Abstract
Catalpa bungei is a deciduous tree native to China. It is characterized as fast growing, being highly adaptable, and having excellent wood qualities. To better understand potential mechanisms involved in adventitious root (AR) formation, we performed transcriptome analysis of softwood cuttings of C. bungei ‘Yu-1’ at three stages of AR formation using the Illumina sequencing method. Following de novo assembly, 62,955 unigenes were obtained, 31,646 (50.26 %) of which were annotated. A total of 11,100 differentially expressed genes (DEGs), including 10,200 unique and 900 common, were identified in four comparisons. Based on the all GO enrichment networks, 46 common and 7 unique GO categories were identified. Cytoskeleton was only significantly enriched in the activation period, while DNA metabolic process was only significantly enriched in the callus formation. Functional annotation analysis revealed that many of these genes were involved in phenylpropanoid biosynthesis, glycolysis, and plant hormone metabolism, suggesting potential contributions to AR formation. Interestingly, the number of DEGs involved in glycolysis decreased while the number of DEGs involved in phenylpropanoid biosynthesis increased following the AR formative process. Overall, our comprehensive transcriptional overview will prove useful, not only in the understanding of molecular networks that regulate AR formation in C. bungei, but also for exploring genes that may improve rooting rates of other trees.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Catalpa bungei is a deciduous tree native to China. It belongs to the Bignoniaceae family and is a member of the Catalpa Scop. genus. C. bungei mainly grows in the Huanghe River and Changjiang River regions of China. It is characterized as fast growing, having excellent wood qualities, and being highly adaptable (Shi et al. 2011). C. bungei propagation is mainly through softwood cuttings; this ensures stability of wood qualities and facilitates the spread of improved cultivars. However, this method is reliant on adventitious root (AR) growth rate. In China, ‘Yu-1’ is largely promoted for growing as it is an easy-to-root C. bungei variety. In our previous study, rooting rate was found to be 85.6 % (Ma et al. 2014). The development process of AR formation in softwood C. bungei ‘Yu-1’ cuttings consists of four key stages: activation, 0–5 days after cutting (DAC); callus formation, 5–20 DAC; root formation, 20–30 DAC; and root elongation, 30–40 DAC (Ma et al. 2014). As a higher rooting rate variety, ‘Yu-1’ provides important material for investigating the molecular mechanisms of AR formation in C. bungei. Presently, the molecular mechanism of AR formation in C. bungei ‘Yu-1’ is poorly understood.
ARs are the key developmental process for asexual propagation in most plants and can either be formed naturally on intact plants in dependence on the developmental program and environmental stimuli or develop in response to injury for example at the wounding site of cuttings (Koyuncu and Balta 2004). However, our knowledge about the mechanisms that enable a somatic differentiated cell to re-differentiate into a meristematic cell to develop into a root and that laid the metabolic basis for this process is fragmentary because of their complexity (Abarca et al. 2011; Birnbaum and Sanchez Alvarado 2008; Ludwig-Müller et al. 2005). Currently, only a limited number of molecular studies into AR formation have been performed. Wei et al. (2013) identified differentially expressed transcripts in single nodal cuttings of Camellia sinensis treated with or without indole-3-butyric acid (IBA) for 24 h by suppressive subtractive hybridization; they found that secondary metabolism, cell rescue, defense, and virulence are involved in AR formation. Thomas et al. (2003) identified two genes encoding proline-rich proteins in Vitis vinifera, and suggested these genes play an important role in the AR formation. S-Adenosylmethionine synthetase (SAMS) catalyzes synthesis of S-adenosylmethionine, a precursor for ethylene and polyamine biosynthesis. Higher expression of the SAMS gene in tetraploid black locust (Robinia pseudoacacia) promotes AR initiation (Quan et al. 2014). A proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins including auxin-related and light-related proteins which positively or negatively correlated with AR formation (Sorin et al. 2006). Presently, few genes are characterized for AR formation of C. bungei, largely due to limited genome information.
