Abstract
A T>G single nucleotide polymorphism (SNP, rs2279744) of the MDM2 gene has been investigated in sarcoma community, but the findings are conflicting. This study was designed to well define the relationship between SNP rs2279744 and sarcoma risk. We did a systematic computerized search of the PubMed, Web of Science, and Science Direct databases to identify the human case–control studies investigating the relationship between SNP rs2279744 and sarcoma risk with complete genetic data. Pooled odds ratios (ORs) were calculated with the Mantel–Haenszel fixed-effect model or the DerSimonian and Laird random effects model to estimate the risk of sarcoma. Overall analysis included five independent studies. On the whole, the T/G genotype or the combined G/G and T/G genotypes appeared to be associated with approximately 1.40-fold higher risk of sarcoma relative to the T/T genotype (T/G vs. T/T: OR 1.33, 95 % CI 1.00–1.77; G/G + T/G vs. T/T: OR 1.42, 95 % CI 1.08–1.85). We noted that the Caucasian populations showed a similarly increased risk of sarcoma ascribed to the carriage of the same genotypes (T/G vs. T/T: OR 1.41, 95 % CI 1.05–1.90; G/G + T/G vs. T/T: OR 1.49, 95 % CI 1.13–1.97). This meta-analysis provides evidence that MDM2 SNP rs2279744 may be significantly associated with increased risk of sarcoma in Caucasian individuals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sarcoma is a type of less common malignancy preferentially occurring in children and adolescents worldwide [1]. Mechanic research in the past decade has established genetic mutations in signaling pathways as a cause of this disease [2]. However, sarcoma is etiologically heterogeneous, and hence, the exact molecular mechanism remains to be elucidated.
The p53 tumor suppressor is one of the most frequently mutated gene, and almost 50 % of all human cancers have described p53 mutations [3, 4]. p53 encodes tumor protein 53, a 53-kDa transcription factor mediating cellular proliferation, senescence, and apoptosis in defense against chromosomal breakage [5, 6]. The murine double minute 2 (MDM2) gene negatively modulates the p53 protein by coding for a ubiquitin protein ligase that acts as a mediator of location, activity, and stability of p53 [7, 8]. Thus, even a small change in MDM2 serum levels may lead to a sharp impairment of the biological function of p53. Altered MDM2 levels have been linked to p53 degradation and inactivation [9, 10], leading to consequent initiation of cancer and poor prognosis [11, 12]. In the promoter region, there locates a T>G single nucleotide polymorphism (SNP) at the 309th nucleotide of the first intron (dbSNP ID rs2279744). The G allele has been reported to upregulate MDM2 protein expression by increasing the binding affinity of the transcriptional activator Sp1 and thereby leads to attenuation in p53 tumor suppression [13, 14], suggesting that the genetic variations in MDM2 promoter may represent a risk factor for various diseases in human. Several epidemiological studies from Brazil and Italy have provided evidence supporting a major role of rs2279744 in modifying the risk of Ewing sarcoma, Kaposi sarcoma, and osteosarcoma [15–17], whereas this sequence variation had no contribution to uterine leiomyosarcoma [18]. To well define the relationship between rs2279744 and sarcoma risk, we decided to perform a meta-analysis of all previously published studies.
Methods
Search strategy
To derive the studies pertaining to MDM2 SNP rs2279744 and sarcoma, we did a systematic computerized search of the PubMed, Web of Science, and Science Direct databases (up to July 20, 2013). The search terms included murine double minute 2, MDM2, sarcoma, polymorphism, and polymorphisms. We also hand searched peer-reviewed journals, known for publishing sarcoma-related articles, and reference lists of all identified records, systematic reviews, and original articles in particular. There were no restrictions for this search.
Inclusion criteria and data extraction
We included human studies that had a case–control or cohort design, which investigated the relationship between SNP rs2279744 and sarcoma risk and that contained genotype counts in detail or provided information from which we could infer the genotype distribution. Two investigators singled out the studies that fulfilled all criteria as previously described and then independently collected the name of the first author, study country, ethnic origin, publication year and journal, source of controls, proportion of male and female in cases and controls, number of genotyped cases and controls, method used to genotype SNP rs2279744, count of genotypes, and P value of Hardy–Weinberg equilibrium (HWE). Discrepancy was resolved via discussion among all investigators.
Statistical analysis
Pooled odds ratios and its 95 % confidence intervals (OR and 95 % CI) were computed to estimate the association of SNP rs2279744 and sarcoma risk. The Z test was used to check the significance of the combined ORs. Between-study heterogeneity was determined by the Q test and the I 2 statistic [19]. We considered the results heterogeneous when P values were higher than 0.10 or I 2 ranged from 0 to 50 %. For the calculation of pooled OR, the Mantel–Haenszel fixed effect model was employed when heterogeneity was insignificantly indicated; otherwise, the DerSimonian and Laird random effects model was used. In addition to overall analysis, stratified analysis was also performed to estimate risk for subgroups according to ethnicity. We performed sensitivity analysis to see whether the overall estimates were substantially affected as a result of the independent studies [20]. Estimation of publication bias was conducted with the aid of Begg’s funnel plots and Egger’s regression test [21]. Statistical data were analyzed using the R Software version 2.15.0 (the R Foundation for Statistical Computing), and P values smaller than .10 were considered significant.
