number of studies indicate that Notch can antagonize Wnt signaling in a Su(H)-independent manner [190]. For example, in the absence of Notch, cells with active of Delta or Su(H) molecules can compensate the loss of the Disheveled mediated Wnt signaling [191, 192], which can lead to the development of tumors associated with high levels of β-catenin in the nucleus [193]. Similarly, mice with inhibition of the Wnt/β-catenin pathway demonstrated decreased levels of Jagged1 expression in ovarian carcinoma cells, suggesting some backward loop interactions between Notch and Wnt pathways [194]. In turn, the Wnt cascade can regulate Gli3 expression and control Shh/Gli patterning of the Hh pathway [195]. It is interesting that Hh signaling can also antagonize Wnt signaling in the colonic epithelial cell cascade through downregulation of the nuclear β-catenin/TCF4 complex [196].
Apart from interactions between the listed pathways, Notch signaling appears to be interconnected with cell proliferation and developmental pathways such as mTOR/PI3K/Akt and Raf/Ras/Erk/Mek pathways in a variety of cell types [197, 198]. In another study, it has been shown that stabilization of Ras molecules in APC mutated mice usually correlates with higher chances of colon cancer development and is controlled by aberrant Wnt or Hh activities [199]. Similarly, the cells with constitutively active mTOR also display elevated levels of Hes1 and NICD expression, probably through STAT3/Jagged/p63 cascades [200]. Treatment of the cells with mTOR inhibitors resulted in decreased levels of both Notch and mTOR signaling [200]. Alternatively, TGF-β/ALK5 activities can also be regulated through the PI3K/Akt-mediated mechanisms [173, 201]. For example, it is known that PI3K/Akt signaling can attenuate the TGF-β-mediated cycle arrest or apoptosis program in response to insulin, interleukins, and other factors, possibly through Smad molecules [173, 201]. TGF-b/BMP pathways are also reciprocally interconnected with Wnt signaling [202]. Downstream components of the pathway, Lef/β-catenin/Smad regulate activities of the shared genes in a synergistic manner. Some studies have suggested connections between p53 molecules and the Hh pathway [203], displaying the complexity of signaling pathway interconnections. Overall, recent studies have indicated that tumor heterogeneity on the molecular level may be a result of the interplay between numerous pathways having an active role in the development of a unique cancer genotype.
Conclusion
The cancer stem cell model suggests that tumors usually evolve from a small population of cancer initiating cells through accumulation of genetic, epigenetic, and somatic alterations, responsible for carcinogenesis, tumor propagation, metastasis, and relapse. According to the model, tumors are composed of cell populations with cancer initiating potential intermixed with the bulk of tumor cells. Therefore, the development of different treatment strategies and therapeutic options to suppress the tumor growth and a possible relapse is very much in need. However, the elaboration of new strategies may be hindered by significant obstacles. First of all, it is the apparent lack of conventional colorectal CSC markers to identify CSC from the rest of the tumor cells. Although putative biomarkers have been determined for different CSC cell populations, it seems that CSCs undergo significant changes in the level of biomarker expression, making it difficult to precisely indicate CSCs on different stages of tumor development based on cell surface biomarkers. In this regard, the investigation focused on elucidation of cell signaling profiles in CSC may provide a possible explanation of spatial and temporal expression of specific cell surface biomarkers. However, investigations related to cell signaling in CSC are complicated by crosstalk between different cell signaling networks and cellular diversity associated with the stem cell development during embryogenesis and normal tissue renewal. This requires further investigations focused on the elaboration of new opportunities in the designing of novel, more effective treatment strategies.
References
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer J Int Cancer. 2014;136(5):E359–86.
Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–43.
Cohnheim J. Congenitales, quergestreiftes muskelsarkom der nieren. Arch Pathol Anat Physiol Klin Med. 1875;65:64–9.
Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–8.
Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–7.
Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–82.
Dick JE. Breast cancer stem cells revealed. Proc Natl Acad Sci U S A. 2003;100:3547–9.
Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401.
Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–5.
Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci U S A. 2007;104:973–8.
Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7.
Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–9.
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–66.
Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15:504–14.
Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–51.
Curley MD, Therrien VA, Cummings CL, Sergent PA, Koulouris CR, Friel AM, et al. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–83.
Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–84.
Dick J. Are cancer stem cells relevant? EJC Suppl. 2010;8:9–9.
O'Brien CA, Pollett A, Gallinger S, Dick JE. Expression of CD133 enriches for colon cancer stem cells. Ann Surg Oncol. 2007;14:22.
Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14:342–56.
Maugeri-Sacca M, Vici P, Di Lauro L, Barba M, Amoreo CA, Gallo E, et al. Cancer stem cells: are they responsible for treatment failure? Future Oncol. 2014;10:2033–44.
Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R. Colon cancer stem cells. J Mol Med Jmm. 2009;87:1097–104.
Choi G, Sammar M, Altevogt P. Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol. 2004;35:255–62.
Blair PA, Norena LY, Flores-Borja F, Rawlings DJ, Isenberg DA, Ehrenstein MR, et al. CD19(+)CD24(hi)CD38(hi) B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity. 2010;32:129–40.
Flores-Borja F, Bosma A, Ng D, Reddy V, Ehrenstein MR, Isenberg DA, et al. CD19(+)CD24(hi)CD38(hi) B cells maintain regulatory T cells while limiting T(H)1 and T(H)17 differentiation. Sci Transl Med. 2013;5:12.
Elghetany MT, Patel J. Assessment of CD24 expression on bone marrow neutrophilic granulocytes: CD24 is a marker for the myelocytic stage of development. Am J Hematol. 2002;71:348–9.
Choi D, Lee HW, Hur KY, Kim JL, Park GS, Jang SH, et al. Cancer stem cell markers CD133 and CD24 correlate with invasiveness and differentiation in colorectal adenocarcinoma. World J Gastroenterol. 2009;15:2258–64.
Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, et al. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783–93.
Papailiou J, Bramis KJ, Gazouli M, Theodoropoulos G. Stem cells in colon cancer. A new era in cancer theory begins. Int J Color Dis. 2011;26:1–11.
Pro B, Dang NH. CD26/dipeptidyl peptidase IV and its role in cancer. Histol Histopathol. 2004;19:1345–51.
Rasmussen HB, Branner S, Wiberg FC, Wagtmann N. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat Struct Biol. 2003;10:19–25.
Dang NH, Torimoto Y, Shimamura K, Tanaka T, Daley JF, Schlossman SF, et al. 1F7 (CD26): a marker of thymic maturation involved in the differential regulation of the CD3 and CD2 pathways of human thymocyte activation. J Immunol. 1991;147:2825–32.
Proost P, De Meester I, Schols D, Struyf S, Lambeir AM, Wuyts A, et al. Amino-terminal truncation of chemokines by CD26/dipeptidyl-peptidase IV. Conversion of RANTES into a potent inhibitor of monocyte chemotaxis and HIV-1-infection. J Biol Chem. 1998;273:7222–7.
Christopherson 2nd KW, Hangoc G, Mantel CR, Broxmeyer HE. Modulation of hematopoietic stem cell homing and engraftment by CD26. Science. 2004;305:1000–3.
Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603–15.
Brizzi MF, Tarone G, Defilippi P. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr Opin Cell Biol. 2012;24:645–51.
Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology. 2002;123:1941–8.
Kemper K, Grandela C, Medema JP. Molecular identification and targeting of colorectal cancer stem cells. Oncotarget. 2010;1:387–95.
Langan RC, Mullinax JE, Ray S, Raiji MT, Schaub N, Xin HW, et al. A pilot study assessing the potential role of non-CD133 colorectal cancer stem cells as biomarkers. J Cancer. 2012;3:231–40.