AR formation is a complex regeneration process with many internal and external factors influencing AR formation and the phytohormone auxin is crucial for AR formation in cuttings (Ahkami et al. 2009; da Costa et al. 2013). Auxin positively regulates AR formation in most plant species. Indole-3-butyric acid (IBA) is widely used in C. bungei propagation to induce rooting. Although indole-3-acetic acid (IAA) is a primarily native auxin in plants, IBA is more effective in promoting ARs (Ludwig-Müller et al. 2005). Brinker et al. (2004) reported that IBA induced expression of genes involved in cell replication and cell wall weakening, while inhibited genes related to auxin transport, photosynthesis and cell wall synthesis during the root initiation of Pinus contorta. Meanwhile, many factors affect AR formation via auxin interactions. Ethylene played a positive role in AR formation with modulation of auxin transport as a central point of ethylene–auxin crosstalk (Negi et al. 2010). It was shown that auxin and light act antagonistically on the development of ARs in Eucalyptus saligna and E. globulus (Fett-Neto et al. 2001). Niemi et al. (2005) showed that light sources with different spectra could affect AR and mycorrhyza formation in Scots pine (Pinus sylvestris) in vitro.
AR formation is a complex quantitative genetic trait regulated by both environmental and endogenous factors. Therefore, RNA-seq technology aimed at describing global changes is particular advantages. Han et al. (2014) investigated genes expression in larch hybrids (Larix spp.) during AR formation using 454 sequencing. Their results suggest the polyamine synthesis pathway and the stress response pathways are the most important signaling pathways for AR formation. To identify specific genes determining initiation and formation of AR in Petunia, a microarray-based transcriptome analysis using the stem base of Petunia hybrida cuttings was performed; this revealed active lipid metabolism in petunia cuttings, especially during the sink establishment phase (first 6 h after excision) (Ahkami et al. 2014). The global transcriptomic data reveal that profound cellular and metabolic reorganization occurs during the root induction stage in mung bean (Li et al. 2015). To better understand potential mechanisms involved in AR formation, transcriptome analysis of single nodal cuttings of C. sinensis treated with or without IBA was performed using the Illumina sequencing method and 1091 differentially expressed unigenes were identified including 656 up- and 435 down-regulated genes (Wei et al. 2014).
In this study, five transcriptomes of softwood cuttings of ARs were initially investigated using RNA-sequencing in C. bungei ‘Yu-1’. In silico bioinformatics analyses was then used to ascertain the dynamics of the genetic molecular network and to annotate transcripts with a standard annotation pipeline. This data provides a valuable platform for understanding the molecular basis of the regulation of AR development in C. bungei.
Materials and methods
Plant material
Softwood cuttings of C. bungei ‘YU-1’ were collected from the Catalpa germplasm nursery at the Institute of Botany, Jiangsu Province & Chinese Academy of Sciences, Nanjing, China. Each softwood cutting was further segmented into smaller cuttings of 1–2 leaves, with 60 % of the leaf blade removed. After excision, the bases of the cuttings were dipped for 60 s in a solution with 2 g/L IBA (Sigma, St. Louis, Missouri, USA). The cuttings of control were without IBA treatment. The cuttings were incubated on a rooting table under 80–90 % relative humidity. Samples were harvested at 0 (p0, control), 1 (p1, activation), 15 (p2, callus formation), 25 (p3, root formation) and 35 (p4, root elongation) DAC. Three independent biological replicates were included for each time point and control tissue. Basal parts (approximately 1.0 cm of the root zone) of the cuttings were taken, immediately frozen in liquid nitrogen, and stored at −80 °C for subsequent analysis.
RNA extraction and library preparation for transcriptome analysis
Total RNAs were extracted using Trizol reagent according to manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). The total RNA concentration was quantified using UV spectrophotometry, and the quality was checked by electrophoresis in a 1 % agarose gel. Equal volumes of RNA from each of the three replications of four time points (1, 15, 25, and 35 DAC) and control were pooled, respectively. Five independent paired-end libraries were subjected to RNA-Seq analysis. Paired-end libraries with approximate average insert lengths of 200 bp were synthesized using a Genomic Sample Prep Kit (Illumina, San Diego, CA, USA) according to manufacturer’s instructions. Prior to cluster generation, library concentration and size were assayed using an Agilent DNA1000 kit (Agilent, Palo Alto, CA, USA) on a 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA). Libraries were sequenced (101 bp paired-end reads) on an Illumina HiSeq 2000 instrument (Illumina, San Diego, CA, USA) by a customer sequencing service (Biomarker Technologies, Beijing, China).
