Abstract
Histone demethylase KDM2A has been reported to be dysregulated in lung cancer. However, its function in gastric cancer remains poorly understood. Here, it was found that the expression level of KDM2A was increased in gastric cancer tissues. Moreover, forced expression of KDM2A in gastric cancer cells promoted cell growth and migration, while the knockdown expression of KDM2A inhibited the tumorigenicity of gastric cancer cells. Mechanistically, KDM2A regulated the growth and motility of gastric cancer cells through downregulating the expression of programmed cell death 4 (PDCD4), a known tumor suppressor in the progression of gastric cancer. Taken together, our study suggested that upregulation of KDM2A was very important in the progression of gastric cancer, and KDM2A might be a promising therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is the fourth most common cancer type worldwide and ranks the second in terms of global cancer-related mortality [1]. Approximately 50 % of cases occur in Eastern Asia (mainly in China) [2]. Although the overall incidence of gastric cancer has declined in the past decades, the incidence remains high in Asian countries [3]. The prognosis of gastric cancer is poor, and the key players in molecular pathogenesis are predominantly unknown at present. Therefore, investigations into the molecular mechanisms involving in gastric cancer progression have major importance and may tend to develop novel avenues for targeted therapy.
Histone lysine methylation is considered as a key chromatin mark that mediates epigenetic and transcriptional regulation of gene expression [4–6]. Altered activity of histone demethylases (HDMs) is emerging as a common defect in the tumorigenesis [7, 8]. H3K27 demethylase KDM6A (also known as UTX) showed tumor suppressor functions and was mutated in the progression of cancer [9, 10]. In addition, amplification or overexpression of the H3K9/H3K36 demethylase KDM4C was very common in cancer samples [11]. Recent studies have demonstrated that the H3K36me2 demethylases KDM2B and KDM7B were also important regulators of cell growth [12].
KDM2A (FBXL11), previously shown to demethylate histone H3K36, contains an F-box, a JmjC domain, a CxxC zinc finger, a PHD domain, and three leucine-rich repeat elements [13–15]. Previous study has revealed a feedback loop between the expression of the KDM2A and the activity of NF-κB signaling [16]. Overexpression of KDM2A inhibited the activity of NF-κB signaling, and the expression of the KDM2A gene was reported to be driven by NF-κB signaling [16–20]. Recently, the biological functions of KDM2A in lung cancer have attracted much interest. KDM2A activated ERK1/2 through epigenetic repression of DUSP3 expression via demethylation of dimethylated H3K36 at the DUSP3 locus [14]. Moreover, KDM2A transcriptionally repressed the histone deacetylase 3 (HDAC3) gene by removing methyl groups from dimethylated H3K36 at the HDAC3 promoter in KDM2A-overexpressing NSCLC cells and released cell cycle-associated genes and cell invasion-related genes from HDAC3 repression [13]. However, the function of KDM2A in the progression of gastric cancer was poorly understood.
In this study, it was found that the expression of KDM2A was significantly upregulated in gastric cancer samples compared to their adjacent tissues. Moreover, in the biological function studies, KDM2A was shown to promote the growth and migration of gastric cancer cells, while the knockdown expression of KDM2A inhibited the growth, migration, and metastasis of gastric cancer cells in vitro and in vivo. Mechanistically, KDM2A promoted cell growth and migration through downregulating the expression of programmed cell death 4 (PDCD4), a well-known tumor suppressor in gastric cancer. Taken together, our study suggested that upregulation of KDM2A played an oncogenic role in the progression of gastric cancer, and KDM2A might be a promising therapeutic target in gastric cancer.
Materials and methods
Primary gastric cancer samples
Primary tissues were collected from patients who received surgery for gastric cancer at Jingjiang People’s Hospital of Jiangsu Province and First Affiliated Hospital of Nanjing Medical University. Patients have no anticancer treatment before tumor resection. All patients have given informed consent. Dissected samples were frozen immediately after surgery and stored at −80 °C until needed. This study and the use of all clinical materials mentioned were approved by individual institutional ethics committees.