Langan RC, Mullinax JE, Raiji MT, Upham T, Summers T, Stojadinovic A, et al. Colorectal cancer biomarkers and the potential role of cancer stem cells. J Cancer. 2013;4:241–50.
Kanwar SS, Yu YJ, Nautiyal J, Patel BB, Majumdar APN. The Wnt/beta-catenin pathway regulates growth and maintenance of colonospheres. Mol Cancer. 2010;9:13.
Sneath RJS, Mangham DC. The normal structure and function of CD44 and its role in neoplasia. J Clin Pathol Mol Pathol. 1998;51:191–200.
Fanali C, Lucchetti D, Farina M, Corbi M, Cufino V, Cittadini A, et al. Cancer stem cells in colorectal cancer from pathogenesis to therapy: controversies and perspectives. World J Gastroenterol. 2014;20:923–42.
Dalerba P, Dylla SJ, Park IK, Liu R, Wang XH, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104:10158–63.
Du L, Wang HY, He LY, Zhang JY, Ni BY, Wang XH, et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res. 2008;14:6751–60.
Lugli A, Iezzi G, Hostettler I, Muraro MG, Mele V, Tornillo L, et al. Prognostic impact of the expression of putative cancer stem cell markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer. Br J Cancer. 2010;103:382–90.
Fan CW, Chen T, Shang YN, Gu YZ, Zhang SL, Lu R, et al. Cancer-initiating cells derived from human rectal adenocarcinoma tissues carry mesenchymal phenotypes and resist drug therapies. Cell Death Dis. 2013;4.
Nagano O, Saya H. Mechanism and biological significance of CD44 cleavage. Cancer Sci. 2004;95:930–5.
Miraglia S, Godfrey W, Yin AH, Atkins K, Warnke R, Holden JT, et al. A novel five-transmembrane hematopoietic stem cell antigen: isolation, characterization, and molecular cloning. Blood. 1997;90:5013–21.
Shaked Y, Ciarrocchi A, Franco M, Lee CR, Man S, Cheung AM, et al. Therapy-induced acute recruitment of circulating endothelial progenitor cells to tumors. Science. 2006;313:1785–7.
Zhang MY, Song T, Yang L, Chen RK, Wu L, Yang ZY, et al. Nestin and CD133: valuable stem cell-specific markers for determining clinical outcome of glioma patients. J Exp Clin Cancer Res. 2008;27:7.
Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer CD133(+) cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–6.
Tang KH, Ma S, Lee TK, Chan YP, Kwan PS, Tong CM, et al. CD133+ liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology. 2012;55:807–20.
Richardson GD, Robson CN, Lang SH, Neal DE, Maitland NJ, Collins AT. CD133, a novel marker for human prostatic epithelial stem cells. J Cell Sci. 2004;117:3539–45.
Horst D, Kriegl L, Engel J, Kirchner T, Jung A. CD133 expression is an independent prognostic marker for low survival in colorectal cancer. Br J Cancer. 2008;99:1285–9.
Grosse-Gehling P, Fargeas CA, Dittfeld C, Garbe Y, Alison MR, Corbeil D, et al. CD133 as a biomarker for putative cancer stem cells in solid tumours: limitations, problems and challenges. J Pathol. 2013;229:355–78.
O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–10.
Zhu LQ, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–U114.
Kemper K, Versloot M, Cameron K, Colak S, Melo FDE, de Jong JH, et al. Mutations in the Ras-Raf Axis underlie the prognostic value of CD133 in colorectal cancer. Clin Cancer Res. 2012;18:3132–41.
Corbo C, Orrù S, Gemei M, Noto RD, Mirabelli P, Imperlini E, et al. Protein cross‐talk in CD133+ colon cancer cells indicates activation of the Wnt pathway and upregulation of SRp20 that is potentially involved in tumorigenicity. Proteomics. 2014;12:2045–59.