De novo assembly and functional annotation
Raw reads were cleaned by removing adaptor sequences, empty reads, and low-quality sequences. Clean reads were assembled into non-redundant transcripts using Trinity, which has been developed specifically for de novo assembly of transcriptomes using short reads (Grabherr et al. 2011). Short sequences (<200 bp) were removed to improve quality. Resulting sequences were used for BLAST searches and annotation against the National Center for Biotechnology Information (NCBI) database Non-redundant protein sequences (NR) (http://www.ncbi.nlm.nih.gov/), Clusters of Orthologous Groups of proteins (COG) (http://www.ncbi.nlm.nih.gov/COG/), Kyoto Encyclopedia of Genes and Genomes Ortholog database (KEGG) (http://www.genome.jp/kegg/kaas/), and the manually annotated and curated protein sequence database (Swiss-Prot) (http://www.expasy.ch/sprot) using an E-value cut-off of 10−5. Functional annotation of Gene Ontology (GO) terms was analyzed using Blast2go software (https://www.blast2go.com/).
Identification of differentially expressed genes
Gene expression levels were estimated by RSEM for each sample (Li and Dewey 2011). Clean data were mapped back onto the assembled transcriptome. The read count for each gene was obtained from the mapping results, and normalized to reads per kilobase of exon model per million mapped reads (RPKM) (Marioni et al. 2008). Statistical comparison of RPKM values between two samples was conducted using the web tool IDEG6 (Romualdi et al. 2003). A false discovery rate (FDR) of 0.01 was used as the P value threshold in multiple tests to evaluate the significance of differences in gene expression. Genes were considered as being differentially expressed in a given library when the P value was <0.01, and a greater than two-fold change in expression across libraries was observed.
GO enrichment analysis was performed with the BiNGO plugin in Cytoscape using the hypergeometric test for statistical analysis (Maere et al. 2005; Smoot et al. 2011). For P value correction, we selected the FDR correction method. GO terms with corrected P value ≤ 0.01 were considered significantly over-represented and shown as colored nodes in the enrichment network. Significantly altered KEGG pathways were identified using a P value based on the hypergeometric distribution. All metabolic pathways with a FDR corrected P value smaller than 0.01 were reported as significantly altered compared with the control. Cluster analysis was performed with the self-organizing tree algorithm (SOTA, Multiple Array Viewer software, MeV 4.9.0; http://www.tm4.org/mev.html).
Gene expression changes were analyzed with MapMan (Thimm et al. 2004; Usadel et al. 2005). The deduced polypeptide sequences were submitted to the Mercator webserver to classify them into MapMan functional plant categories (Lohse et al. 2014). For color-coded representation (heat map) in MapMan, the log2 transformed fold-change value for each DEG was used. Deduced polypeptide sequences shorter than 30 amino acids were excluded when generating the MapMan metabolic pathway maps.
Real-time quantitative RT-PCR (qRT-PCR) analysis
Total RNA was prepared as described above. For each sample, 2 μg of total RNA was reverse transcribed into cDNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA). Reverse transcription was performed at 37 °C for 60 min in a 20 μl reaction with a final denaturation step at 95 °C for 5 min. Primers were designed using Primer premier 5 (http://www.premierbiosoft.com/primerdesign/) (Table S1) and synthesized by SIPUJIN (Nanjing, China). Reactions were carried out using SYBR Green PCR Master Mix (Roche Applied Science, Mannheim, Germany) on an ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with three replicates, the Actin gene from C. bungei was amplified as an internal control for data normalization. Reaction volumes of 25 μl contained 12.5 μl SYBR Green PCR Master Mix, 9.5 μl deionized H2O, 1 μl primers, and 2 μl cDNA. Amplification reactions were initiated with a pre-denaturing step (95 °C for 10 min); followed by 40 cycles of denaturing (95 °C for 10 s), annealing (60 °C for 35 s), and extension (72 °C for 35 s). Three replicates of the qPCR assay were performed for each gene. Data were processed using the \(2^{{ - \varDelta \varDelta {\text{C}}_{\text{t}} }}\) method (Livak and Schmittgen 2001).