Results
Characteristics of studies
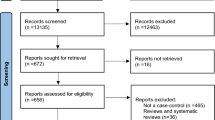
We primarily retrieved 56 articles, of which 52 articles were discarded due to multiple reasons, such as obviously irrelevant research, expression-based studies, and lack of required data. As a result, four articles remained for further analysis [15–18]. As an Italian article reported two different case–control studies, our final pooling data included five independent populations. The flow chart of study selection is presented in Fig. 1.
The studies eligible for this quantitative assessment were published from 2005 to 2013, involving four Caucasian populations and one African population. Control subjects were all chosen from general population. Most of the studies selected the classical polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) to determine the genotypes of SNP rs2279744. Both male and female subjects were analyzed in our analysis. All genotype distributions in control population were in HWE (P > 0.10) (Table 1).
Quantitative synthesis
The meta-analysis results are summarized in Table 2.
Data from five studies were analyzed in overall analysis. On the whole, the T/G genotype of T/G vs. T/T comparison appeared to be associated with 1.33-fold higher risk of sarcoma relative to the T/T genotype (T/G vs. T/T: OR 1.33, 95 % CI 1.00–1.77, Fig. 2). Similarly, the combined G/G and T/G genotypes showed a significantly 42 % increased risk of sarcoma (G/G + T/G vs. T/T: OR 1.42, 95 % CI 1.08–1.85, Fig. 3).
Such a significant increase in the risk of sarcoma was also revealed in stratified analysis according to ethnicity. There was nearly 50 % increased risk of sarcoma associated with the T/G or G/G and T/G combined genotypes (T/G vs. T/T: OR 1.41, 95 % CI 1.05–1.90, Fig. 2; G/G + T/G vs. T/T: OR 1.49, 95 % CI 1.13–1.97, Fig. 3).
Heterogeneity test
As shown in Table 2, the overall estimates of three comparisons were found heterogeneous (P < 0.10 or I 2 > 50 %). Subgroup analysis revealed that ethnicity was a cause of the moderate heterogeneity.
Sensitivity analysis and publication bias
We did not see any substantial alternation of overall estimates using the sensitivity analysis, suggesting our results were robust (data not shown). Using the analytic tools described by Begg and Egger, we found shapes of the funnel plots symmetrical, and there was no statistical evidence of significant publication bias in the literature (P > 0.10).
Discussion
Multiple lines of evidence have associated enhanced expression of MDM2 with the initiation of human cancer [12, 22]. Several groups further provide data that overexpressed MDM2 increases the likelihood of cancer by accelerating cancerous progression and blocking response to conventional chemotherapy [23, 24]. Much effort diverted to the complicated etiology of sarcoma more than 20 years ago also revealed a relevance of MDM2 overexpression to apoptosis, cell-cycle control, and resistance to traditional treatments [25, 26]. Genetic alternations in the promoter region of MDM2 are reported to induce higher levels of the protein, malfunction of the p53 pathway, and formation of chromatin-associated MDM2–p53 complexes [27–29], thus initiating the onset of aggressive diseases, such as sarcoma. The data demonstrated across the last two decades suggest that MDM2 may possibly have an involvement in human malignancies.
The MDM2 SNP rs2279744 is of special interest in recent years due to its pivotal role played in the regulation of its promoter protein, which has been implicated in sarcoma. Alhopuro et al., who included 68 Finnish patients with uterine leiomyosarcoma, reported that SNP rs2279744 had no significant contribution to the formation of this sarcoma [18]. However, a study examining the association between SNP rs2279744 and risk of osteosarcoma demonstrated that the G allele was related to a significantly increased risk only for female patients [17]. Similarly, an almost 2.38-fold increased risk of human immunodeficiency virus-associated malignancy (Kaposi sarcoma) was seen in a Caucasian population [16], a discovery in accord with the most recent study of Brazilian patients with Ewing sarcoma [15]. It is important to note that each of the published studies employed a small population that may affect the statistical power and lead to biased estimations as a consequence. Therefore, a larger study is required to strengthen the detection power and provide compelling evidence for the association under evaluation.