Mohammadi M, Bzorek M, Bonde JH, Nielsen HJ, Holck S. The stem cell marker CD133 is highly expressed in sessile serrated adenoma and its borderline variant compared with hyperplastic polyp. J Clin Pathol. 2013;66:403–8.
Arena V, Caredda E, Cufino V, Stigliano E, Scaldaferri F, Gasbarrini A, et al. Differential CD133 expression pattern during mouse colon tumorigenesis. Anticancer Res. 2011;31:4273–5.
Sgambato A, Corbi M, Svelto M, Caredda E, Cittadini A. New insights into the cd133 (prominin-1) expression in mouse and human colon cancer cells; in Corbeil D (ed) Prominin-1. New York, Springer. 2013. vol 777, pp 145–166
Yi JM, Tsai HC, Glockner SC, Lin S, Ohm JE, Easwaran H, et al. Abnormal DNA methylation of CD133 in colorectal and glioblastoma tumors. Cancer Res. 2008;68:8094–103.
Ong CW, Kim LG, Kong HH, Low LY, Iacopetta B, Soong R, et al. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod Pathol. 2010;23:450–7.
Kojima M, Ishii G, Atsumi N, Fujii S, Saito N, Ochiai A. Immunohistochemical detection of CD133 expression in colorectal cancer: a clinicopathological study. Cancer Sci. 2008;99:1578–83.
Kawamoto A, Tanaka K, Saigusa S, Toiyama Y, Morimoto Y, Fujikawa H, et al. Clinical significance of radiation-induced CD133 expression in residual rectal cancer cells after chemoradiotherapy. Exp Ther Med. 2012;3:403–9.
Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133− metastatic colon cancer cells initiate tumors. J Clin Investig. 2008;118:2111–20.
Feng JM, Miao ZH, Jiang Y, Chen Y, Li JX, Tong LJ, et al. Characterization of the conversion between CD133(+) and CD133(−) cells in colon cancer SW620 cell line. Cancer Biol Ther. 2012;13:1396–406.
Mao Q, Zhang Y, Fu XY, Xue JX, Guo WH, Meng MB, et al. A tumor hypoxic niche protects human colon cancer stem cells from chemotherapy. J Cancer Res Clin Oncol. 2013;139:211–22.
Weidle UH, Eggle D, Klostermann S, Swart GW. ALCAM/CD166: cancer-related issues. Cancer Genomics Proteomics. 2010;7:231–43.
Ohneda O, Ohneda K, Arai F, Lee J, Miyamoto T, Fukushima Y, et al. ALCAM (CD166): its role in hematopoietic and endothelial development. Blood. 2001;98:2134–42.
Halfon S, Abramov N, Grinblat B, Ginis I. Markers distinguishing mesenchymal stem cells from fibroblasts are downregulated with passaging. Stem Cells Dev. 2011;20:53–66.
Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma SM, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72.
Burkhardt M, Mayordomo E, Winzer KJ, Fritzsche F, Gansukh T, Pahl S, et al. Cytoplasmic overexpression of ALCAM is prognostic of disease progression in breast cancer. J Clin Pathol. 2006;59:403–9.
Mezzanzanica D, Fabbi M, Bagnoli M, Staurengo S, Losa M, Balladore E, et al. Subcellular localization of activated leukocyte cell adhesion molecule is a molecular predictor of survival in ovarian carcinoma patients. Clin Cancer Res. 2008;14:1726–33.
Rajasekhar VK, Studer L, Gerald W, Socci ND, Scher HI. Tumour-initiating stem-like cells in human prostate cancer exhibit increased NF-kappa B signalling. Nat Commun. 2011;2:13.
Levin TG, Powell AE, Davies PS, Silk AD, Dismuke AD, Anderson EC, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072–U2378.
Weichert W, Knosel T, Bellach J, Dietel M, Kristiansen G. ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol. 2004;57:1160–4.
Horst D, Kriegl L, Engel J, Kirchner T, Jung A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Investig. 2009;27:844–50.
Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67.