Results
De novo assembly and function annotation
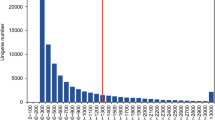
To gain a general overview of the C. bungei transcriptome, five libraries representing control (p0), activation (p1), callus formation (p2), root formation (p3) and root elongation (p4) stages were designed for RNA-seq. A total of 165,977,661 high-quality reads, comprising approximately 33.52 Gb of sequence data, were generated from the five cDNA libraries (Table 1). The distribution of reads in the five libraries was 30.16 million for p0, 32.08 million for p1, 35.25 million for p2, 33.67 million for p3, and 34.82 million for p4 (Table 1). The GC percentage in the five libraries ranged from 45.40 % (p2) to 46.48 % (p3) (Table 1). The Q30 percentage of the five libraries was more than 82 %, suggesting that the quality of the sequence data was high (Table 1). The clean sequence data has been deposited in the Short Read Archive database of the National Center for Biotechnology Information under the accession numbers SRP059272. Using the Trinity method, the high-quality reads were assembled into 6,315,145 contigs (Table 1). All contigs together produced 116,941 transcripts and 62,955 unigenes with an average length of 1162.61 and 783.84 bp, respectively (Table 2). The N50 length of the transcripts and unigenes was 1945 and 1417 bp, respectively (Table 2). The majority of unigenes, accounting for 57.89 % of sequence data, ranged from 201 to 500 bp. Of the remaining unigenes, 11,877 (18.87 %) ranged in length from 501 to 1000 bp, 8926 (14.18 %) from 1001 to 2000 bp, while 5708 (9.07 %) were longer than 2000 bp (Table 2).
Multiple databases were interrogated to assign accurate annotation information to the unigenes. 31,646 unigenes (50.27 %) were annotated using BLASTx with an E-value threshold of 10−5 (Table 3). Among the five databases used, most unigenes were annotated to NR (31,184, 49.53 %), GO (24,982, 39.68), and Swiss-Prot (20,973, 33.31 %) (Table 3). According to the E-value distribution of the top hits in the NR databases, 22.12 % of the matched sequences showed complete homology (=0), while 37.07 % and 40.82 % of the matched sequences showed strong homology (between 0 and 1.0e−50) and moderate homology (between 1.0e−5 and 1.0e−50), respectively (Table 4). The nine top-hit species based on NR annotation indicated that 21.89 % of the unigenes were annotated with sequences from Solanum lycopersicum, while 58.15 % of the unigenes were annotated with sequences from the five top-hit species, including Solanum lycopersicum, V. vinifera, Genlisea aurea, Theobroma cacao and Populus trichocarpa (Table 4).
GO annotated unigenes could be divided into cellular component (90,853), molecular function (33,709), and biological process (130,429) clusters. Among the cellular components category, cell part (20,345; 22.39 %), cell (20,063; 22.08 %) and organelle (18,005; 19.82 %) were the dominant groups (Fig. 1a). For the molecular function terms, binding (14,282; 42.37 %) and catalytic activity (12,764; 37.87 %) were the dominant groups (Fig. 1b). Most biological process genes were involved in cellular processes (19,224; 14.74 %) and metabolic processes (18,842; 14.45 %) (Fig. 1c). 11,695 unigenes were annotated to the 25 COG categories, with most unigenes classified into the cluster for ‘general function prediction only’ (2939; 18.03 %), followed by ‘translation, ribosomal structure and biogenesis’ (1438; 8.83 %), and ‘transcription’ (1414; 8.68 %) (Fig. S1). For KEGG analysis, 7466 unigenes were assigned to 117 KEGG pathways (Table S3). The largest group of unigenes identified from KEGG analysis was Ribosomes (766 unigenes, ko03010) while the smallest group was glucosinolate biosynthesis (one unigene, ko00966) (Table S3). 210 unigenes were annotated to plant hormone signal transduction (ko04075), while phenylpropanoid biosynthesis (ko00940) was annotated with 104 unigenes (Table S3).
Identification of differentially expressed genes (DEGs)
More than 30 million clean reads were obtained from each library and then aligned to the reference transcriptome separately. At each AR formation stage, we detected both unique and common sets of DEGs. Approximately 83.66 % (p0), 85.11 % (p1), 84.18 % (p2), 82.94 % (p3) and 84.55 % (p4) of the total reads mapped to the reference transcriptome assembly, 57.97 % (p2) to 66.36 % (p3) of reads mapped uniquely to the reference transcriptome assembly, and 33.64–42.03 % of reads were filtered as multiple-aligned (Table 5). A total of 11,100 DEGs, including 10,200 unique and 900 common, were identified by comparing libraries, including 1203, 1312, 2341 and 1318 unique DEGs from p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Fig. 2a).