Cell culture and transfection
Gastric epithelial cell lines (GES-1) and gastric cancer cell lines (AGS, BGC803, MGC823, and SGC7901) were obtained from the American Type Culture Collection and cultured in DMEM medium (Invitrogen) containing 10 % heat-inactivated fetal bovine serum, 10 units/ml penicillin G, and 10 mg/ml streptomycin. Cells were incubated at 37 °C in humidified air containing 5 % CO2. To generate the KDM2A expression vector, the open reading frame of human KDM2A cDNA was cloned into the eukaryotic expression vector pcDNA3.1. The KDM2A expression vector and empty pcDNA3.1 vector were transfected into GES-1 and AGS cells using Lipofectamine 2000 reagent (Invitrogen). The transfected cells were selected in the presence of 600 μg/ml G418, and the resistant cells were pooled and used for further study.
Immunohistochemistry
Paraffin-embedded gastric cancer tissues and paired adjacent tissues were obtained from the Jingjiang People’s Hospital of Jiangsu Province and First Affiliated Hospital of Nanjing Medical University. Five-micrometer-thick consecutive sections were cut and mounted on glass slides. After deparaffinizing, rehydrating, antigen retrieval, and blocking endogenous peroxidases, the sections were washed thrice in 0.01 mol/l phosphate-buffered saline (PBS) (8 mmol/l Na2HPO4, 2 mmol/l NaH2PO4, and 150 mmol/l NaCl) for 5 min each and blocked for 1 h in 0.01 mol/l PBS supplemented with 0.3 % Triton X-100 and 5 % normal goat serum, followed by addition of anti-KDM2A antibody (1:200, Abcam) at 4 °C overnight. After brief washes in 0.01 mol/l PBS, sections were exposed for 2 h to 0.01 mol/l PBS containing horseradish peroxidase-conjugated mouse anti-goat IgG (1:500), followed by development with 0.3 % H2O2 and 0.03 % 3,30-diaminobenzidine in 0.05 mol/l Tris–HCl (pH 7.6). Immunohistochemistry was performed at least three times, and the sections were counterstained with hematoxylin.
Immunoblotting
Western blot analysis was performed according to the standard protocols. Anti-KDM2A antibody was purchased from Abcam. Antibodies to proliferating cell nuclear antigen (PCNA), CyclinD1, GAPDH, myc tag, and PDCD4 were purchased from Santa Cruz. Antibody to phosphorylated ERK was purchased from Cell Signaling Technology. Secondary antibodies/rabbit anti-mouse IgG (Sigma) and goat anti-rabbit IgG (Cell Signaling Technology) were used at a dilution of 1:1,500. Primary antibodies were diluted in TBST containing 1 % BSA. The immunoreactive protein bands were visualized by ECL kit (Pierce).
RT-PCR analysis
TRIzol reagent (Invitrogen) was used to isolate total RNA from tissues. Two micrograms of total RNA with high quality was processed directly to cDNA with the reverse transcription kit (Promega, Madison, WI), following the manufacturer’s instructions, in a total volume of 25 μl. The primer pair used for amplification of the human KDM2A gene was as follows: forward primer 5′-TGTATCTGCAATGAGATTGTT-3′ and reverse primer 5′-TATGGTGACTCGG TCATCCC-3′. As an internal standard, a fragment of human beta-actin was amplified by PCR using the following primers: forward primer 5′-GATCATTGCTCCTCCTGAGC-3′ and reverse primer 5′-ACTCCT GCTTGCTGATCCAC-3′. Amplification reactions were performed in a 20-μl volume of the LightCycler-DNA Master SYBR Green I mixture from Roche Applied Science as follows: with 10 pmol of primer, 2 mM MgCl2, 200 μM dNTP mixture, and 0.5 units of Taq DNA polymerase and universal buffer. All of the reactions were performed in triplicate in an iCycler iQ System (Bio-Rad), and the thermal cycling conditions were as follows: 95 °C for 3 min; 40 cycles of 95 °C for 30 s, 58 °C for 20 s, and 72 °C for 30 s; and 72 °C for 10 min. To confirm the specificity of amplification, the PCR products from each primer pair were subjected to a melting curve analysis and electrophoresis in 2 % agarose gel.