Subramaniam D, Ramalingam S, Houchen CW, Anant S. Cancer stem cells: a novel paradigm for cancer prevention and treatment. Mini-Rev Med Chem. 2010;10:359–71.
Deng S, Yang XJ, Lassus H, Liang S, Kaur S, Ye QR, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLos ONE. 2010;5(4):e10277.
Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–9.
Shenoy A, Butterworth E, Huang EH. ALDH as a marker for enriching tumorigenic human colonic stem cells. Methods Mol Biol (Clifton, NJ). 2012;916:373–85.
Park IK, Qian DL, Kiel M, Becker MW, Pihalja M, Weissman IL, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–5.
Molofsky AV, Pardal R, Iwashita T, Park I-K, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425:962–7.
Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–60.
Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313.
Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–56.
Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu R-M. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–9.
Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–U148.
Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–20.
Dovey JS, Zacharek SJ, Kim CF, Lees JA. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci U S A. 2008;105:11857–62.
Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7:86–95.
Lukacs RU, Memarzadeh S, Wu H, Witte ON. Bmi-1 is a crucial regulator of prostate stem cell self-renewal and malignant transformation. Cell Stem Cell. 2010;7:682–93.
Bruggeman SWM, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, et al. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328–41.
Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, et al. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–41.
Maynard MA, Ferretti R, Hilgendorf KI, Perret C, Whyte P, Lees JA. Bmi1 is required for tumorigenesis in a mouse model of intestinal cancer. Oncogene. 2014;33:3742–7.
Kim JH, Yoon SY, Kim CN, Joo JH, Moon SK, Choe IS, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004;203:217–24.
Du J, Li Y, Li J, Zheng J. Polycomb group protein Bmi1 expression in colon cancers predicts the survival. Med Oncol. 2010;27:1273–6.
Tateishi K, Ohta M, Kanai F, Guleng B, Tanaka Y, Asaoka Y, et al. Dysregulated expression of stem cell factor Bmi1 in precancerous lesions of the gastrointestinal tract. Clin Cancer Res. 2006;12:6960–6.
Kreso A, Galen PV, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2013;20:29–36.
Kreso A, van Galen P, Pedley NM, Lima-Fernandes E, Frelin C, Davis T, et al. Self-renewal as a therapeutic target in human colorectal cancer. Nat Med. 2014;20:29.
Denzel S, Maetzel D, Mack B, Eggert C, Barr G, Gires O. Initial activation of EpCAM cleavage via cell-to-cell contact. BMC Cancer. 2009;9.
Trzpis M, McLaughlin PMJ, de Leij L, Harmsen MC. Epithelial cell adhesion molecule—more than a carcinoma marker and adhesion molecule. Am J Pathol. 2007;171:386–95.
Baeuerle PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–23.
Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLos ONE. 2008;3(6):e2428.
van der Gun BT, Melchers LJ, Ruiters MH, de Leij LF, McLaughlin PM, Rots MG. EpCAM in carcinogenesis: the good, the bad or the ugly. Carcinogenesis. 2010;31:1913–21.
Sato T, Vries RG, Snippert HJ, Wetering MVD, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5.
Kemper K, Prasetyanti PR, De Lau W, Rodermond H, Clevers H, Medema JP. Monoclonal antibodies against Lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–86.
Walker F, Zhang HH, Odorizzi A, Burgess AW. Lgr5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLos ONE. 2011;6(7):e22733.
Takahashi H, Ishii H, Nishida N, Takemasa I, Mizushima T, Ikeda M, et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol. 2011;18:1166–74.
Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24.
Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, Okano H. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eiF4G for PABP. J Cell Biol. 2008;181:639–53.
Battelli C, Nikopoulos GN, Mitchell JG, Verdi JM. The RNA-binding protein Musashi-1 regulates neural development through the translational repression of p21(WAF-1). Mol Cell Neurosci. 2006;31:85–96.
Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, Weinmaster G, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–900.
Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, Ramalingam S, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–58.