In this study, we gathered all published literature and conducted a meta-analysis to evaluate the relation of SNP rs2279744 and sarcoma risk. The analysis of total populations showed approximately 40 % increased risk of sarcoma in relation to the T/G genotype or the combined G/G and T/G genotypes. Risk estimates for subgroups according to ethnicity implicated that the T/G or G/G and T/G combined genotypes appeared to be associated with an almost 1.50-fold risk of sarcoma among Caucasian subjects. These findings are consistent with most of the published studies reporting the relationship of SNP rs2279744 and risk of sarcoma and the experimental data as previously detailed. An early meta-analysis of four independent studies showed a 34 % elevated risk to develop sarcoma ascribed to the TG and GG genotypes, a discovery similar to that revealed in the current study [30]. Nevertheless, Cai et al. failed to include a subsequently published study and assumed one comparison model (T/T vs T/G + G/G) only, and it is the relatively smaller sample and the effect size estimation for a single model may likely lead consequently to an underestimated association. By adding an additional dataset, we observed a stronger overall association and statistical evidence of a significantly elevated risk of sarcoma among Caucasians, which were not found in the previous analysis.
Although this is the largest and most comprehensive analysis to date, several possible limitations should be addressed. Firstly, we did not consider unpublished studies and the studies published in databases rather than those searched in the present analysis, so publication bias may have occurred. Secondly, our study probably has non-differential misclassification bias, because of the nonstandardized selection of control subjects. For example, some studies randomly selected healthy volunteers as controls, and for some studies, healthy blood donors served as a reference group. Thirdly, as sarcoma is heterogeneous in nature, common confounding factors, such as age, gender, and carcinogenic exposure that may modify risk of sarcoma by interaction with candidate genes, should be considered.
In conclusion, we reported a significant association between SNP rs2279744 in the promoter of MDM2 and sarcoma risk. Our findings also revealed that SNP rs2279744 increased the risk of sarcoma among Caucasians. These associations, however, remain to be examined by a future study involving a large number of samples.
References
Dorfman HD, Czerniak B. Bone cancers. Cancer. 1995;75(1 Suppl):203–10.
Borden EC et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res. 2003;9(6):1941–56.
Hollstein M et al. p53 mutations in human cancers. Science. 1991;253(5015):49–53.
Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet. 2002;3:101–28.
Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1(14):1001–8.
Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9(2):95–107.
Huang SF et al. Combined effects of MDM2 SNP 309 and p53 mutation on oral squamous cell carcinomas associated with areca quid chewing. Oral Oncol. 2009;45(1):16–22.
Michael D, Oren M. The p53-Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13(1):49–58.
Poyurovsky MV, Prives C. Unleashing the power of p53: lessons from mice and men. Genes Dev. 2006;20(2):125–31.
Greenblatt MS et al. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54(18):4855–78.
Jones SN et al. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci U S A. 1998;95(26):15608–12.
Freedman DA, Levine AJ. Regulation of the p53 protein by the MDM2 oncoprotein–thirty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 1999;59(1):1–7.
Bond GL et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119(5):591–602.
Bond GL, Hu W, Levine A. A single nucleotide polymorphism in the MDM2 gene: from a molecular and cellular explanation to clinical effect. Cancer Res. 2005;65(13):5481–4.
Thurow HS et al. Ewing sarcoma: influence of TP53 Arg72Pro and MDM2 T309G SNPs. Mol Biol Rep. 2013;40(8):4929–34.
Tornesello ML et al. MDM2 and CDKN1A gene polymorphisms and risk of Kaposi’s sarcoma in African and Caucasian patients. Biomarkers. 2011;16(1):42–50.
Toffoli G et al. Effect of TP53 Arg72Pro and MDM2 SNP309 polymorphisms on the risk of high-grade osteosarcoma development and survival. Clin Cancer Res. 2009;15(10):3550–6.
Alhopuro P et al. The MDM2 promoter polymorphism SNP309T–>G and the risk of uterine leiomyosarcoma, colorectal cancer, and squamous cell carcinoma of the head and neck. J Med Genet. 2005;42(9):694–8.
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–58.
Tobias A. Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull. 1999;47:15–7.
Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–57.
Leach FS et al. p53 mutation and MDM2 amplification in human soft tissue sarcomas. Cancer Res. 1993;53(10 Suppl):2231–4.
Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55(1):96–107.
Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2(1):1–8.
Oliner JD et al. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358(6381):80–3.
Cordon-Cardo C et al. Molecular abnormalities of mdm2 and p53 genes in adult soft tissue sarcomas. Cancer Res. 1994;54(3):794–9.
Birgander R et al. The codon 31 polymorphism of the p53-inducible gene p21 shows distinct differences between major ethnic groups. Hum Hered. 1996;46(3):148–54.
Arva NC et al. A chromatin-associated and transcriptionally inactive p53-Mdm2 complex occurs in mdm2 SNP309 homozygous cells. J Biol Chem. 2005;280(29):26776–87.
Ruijs MW et al. The single-nucleotide polymorphism 309 in the MDM2 gene contributes to the Li-Fraumeni syndrome and related phenotypes. Eur J Hum Genet. 2007;15(1):110–4.
Cai X, Yang M. The functional MDM2 T309G genetic variant but not P53 Arg72Pro polymorphism is associated with risk of sarcomas: a meta-analysis. J Cancer Res Clin Oncol. 2012;138(4):555–61.
Author information
Authors and Affiliations
Corresponding author
Additional information
Dawei Zhang and Yuanyuan Ding are contributed equally to this work.