Silencing the expression of KDM2A in the gastric cancer cells
Target sequences for human KDM2A small interfering RNA were as listed: 5′-AATGCCAAATTGCTGGGAAT-3′ and 5′-AAGGAGCTGTCTGAAGTTGAG-3′. The control nucleotide sequence of small interfering RNA was 5′-AAGGTCCAACCAGGGGGTAA-3′, which was the random sequence that was not related to KDM2A mRNA. In our experiments, FG12 lentiviral vector, which has an independent open reading frame of green fluorescence protein (GFP), was used to produce small, double-stranded RNA (siRNA) to inhibit target gene expression in gastric cancer cells. To construct the hairpin siRNA expression cassette, complementary DNA oligonucleotides for siRNA of KDM2A (si KDM2A) or mutated sequence of KDM2A siRNA as control (si con) were synthesized, annealed, and inserted into FG12 according to the manufacturer’s protocol. FG12 vector with si KDM2A or si con was transfected into HEK293T cells, and the virus was harvested from culture medium. The harvested virus was purified by centrifugation at 25,000×g (4 °C, 150 min), and appropriate amounts of virus were used to infect cells. After 3 days of infection, the GFP-positive cells were sorted by flow cytometry (BD Biosciences), which all stably expressed KDM2A siRNA or control siRNA.
Crystal violet assay
For cell growth assay, equal numbers of cells were seeded in 12-well plates and cultured in medium supplemented with 10 % fetal bovine serum (FBS) for 7 days. Medium was changed every other day. Cell growth was stopped after 7 days in culture by removing the medium and adding 0.5 % crystal violet solution in 20 % methanol. After staining for 5 min, the fixed cells were washed with PBS and photographed.
Soft agar assay
The soft agar assay was performed to evaluate the effect of KDM2A on the tumorigenesis in vitro. Briefly, cells (1 × 104) were resuspended in medium containing 10 % FBS with 0.3 % agarose and layered on the top of 0.6 % agar in medium supplemented with 20 % FBS on 60-mm plates. After 14 days of culture at 37 °C, plates were stained with 0.005 % crystal violet for 1 h. Colonies were photographed.
Boyden chamber assay
Boyden chamber (polycarbonate membrane with 8 μm pore size) was obtained from Neuroprobe Corporation, Bethesda, MD, USA. Cells (2 × 105) in 0.05 ml medium containing 1 % FBS were placed in the upper chamber, and the lower chamber was loaded with 0.152 ml medium containing 10 % FBS. After the incubation for 8 h, cells that migrated to the lower surface of the filter were detected with traditional staining and five fields of each well were counted. Three wells were examined for each cell type, and the experiments were repeated for at least three times.
In vivo metastasis assay
The SGC7901-Luc stable cell line (overexpressing luciferase) was established with G418 selection. Luciferase expression was determined using luciferin (Xenogen) and an in vivo imaging system (Xenogen). The luciferase-expressing SGC7901/si con cells and luciferase-expressing SGC7901/si KDM2A cells (1 × 106 cells in 100 μl PBS) were injected into the nude mice (six nude mice for each group). The metastatic lesions were monitored every week. Before mice were anesthetized with Forane (Abbott), the aqueous solution of luciferin (150 mg/kg intraperitoneally) was injected 10 min before imaging. The animals were placed into a light-tight chamber of the CCD camera system (Xenogen), and the photons emitted from the luciferase-expressing cells within the animal were quantified for 1 min, using the software program Living Image (Xenogen) as an overlay on Igor (Wavemetrics).
Results
KDM2A is upregulated in gastric cancer
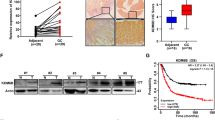
KDM2A has been reported to be upregulated in lung cancer [13]. However, the expression pattern of KDM2A in gastric cancer still remains unknown. To further understand the expression of KDM2A in gastric cancer, we determined the expression level of KDM2A in 61 pairs of primary gastric cancer and their corresponding nontumor tissues by real-time PCR. We found that KDM2A expression was elevated by more than twofold in gastric cancer compared with their corresponding nontumor tissues (Fig. 1a). Moreover, the immunohistochemistry assay demonstrated strong KDM2A nuclear staining in the gastric cancer samples (Fig. 1b). In addition, elevated protein level of KDM2A was detected in gastric cancer samples using Western blot analysis (Fig. 1c). In series of gastric epithelial cells (GES-1) and gastric cancer cells (AGS, BGC803, SGC7901, and MGC823), the expression of KDM2A was dramatically upregulated in gastric cancer cells (Fig. 1d). Collectively, the above findings suggested that upregulation of KDM2A might play an important role in the progression of gastric cancer.