Expression profilings of DEGs in C. bungei. a Venn diagrams representing the numbers of DEGs and the overlaps of sets obtained across four comparisons; b Numbers of up- and down-regulated DEGs among four libraries (between p1 and p0, between p2 and p0, between p3 and p0, between p4 and p0, respectively); c Overall expression profiles for DEGs expressed in the four stages of AR formation; d Hierarchical cluster heat map and cluster tree from the common DEGs. Red indicates up-regulated genes and green indicates down-regulated genes
There were 4551 DEGs identified between p1 and p0, with 2383 genes up-regulated and 2168 genes down-regulated; 5074 DEGs were identified between p2 and p0, with 2565 genes up-regulated and 2509 genes down-regulated; 5182 DEGs were identified between p3 and p0, with 3440 genes up-regulated and 1742 genes down-regulated; 4505 DEGs were identified between p4 and p0, with 2269 genes up-regulated and 2236 genes down-regulated (Fig. 2b). The ranking of up-regulated DEGs (from low to high) was p4/p0, p1/p0, p2/p0 and p3/p0 while the ranking of down-regulated DEGs was p3/p0, p1/p0, p4/p0 and p2/p0. Interestingly, p3/p0 had the most up-regulated DEGs while p2/p0 had the most down-regulated DEGs. These results suggested that root formation was associated with the accumulation of novel transcripts, and that some transcripts were more abundant immediately after callus formation.
According to the number of aligned reads, each DEG was assigned with a RPKM value, which could normalize for the total reads obtained in each individual library. Based on this analysis, the gene expression levels in four libraries were classified into five categories (rare, low, moderate, high and extremely high) (Fig. 2c; Table S3). The RPKM value of each DEG ranged from 0 to 10,541.32, with a mean of 25.61 for p1, 19.46 for p2, 29.15 for p3 and 22.87 for p4 (Table S3). The largest portion of DEGs in p2 and p3 libraries exhibited rare expression (RPKM < 3), and the smallest portion was extremely high expression. However, the largest portion of DEGs in p1 and p4 libraries exhibited rare expression (RPKM < 3), and the smallest portion was high expression (Fig. 2c; Table S3).
In addition, 900 common DEGs were detected at all four stages. Cluster analysis of the 900 common DEGs was performed to examine their expression patterns. The common DEGs were grouped into two clusters, designated cluster A-B (Fig. 3d). There were 406genes (45.11 %) in the cluster A category and 494 genes (54.89 %) in the cluster B.
GO enrichment and cluster analysis of DEGs
To explore the biological functions of AR formative genes, a total of 10,849 sequences were used as query to perform an alignment with 30,467 Arabidopsis proteins at an E value ≤1e−5. Of these, 7714 DEGs (71.10 %) were annotated. GO functional enrichment analysis for all DEGs were analyzed using BiNGO, a Cytoscape plugin assessing overrepresentation of ontologies in biological networks (Maere et al. 2005). In order to clearly depict the GO enrichment network, we selected the collapsed GO-slim plant terms as namespace (Consortium 2004). The functional enrichment of DEGs indicated that, at FDR ≤ 0.01, 57 GOs were enriched in the p1/p0, 60 GOs were enriched in the p2/p0, 61 GOs were enriched in the p3/p0, and 56 GOs were enriched in p4/p0 (Fig. 3; Table S4). Based on the all GO enrichment networks, 46 common and 7 unique GO categories were identified. Among 7 unique enrichment GO terms, four GO categories, including Golgi apparatus, lysosome, nucleolus, nucleus and protein metabolic process, were significantly enriched in root formative period. Additionally, cytoskeleton was only significantly enriched in the activation period, while DNA metabolic process was only significantly enriched in the callus formation (Fig. 3; Table S4). These results suggested that GO terms experienced a dynamic change upon hormone stimulus to occur the corresponding AR formation. Among 46 common GO terms, response to stress, secondary metabolic process, transport, biosynthetic process, catalytic activity and metabolic process were the top-10 GO terms in four AR formative stages (Fig. 3; Table S4). These results suggested that these GO terms were important for the AR formation.