Schulenburg A, Cech P, Herbacek I, Marian B, Wrba F, Valent P, et al. CD44-positive colorectal adenoma cells express the potential stem cell markers musashi antigen (msi1) and ephrin B2 receptor (EphB2). J Pathol. 2007;213:152–60.
Ishizuya-Oka A, Shimizu K, Sakakibara S, Okano H, Ueda S. Thyroid hormone-upregulated expression of Musashi-1 is specific for progenitor cells of the adult epithelium during amphibian gastrointestinal remodeling. J Cell Sci. 2003;116:3157–64.
Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, et al. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41.
Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, et al. RNA-binding protein Musashi family: roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. Proc Natl Acad Sci U S A. 2002;99:15194–9.
Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75.
Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–98.
Seifert JRK, Mlodzik M. Frizzled/PCP signalling: a conserved mechanism regulating cell polarity and directed motility. Nat Rev Genet. 2007;8:126–38.
Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10:468–77.
Burgess AW, Faux MC, Layton MJ, Ramsay RG. Wnt signaling and colon tumorigenesis—a view from the periphery. Exp Cell Res. 2011;317:2748–58.
Nemeth MJ, Topol L, Anderson SM, Yang YZ, Bodine DM. Wnt5a inhibits canonical Wnt signaling in hematopoietic stem cells and enhances repopulation. Proc Natl Acad Sci U S A. 2007;104:15436–41.
MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26.
de Sousa EMF, Vermeulen L, Richel D, Medema JP. Targeting Wnt signaling in colon cancer stem cells. Clin Cancer Res. 2011;17:647–53.
Takemaru KI, Moon RT. The transcriptional coactivator cbp interacts with beta-catenin to activate gene expression. J Cell Biol. 2000;149:249–54.
Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 2000;19:1839–50.
Crosnier C, Stamataki D, Lewis J. Organizing cell renewal in the intestine: stem cells, signals and combinatorial control. Nat Rev Genet. 2006;7:349–59.
van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–50.
Haegebarth A, Clevers H. Wnt signaling, lgr5, and stem cells in the intestine and skin. Am J Pathol. 2009;174:715–21.
Zeilstra J, Joosten SPJ, Dokter M, Verwiel E, Spaargaren M, Pals ST. Deletion of the WNT target and cancer stem cell marker CD44 in Apc (Min/+) mice attenuates intestinal tumorigenesis. Cancer Res. 2008;68:3655–61.
van der Flier LG, van Gijn ME, Hatzis P, Kujala P, Haegebarth A, Stange DE, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–12.
Munz M, Baeuerle PA, Gires O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res. 2009;69:5627–9.
Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–U117.
Mak AB, Nixon AML, Kittanakom S, Stewart JM, Chen GI, Curak J, et al. Regulation of CD133 by HDAC6 promotes beta-catenin signaling to suppress cancer cell differentiation. Cell Rep. 2012;2:951–63.
Cho J-H, Dimri M, Dimri GP. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J Biol Chem. 2013;288:3406–18.
Krause U, Ryan DM, Clough BH, Gregory CA. An unexpected role for a Wnt-inhibitor: Dickkopf-1 triggers a novel cancer survival mechanism through modulation of aldehyde-dehydrogenase-1 activity. Cell Death Dis. 2014;5:e1093.
Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–612.
Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106.
Artavanis-Tsakonas S, Rand M, Lake R. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Gray GE, Mann RS, Mitsiadis E, Henrique D, Carcangiu ML, Banks A, et al. Human ligands of the notch receptor. Am J Pathol. 1999;154:785–94.
Kopan R, Ilagan MXG. The canonical notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33.
Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian XL, Pan DJ, et al. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206.
Qiao L, Wong BCY. Role of notch signaling in colorectal cancer. Carcinogenesis. 2009;30:1979–86.
Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–8.
van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–63.