The expression of KDM2A was increased in gastric cancer. a Relative mRNA level of KDM2A in human gastric cancer samples and adjacent tissues. Real-time PCR was performed on 61 gastric cancer samples and 61 adjacent tissues. The KDM2A expression was normalized to that of beta-actin. Each bar was the log2 value of the ratio of KDM2A expression levels between gastric cancer tissues (T) and matched adjacent tissues (N) from the same patient. Data was calculated from triplicates. *P = 0.019. b The protein level of KDM2A in gastric cancer samples and paired adjacent tissues was examined by immunohistochemistry. c The protein level of KDM2A in gastric cancer samples and paired adjacent tissues was examined by Western blot analysis. d The protein level of KDM2A in gastric cancer cell lines
Ectopic expression of KDM2A in GES-1 and AGS cells promotes cell growth and migration
Because we observed that upregulation of KDM2A in gastric cancer was a frequent event, we postulated that overexpression of KDM2A in gastric cancer cells might exert promoting effects on the progression of gastric cancer. To confirm whether KDM2A was associated with the growth and migration of gastric cancer cells, we established stable KDM2A-expressing sublines using GES-1 and AGS cells, as GES-1 and AGS cells had low basal levels of KDM2A. The KDM2A expression vector was introduced into GES-1 and AGS cells. Successful overexpression of KDM2A was confirmed by Western lot analysis (Fig. 2a). All cells overexpressing KDM2A displayed growth advantage compared with control cells in the crystal violet assay (Fig. 2b). Also, we found that KDM2A significantly promoted the anchorage-independent growth of AGS cells on soft agar (Fig. 2c). Moreover, overexpression of KDM2A significantly enhanced the migratory abilities of GES-1 and AGS cells (Fig. 2d). In summary, our results clearly indicated that KDM2A was involved in the migration and growth of gastric cancer cells.
Overexpression of KDM2A promoted the growth, colony formation, and migration of GES-1 and AGS cells. a The GES-1 and AGS cells were stably transfected with either the pcDNA3.1 vector or the KDM2A expression vector. G418-resistant cells were pooled and confirmed the overexpression of KDM2A by Western blot analysis. b The effects of KDM2A on the growth of GES-1 and AGS cells were measured by crystal violet assay. c The effects of KDM2A on the anchorage-independent growth of AGS cells were measured by soft agar assay. Data shown was the representative results from three independent experiments. **P < 0.01 compared to the control group. d The effects of KDM2A on the migration of GES-1 and AGS cells. **P < 0.01
Knockdown of KDM2A exerts strong tumor-suppressive effects on gastric cancer cells
To determine the role of endogenous KDM2A in the progression of gastric cancer, the expression of KDM2A was knocked down in SGC7901 and GES-1 cells, which was confirmed by Western blot analysis (Fig. 3a). The tumor-suppressive function of silencing KDM2A was studied by cell growth assay, foci formation assay, and cell migration assay. Crystal violet assay demonstrated that downregulation of KDM2A inhibited the growth of SGC7901 and GES-1 cells in liquid culture (Fig. 3b). In addition, foci formation assay showed that the efficiency of foci formation was significantly inhibited in cells that knockdown the expression of KDM2A (Fig. 3c). Cell migration assay also revealed that the migration rates of SGC7901 and GES-1 cells were significantly inhibited after knockdown of the expression of KDM2A (Fig. 3d).