Concurrently, a hierarchical clustering was used to group the matched 7714 DEGs into clusters, based on their RPKM values. We identified 11 distinct clusters in which four main clusters (K1-K4) accounted for 65.95 % of DEGs in four libraries (approx. 5087 genes; Fig. 4a). Compared to control, 1377 genes in the K1 category had highest expression levels in the p1 stage, while 353 genes in K2 had highest expression levels in the p4 stage, 1043 genes in K3 had highest expression levels in the p2 stage, and 2314 genes in the K4 category had highest expression levels in the p3 stage (Fig. 4a). All DEGs were categorized into 35 Mapman bins (Fig. 4b). Most of the Mapman bins showed enrichment for particular clusters of gene expression (Fig. 3b). Genes that encode enzymes for glycolysis, lipid metabolism, hormone metabolism (e.g., ABA, AUX, CK and JA), misc, signaling and development were greatly enriched in clusters K1 and K2, representing genes that were expressed at the lowest levels in the p3 stage. However, genes (cluster K4) encoding PS, OPP, cell wall, hormone metabolism (e.g., ET and GA) and polyamine metabolism were in a highest expression levels of p3 stage. Genes that encode enzymes for fermentation, gluconeogenesis/glyoxylate cycle and hormone metabolism (e.g., BRs) were greatly enriched in clusters K3, representing genes that were expressed at the highest levels in the p2 stage. These results indicated that DEG experienced a dynamic change upon hormone stimulus to occur the corresponding AR formation.
Cluster and functional enrichment analysis of DEGs in four libraries. a Four major clusters were identified in the four stages of AR formation. The SOTA module embedded in the MeV software was used to produce clusters. Error bars show standard deviation; b Functional category enrichment among the four major clusters based on Mapman annotation
KEGG enrichment analysis
Pathway enrichment analysis revealed that 6, 4, 4 and 8 pathways (corrected P value ≤ 0.01) were the significant difference pathways enriched in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Fig. 5). Phenylalanine metabolism and phenylpropanoid biosynthesis were the common enrichment pathways and more DEGs were up-regulated than down-regulated (Fig. 5). However, photosynthesis and photosynthesis - antenna proteins were enriched in p1/p0, p2/p0 and p4/p0, respectively and more DEGs were down-regulated than up-regulated (Fig. 5). These results indicated that phenylalanine metabolism and phenylpropanoid biosynthesis were continuously up-regulated while photosynthesis and photosynthesis - antenna proteins were down-regulated on different AR formative stages. Additionally, plant hormone signal transduction were enriched in p3/p0 and p4/p0, respectively and more DEGs were up-regulated than down-regulated (Fig. 5). These results suggested that plant hormone signal transduction might play important role in the last two stages of AR formation.
KEGG pathway network analysis showed that glycolysis/gluconeogenesis and glycosphingolipid biosynthesis - globo series were only significantly enriched in p1/p0 and more DEGs were up-regulated than down-regulated. Ribosome was only significantly enriched in p3/p0 and more DEGs were up-regulated than down-regulated. Terpenoid backbone biosynthesis and ubiquinone and other terpenoid-quinone biosynthesis were only significantly enriched in p4/p0 and more DEGs were up-regulated than down-regulated while carotenoid biosynthesis were only significantly enriched in p4/p0 and more DEGs were down-regulated than up-regulated. These results indicated that pathways experienced a dynamic change upon hormone stimulus to occur the corresponding AR formation.
Candidate functional genes related to AR formation
Glycolysis
Based on the KEGG annotations, there were 69 DEGs involved in glycolysis (Table S5). There were 43 DEGs with 30 up-regulated and 13 down-regulated, 32 DEGs with 23 up-regulated and 9 down-regulated, 22 DEGs with 18 up-regulated and 4 down-regulated, and 17 DEGs with 9 up-regulated and 8 down-regulated in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S5). It is worth noting that the number of DEGs decreased following the AR formative process. Further analysis indicated that 20 DEGs, 9 DEGs, 8 DEGs, and 5 DEGs were specifically induced in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S5). For the unique DEGs in p1/p0, 6-phosphofructokinase 3 (PFK3) was up-regualted while hexokinase-2 (HXK2) was down-regulated in the activation period (Table S5). Among 900 common DEGs, four DEGs, including phosphoenolpyruvate carboxykinase (PCKA, comp100533_c0), fructose-bisphosphate aldolase 2 (FBA2, comp107097_c0), non-cell-autonomous protein pathway1 (NCAPP1, comp45220_c0), and aldehyde dehydrogenase family 3 member F1-like (ALDH3F1, comp74070_c0), were involved in glycolysis (Table 6). Interestingly, the PCKA and NCAPP1 were simultaneously up-regulated while FBP and ALDH3F1 were simultaneously down-regulated during AR formation (Table 6).