Miyamoto Y, Maitra A, Ghosh B, Zechner U, Argani P, Iacobuzio-Donahue CA, et al. Notch mediates TGF alpha-induced changes in epithelial differentiation during pancreatic tumorigenesis. Cancer Cell. 2003;3:565–76.
Leong KG, Gao WQ. The notch pathway in prostate development and cancer. Differentiation. 2008;76:699–716.
Baliko F, Bright T, Poon R, Cohen B, Egan S, Alman B. Inhibition of notch signaling induces neural differentiation in Ewing sarcoma. Am J Pathol. 2007;170:1686–94.
Ramdass B, Maliekal TT, Lakshmi S, Rehman M, Rema P, Nair P, et al. Coexpression of Notch1 and NF-kappaB signaling pathway components in human cervical cancer progression. Gynecol Oncol. 2007;104:352–61.
Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–7.
Meng RD, Shelton CC, Li YM, Qin LX, Notterman D, Paty PB, et al. Gamma-secretase inhibitors abrogate oxaliplatin-induced activation of the notch-1 signaling pathway in colon cancer cells resulting in enhanced chemosensitivity. Cancer Res. 2009;69:573–82.
Zhao D, Mo Y, Li M-T, Zou S-W, Cheng Z-L, Sun Y-P, et al. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J Clin Investig. 2014;124:5453–65.
Rezza A, Skah S, Roche C, Nadjar J, Samarut J, Plateroti M. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123:3256–65.
Fan X, Khaki L, Zhu TS, Soules ME, Talsma CE, Gul N, et al. NOTCH pathway blockade depletes CD133-positive glioblastoma cells and inhibits growth of tumor neurospheres and xenografts. Stem Cells. 2010;28:5–16.
Veenendaal LM, Kranenburg O, Smakman N, Klomp A, Rinkes I, van Diest PJ. Differential Notch and TGF beta signaling in primary colorectal tumors and their corresponding metastases. Cell Oncol. 2008;30:1–11.
Ghaleb AM, McConnell BB, Nandan MO, Katz JP, Kaestner KH, Yang VW. Haploinsufficiency of Kruppel-like factor 4 promotes adenomatous polyposis coli-dependent intestinal tumorigenesis. Cancer Res. 2007;67:7147–54.
Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-|[beta]| family signalling. Nature. 2003;425:577–84.
Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–84.
Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–21.
Akhurst RJ. TGF beta signaling in health and disease. Nat Genet. 2004;36:790–2. United States.
Elliott RL, Blobe GC. Role of transforming growth factor beta in human cancer. J Clin Oncol. 2005;23:2078–93.
Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700.
Moustakas A, Souchelnytskyi S, Heldin CH. Smad regulation in TGF-beta signal transduction. J Cell Sci. 2001;114:4359–69.
Izzi L, Attisano L. Regulation of the TGF|[beta]| signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–8.
Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309.
Siegel PM, Shu W, Massague J. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J Biol Chem. 2003;278:35444–50.
Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-beta-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci. 2012;125:1259–73.
McQualter JL, McCarty RC, Van der Velden J, O'Donoghu RJJ, Asselin-Labat M-L, Bozinovski S, et al. TGF-beta signaling in stromal cells acts upstream of FGF-10 to regulate epithelial stem cell growth in the adult lung. Stem Cell Res. 2013;11:1222–33.
Yamashita M, Fatyol K, Jin C, Wang X, Liu Z, Zhang YE. TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol Cell. 2008;31:918–24.
Javelaud D, Mauviel A. Crosstalk mechanisms between the mitogen-activated protein kinase pathways and Smad signaling downstream of TGF-|[beta]|: implications for carcinogenesis. Oncogene. 2005;24:5742–50.
Takayama T, Miyanishi K, Hayashi T, Sato Y, Niitsu Y. Colorectal cancer: genetics of development and metastasis. J Gastroenterol. 2006;41:185–92.
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60.