The knockdown expression of KDM2A inhibited the growth and migration of SGC7901 and GES-1 cells. a The knockdown expression of KDM2A in SGC7901 and GES-1 cells. b The knockdown expression of KDM2A inhibited the growth of SGC7901 and GES-1 cells measured by crystal violet assay. Data shown was the representative results from three independent experiments. The OD600nm value was measured. **P < 0.01. c The knockdown expression of KDM2A inhibited the anchorage-independent growth of SGC7901 and GES-1 cells measured by soft agar assay. Data shown was the representative results from three independent experiments. d The knockdown expression of KDM2A inhibited the migration of SGC7901 and GES-1 cells
KDM2A promotes the growth and migration of gastric cancer cells by downregulating the expression of PDCD4
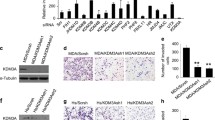
Recently, KDM2A was found to inhibit the expression of PDCD4 in the lung cancer cells in a screening using microarray [14]. The human PDCD4 gene was found to be localized on chromosome 10q24 [21]. Its encoding product, a 64-kDa protein, was found to be involved in the apoptotic machinery and suppressed cell transformation, tumorigenesis, and invasion [22–24]. PDCD4 is expressed in many adjacent tissues, such as normal mammary gland and normal human lung tissue [25–27]. Its levels were found to be markedly decreased in primary patient tumor samples from lung cancer, breast carcinoma, colon cancer, gastric cancer, and hepatocellular carcinoma [24, 26, 28–32]. In the next study, we investigated whether KDM2A promoted the growth and migration of gastric cancer cells through negatively regulating the expression of PDCD4. Forced expression of KDM2A in GES-1 and AGS cells inhibited the expression of PDCD4, while the knockdown expression of KDM2A upregulated the expression of PDCD4 (Fig. 4a, b). Moreover, KDM2A positively regulated the expression of CyclinD1 and PCNA in GES-1 and AGS cells, two molecules associated with cell proliferation (Fig. 4a, b). Although KDM2A was reported to activate ERK signaling in lung cancer, KDM2A showed marginal effects on the phosphorylation of ERK in gastric cancer cells (Fig. 4a, b), indicating KDM2A promoted the tumorigenesis of gastric cancer cells independent of ERK signaling. These observations suggested that downregulation of PDCD4 might mediate the function of KDM2A in gastric cancer cells. To further address this issue, we explored the function of PDCD4 in gastric cancer cells overexpressing KDM2A. Boyden Chamber assay and foci formation assay revealed that the elevated expression of PDCD4 partially rescued cell growth and migration (Fig. 4c, d).
KDM2A negatively regulated the expression of PDCD4. a Overexpression of KDM2A downregulated the expression of PDCD4 and upregulated the expression of PCNA, phosphorylated ERK, and CyclinD1. b The knockdown expression of KDM2A upregulated the expression of PDCD4 and downregulated the expression of PCNA, phosphorylated ERK, and CyclinD1. c Overexpression of PDCD4 attenuated the migration of GES-1 cells. d Overexpression of PDCD4 attenuated the anchorage-independent growth of AGS cells
The knockdown expression of KDM2A inhibits the metastasis of gastric cancer cells in vivo
Our in vitro studies suggested that the knockdown expression of KDM2A suppressed the migration of gastric cancer cells. Therefore, we evaluated whether the knockdown expression of KDM2A in gastric cancer cells affected the metastasis of gastric cancer cells in vivo by utilizing a tumor metastasis mouse model. Consistent with the in vitro study, bioluminescent signals appeared in mice injected with SGC7901/pcDNA3.1 cells 2 weeks later and developed progressively stronger in the following days. In contrast, much weaker bioluminescent signals and fewer metastatic lesions were detected in mice that received SGC7901/si KDM2A cells (Fig. 5a, b). Therefore, downregulation of KDM2A suppressed the metastasis of gastric cancer cells in vivo.
The knockdown expression of KDM2A inhibited the metastasis of SGC7901 cells in vivo. a Monitoring metastasis of bioluminescent SGC7901/si con and SGC7901/si KDM2A cells. Images were obtained every week after injection, respectively. b Mean photon counts of each group of mice were quantified and were displayed over time. Each point represented the mean ± SD. *P < 0.05; **P < 0.01
Discussion
In this study, we defined critical roles for KDM2A in gastric cancer pathogenesis and provided insights into the functions of this chromatin-modifying enzyme. Examination of human tumor specimens and biological functional studies revealed that KDM2A was an important driver of gastric cancer through modulating the growth, migration, and metastasis of gastric cancer cells.