Phenylpropanoid biosynthesis
According to the KEGG annotations, there were 54 DEGs involved in phenylpropanoid biosynthesis (Table S6). There were 28 DEGs with 13 up-regulated and 15 down-regulated, 30 DEGs with 17 up-regulated and 13 down-regulated, 32 DEGs with 23 up-regulated and 9 down-regulated, and 36 DEGs with 27 up-regulated and 9 down-regulated in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S6). It is worth noting that the number of DEGs increased following the AR formative process. Further analysis indicated that 2 DEGs, 6 DEGs, 7 DEGs, and 9 DEGs were specifically expressed in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S6). Among those unigenes involved in phenylpropanoid biosynthesis, 14 DEGs were involved in the response to wounding and lignin biosynthesis. Seven DEGs belonged to 4-coumarate–CoA ligase (4CL) gene family were detected in our transcriptome (Table S6). The four unigene homologous with Gm4CL2 (comp100601_c1, comp117634_c0, comp117634_c1 and comp117634_c2) were the common DEG, being down-regulated at all AR formative stages (Table 6). The two common DEG encoding caffeoyl-CoA O-methyltransferase (CCoAOMT, comp124594_c0 and comp110020_c1) were involved in the response to wounding and lignin biosynthesis; they were up-regulated at all AR formative stages (Table 6). Twenty three DEGs encoding peroxidases (POD) were detected in the phenylpropanoid biosynthesis category and six DEGs were common to all comparisons (Table 6). Among the six POD unigenes, four uingenes, including POD2, POD61, POD47 and poxN1, were simultaneously up-regulated while two POD72 were simultaneously down-regulated during AR formation (Table 6).
Hormone metabolism
Based on the GO annotations and KEGG annotation, there were 74 DEGs involved in the auxin (AUX) metabolism, 17 DEGs involved in the abscisic acid (ABA) metabolism, 34 DEGs involved in the ethylene (ET) metabolism, 18 DEGs involved in the cytokinin (CTK) metabolism, and 44 DEGs involved in the brassinosteroid (BR) metabolism (Table S7). The number of DEGs involved in ET metabolism and CTK metabolism was highest during AR callus formation; that involved in ABA metabolism was highest during AR root formation; that involved in AUX metabolism and BR metabolism was highest during AR root elongation (Table S7). The similar trend of unique DEGs was observed during AR formation. The number of up-regulated genes involved in AUX metabolism and BR metabolism was less than that of down-regulated during AR activation (Table S7). The number of up-regulated genes involved in CTK metabolism was less than that of down-regulated during AR callus formation (Table S7). These results indicated that the type of plant hormone expressed a dynamic change to occur the corresponding AR formation.
Among those unigenes involved in hormone metabolism, a total of 30 common unigenes with 13 up-regulated and 17 down-regulated were identified (Table 6). The crucial genes for hormone biosynthesis of ET, CTK and BRs such as 1-aminocyclopropane-1-carboxylate oxidase 1 (ACO1), ACO5, cytokinin hydroxylase (CYP735A1) and cytochrome P450 85A1 (CYP85A1) were simultaneously up-regulated during AR formation (Table 6). Besides genes involved in hormone biosynthesis, we also identified several genes responsible for hormone catabolism and conjugation, which function in hormone balance. The crucial genes for hormone conjugation including indole-3-acetic acid-amido synthetase 3.1 (GH3.1), GH3.6, auxin response factor 5 (ARF5), and ethylene-responsive transcription factor 3 (ERF3) were simultaneously up-regulated while auxin efflux carrier component 3 (PIN3), PIN8, and ERF098 were simultaneously down-regulated during AR formation (Table 6). Additionally, the crucial genes for hormone biosynthesis of AUX and ET such as indole-3-pyruvate monooxygenase YUCCA7 (YUC7), and S-adenosylmethionine synthase (SAM) were special up-regulated in the AR root formation (Table S7). The unigene CYP724B1 involved in BRs biosynthesis was special up-regulated in the AR callus formation (Table S7).