Wang J, Sun L, Myeroff L, Wang X, Gentry LE, Yang J, et al. Demonstration that mutation of the type ii transforming growth factor beta receptor inactivates its tumor suppressor activity in replication error-positive colon carcinoma cells. J Biol Chem. 1995;270:22044–9.
Thiagalingam S, Lengauer C, Leach FS, Schutte M, Hahn SA, Overhauser J, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet. 1996;13:343–6.
Takaku K, Oshima M, Miyoshi H, Matsui M, Seldin MF, Taketo MM. Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell. 1998;92:645–56.
Becker C, Fantini MC, Schramm C, Lehr HA, Wirtz S, Nikolaev A, et al. TGF-beta suppresses tumor progression in colon cancer by inhibition of IL-6 trans-signaling. Immunity. 2004;21:491–501.
Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes Dev. 2001;15:3059–87.
Varjosalo M, Taipale J. Hedgehog: functions and mechanisms. Genes Dev. 2008;22:2454–72.
Micchelli CA, The I, Selva E, Mogila V, Perrimon N. Rasp, a putative transmembrane acyltransferase, is required for hedgehog signaling. Development. 2002;129:843–51.
Merchant AA, Matsui W. Targeting hedgehog—a cancer stem cell pathway. Clin Cancer Res. 2010;16:3130–40.
Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. Embo Mol Med. 2009;1:338–51.
Tanaka H, Nakamura M, Kameda C, Kubo M, Sato N, Kuroki S, et al. The Hedgehog signaling pathway plays an essential role in maintaining the CD44(+)CD24(−/low) subpopulation and the side population of breast cancer cells. Anticancer Res. 2009;29:2147–57.
Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67:2187–96.
Hayward P, Kalmar T, Arias AM. Wnt/Notch signalling and information processing during development. Development. 2008;135:411–24.
Lawrence N, Langdon T, Brennan K, Arias AM. Notch signaling targets the Wingless responsiveness of a Ubx visceral mesoderm enhancer in Drosophila. Curr Biol. 2001;11:375–85.
Brennan K, Klein T, Wilder E, Arias AM. Wingless modulates the effects of dominant negative notch molecules in the developing wing of Drosophila. Dev Biol. 1999;216:210–29.
Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–21.
Chen X, Stoeck A, Lee SJ, Shih Ie M, Wang MM, Wang TL. Jagged1 expression regulated by Notch3 and Wnt/beta-catenin signaling pathways in ovarian cancer. Oncotarget. 2010;1:210–8.
Alvarez-Medina R, Cayuso J, Okubo T, Takada S, Marti E. Wnt canonical pathway restricts graded Shh/Gli patterning activity through the regulation of Gli3 expression. Development. 2008;135:237–47.
van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, et al. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277–82.
Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, et al. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–6.
Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–64.
van den Brink GR, Hardwick JCH. Hedgehog Wnteraction in colorectal cancer. Gut. 2006;55:912–4.
Ma J, Meng Y, Kwiatkowski DJ, Chen X, Feng H, Sun Q, et al. Mammalian target of rapamycin regulates murine and human cell differentiation through STAT3/p63/Jagged/Notch cascade. J Clin Investig. 2010;120:103–14.
Song KY, Wang H, Krebs TL, Danielpour D. Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. EMBO J. 2006;25:58–69.
Nishita M, Hashimoto MK, Ogata S, Laurent MN, Ueno N, Shibuya H, et al. Interaction between Wnt and TGF-beta signalling pathways during formation of Spemann’s organizer. Nature. 2000;403:781–5.
Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, et al. Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci U S A. 2008;105:4838–43.
Acknowledgments
The authors are thankful for the financial support provided through the grant “Analysis of gene expression for different stages of colorectal cancer” (“Programme-targeted funding 2014-2017”; Government of the Republic of Kazakhstan).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Abetov, D., Mustapova, Z., Saliev, T. et al. Biomarkers and signaling pathways of colorectal cancer stem cells. Tumor Biol. 36, 1339–1353 (2015). https://doi.org/10.1007/s13277-015-3198-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3198-4