Dysregulated expression of histone methyltransferases and demethylases is an emerging epigenetic mechanism underlying cancer development and metastasis [14, 33]. Recently, it showed that KDM2A was necessary for the tumorigenic and metastatic capabilities of lung cancer cells through transcriptionally repressing the expression of histone deacetylase 3 (HDAC3) and dual-specificity phosphatase 3 (DUSP3), which led to activation of ERK signaling and the upregulation of cell cycle-associated genes (e.g., CDK6) and cell invasion-related genes (e.g., NANOS1) [13, 14, 34]. Here, we reported that KDM2A repressed the expression of PDCD4 in the tumorigenesis of gastric cancer. Numerous studies have reported that PDCD4 was downregulated in gastric cancer and sensitized the gastric cancer cells to apoptosis [31, 35–38]. In addition, downregulation of PDCD4 by microRNA-21 promoted the tumorigenesis of gastric cancer cells [39–41]. Data reported here showed that PDCD4 overexpression significantly attenuated the tumorigenic and invasive abilities of KDM2A-overexpressing cells. Thus, this work supported the notion that KDM2A contributed to gastric cancer tumorigenesis and invasion at least in part by repressing the expression of PDCD4.
Although our data are suggestive, elucidation of the exact role and underlying mechanism of action for KDM2A in the development and progression of gastric cancer will require additional investigations using KDM2A knockout mice model. Also, additional studies and validation of KDM2A as a prognostic marker in malignant gastric cancer are needed. Collectively, our study highlighted the oncogenic role of KDM2A in the gastric cancer, and KDM2A might be a promising therapeutic target for gastric cancer.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30.
Lu YF, Liu ZC, Li ZH, et al. Esophageal/gastric cancer screening in high-risk populations in Henan Province, China. Asian Pac J Cancer Prev. 2013;15(3):1419–22.
Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47(12):1077–83.
Mason MJ, Fan G, Plath K, Zhou Q, Horvath S. Signed weighted gene co-expression network analysis of transcriptional regulation in murine embryonic stem cells. BMC Genomics. 2009;10:327.
Shehin SE, Stephenson RO, Greenlee WF. Transcriptional regulation of the human CYP1B1 gene. Evidence for involvement of an aryl hydrocarbon receptor response element in constitutive expression. J Biol Chem. 2000;275(10):6770–6.
Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol. 1999;199(6):471–87.
Grant S. Targeting histone demethylases in cancer therapy. Clin Cancer Res. 2009;15(23):7111–3.
Kampranis SC, Tsichlis PN. Histone demethylases and cancer. Adv Cancer Res. 2009;102:103–69.
Kim JH, Sharma A, Dhar SS, et al. UTX and MLL4 coordinately regulate transcriptional programs for cell proliferation and invasiveness in breast cancer cells. Cancer Res. 2012;74(6):1705–17.
Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2013;42(2):181–5.
Yamamoto S, Tateishi K, Kudo Y, et al. Histone demethylase KDM4C regulates sphere formation by mediating the cross talk between Wnt and Notch pathways in colonic cancer cells. Carcinogenesis. 2013;34(10):2380–8.
Suzuki T, Ozasa H, Itoh Y, et al. Identification of the KDM2/7 histone lysine demethylase subfamily inhibitor and its antiproliferative activity. J Med Chem. 2013;56(18):7222–31.
Dhar SS, Alam H, Li N, et al. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2012;289(11):7483–96.
Wagner KW, Alam H, Dhar SS, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2014;123(12):5231–46.
Tanaka Y, Okamoto K, Teye K, et al. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2013;29(9):1510–22.
Lu T, Jackson MW, Singhi AD, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A. 2009;106(38):16339–1644.
Lu T, Yang M, Huang DB, et al. Role of lysine methylation of NF-kappaB in differential gene regulation. Proc Natl Acad Sci U S A. 2012;110(33):13510–5.
Du J, Ma Y, Ma P, Wang S, Fan Z. Demethylation of epiregulin gene by histone demethylase FBXL11 and BCL6 corepressor inhibits osteo/dentinogenic differentiation. Stem Cells. 2012;31(1):126–36.
Ge R, Wang Z, Zeng Q, Xu X, Olumi AF. F-box protein 10, an NF-kappaB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 2012;18(7):1184–95.
Lu T, Jackson MW, Wang B, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2012;107(1):46–51.
Soejima H, Miyoshi O, Yoshinaga H, et al. Assignment of the programmed cell death 4 gene (PDCD4) to human chromosome band 10q24 by in situ hybridization. Cytogenet Cell Genet. 1999;87(1–2):113–4.
Wei NA, Liu SS, Leung TH, et al. Loss of programmed cell death 4 (Pdcd4) associates with the progression of ovarian cancer. Mol Cancer. 2009;8:70.
Wei ZT, Zhang X, Wang XY, et al. PDCD4 inhibits the malignant phenotype of ovarian cancer cells. Cancer Sci. 2009;100(8):1408–13.
Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283(2):1026–33.
Yoshinaga H, Matsuhashi S, Fujiyama C, Masaki Z. Novel human PDCD4 (H731) gene expressed in proliferative cells is expressed in the small duct epithelial cells of the breast as revealed by an anti-H731 antibody. Pathol Int. 1999;49(12):1067–77.
Kalinichenko SV, Kopantzev EP, Korobko EV, et al. Pdcd4 protein and mRNA level alterations do not correlate in human lung tumors. Lung Cancer. 2008;62(2):173–80.
Chen Y, Knosel T, Kristiansen G, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200(5):640–6.
Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene. 2004;23(49):8135–45.
Goke R, Barth P, Schmidt A, Samans B, Lankat-Buttgereit B. Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21 (Waf1/Cip1). Am J Physiol Cell Physiol. 2004;287(6):C1541–6.
Zhang Z, DuBois RN. Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene. 2001;20(33):4450–6.
Wang W, Zhao J, Wang H, et al. Programmed cell death 4 (PDCD4) mediates the sensitivity of gastric cancer cells to TRAIL-induced apoptosis by down-regulation of FLIP expression. Exp Cell Res. 2012;316(15):2456–64.
Zhang S, Li J, Jiang Y, Xu Y, Qin C. Programmed cell death 4 (PDCD4) suppresses metastastic potential of human hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2009;28:71.
Meng F, Sun G, Zhong M, Yu Y, Brewer MA. Inhibition of DNA methyltransferases, histone deacetylases and lysine-specific demethylase-1 suppresses the tumorigenicity of the ovarian cancer ascites cell line SKOV3. Int J Oncol. 2012;43(2):495–502.
Lu Y, Chang Q, Zhang Y, et al. Lung cancer-associated JmjC domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle. 2009;8(13):2101–9.
Tu H, Sun H, Lin Y, et al. Oxidative stress upregulates PDCD4 expression in patients with gastric cancer via miR-21. Curr Pharm Des. 2012;20(11):1917–23.
Guo PT, Yang D, Sun Z, Xu HM. PDCD4 functions as a suppressor for pT2a and pT2b stage gastric cancer. Oncol Rep. 2012;29(3):1007–12.
Wang WQ, Zhang H, Wang HB, et al. Programmed cell death 4 (PDCD4) enhances the sensitivity of gastric cancer cells to TRAIL-induced apoptosis by inhibiting the PI3K/Akt signaling pathway. Mol Diagn Ther. 2012;14(3):155–61.
Motoyama K, Inoue H, Mimori K, et al. Clinicopathological and prognostic significance of PDCD4 and microRNA-21 in human gastric cancer. Int J Oncol. 2012;36(5):1089–95.
Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388(3):539–42.
Chen Y, Liu W, Chao T, et al. MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett. 2008;272(2):197–205.
Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
Yufeng Huang and Yiqian Liu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Huang, Y., Liu, Y., Yu, L. et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumor Biol. 36, 271–278 (2015). https://doi.org/10.1007/s13277-014-2630-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2630-5