DNA metabolism
Based on the GO annotations and KEGG annotation, there were 52 DEGs involved in the DNA metabolism. There were 20 DEGs with 5 up-regulated and 15 down-regulated, 27 DEGs with 19 up-regulated and 8 down-regulated, 26 DEGs with 7 up-regulated and 19 down-regulated, and 14 DEGs with 3 up-regulated and 11 down-regulated in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S8). Further analysis indicated that 2 DEGs, 16 DEGs, 8 DEGs, and 4 DEGs were specifically induced in p1/p0, p2/p0, p3/p0 and p4/p0, respectively (Table S8). It is worth noting that the number of total DEGs/up-regulated DEGs/unique DEGs was all highest in the AR callus formation. Among those unigenes involved in DNA metabolism, 20 DEGs were involved in the cell division and cell cycle. The number of up-regulated genes in this category was highest in the callus formation in comparison to other three stages of AR formation (Table S8). The unigenes homologous with Acyl-CoA-binding domain-containing protein 5 (ACBP5) involved in plant development was simultaneously down-regulated during AR formation (Table 6).
Validation of gene expression
We randomly selected 15 differentially expressed unigenes for qRT-PCR validation of our RNA-seq data. The qRT-PCR profiles of 12 genes were completely in agreement with those obtained by using RNA-seq (Fig. 6). The qPCR analysis of the transcripts confirmed that PCKA (comp100533_c0), CCoAOMT (comp110020_c1), poxN1 (comp110105_c0), and ERF110 (comp114889_c1) were simultaneously up-regulated during AR formation; ARF6 (comp122156_c1) and s-adenosylmethionine synthase 2 (SAMS2, comp96145_c0) were simultaneously down-regulated during AR formation (Fig. 6).
Discussion
A first insight into AR formation induced by IBA in C. bungei
ARs refer to roots that form from any tissue that is not a root, such as leaves and stems. Adventitious rooting is one of the most important mechanisms of vegetative propagation in plants and one of the most important methods for the commercial production of woody plants throughout the world (Legué et al. 2014). However, lack of extensive genomic sequences and information on functional genes, especially gene expression profiles and functional assignment of genes, had hindered our understanding of the molecular processes of AR formation in C. bungei and related species. Based on the physiological and anatomical analysis of C. bungei, AR formation should be considered a complex multi-step process (Liu et al. 2008). Transcriptomic data can reveal gene expression profiles and give fundamental insights into biological processes. As a high-throughput, accurate and low-cost method, RNA-Seq, a new next-generation sequencingmethod, has been widely applied to analyze transcriptomes qualitatively and quantitatively, especially for species without available genomic information. To illuminate the network of genes involved in AR formation in C. bungei, we performed comparative transcriptome analysis of softwood cuttings at various stages of AR formation. In the present study, the application of the RNA-Seq technology generated a total of 116,941 transcriptome sequences. These sequences provide a valuable resource for AR formative specific gene discovery and for the assembly of a C. bungei reference at the genomic level. Our data, which was generated from different AR formation, may facilitate in the analysis of transcriptomic changes associated with AR development. Moreover, it may help in the genetic manipulation of varieties to control the AR development of C. bungei. This RNA-seq based transcriptome analysis provides an initial view of the C. bungei AR development process, assisting in the identification of candidate genes related to glycolysis, phenylpropanoid biosynthesis, hormone metabolism and DNA metabolism, thus paving the way for further elucidation of the molecular mechanism underlying AR formation.
The relationship of glycolysis and AR formation
Glycolysis is the metabolic pathway that converts glucose into pyruvate and adenosine triphosphate (ATP). It involves in various developmental events such as seed germination, flowering, and AR formation. Additionally, sucrose, glucose and fructose converted to each other and normally metabolized through glycolysis (Wang and Weathers 2007). Sucrose affects the formation of sink organs and is a notable inducer of AR formation in P. hybrida (Ahkami et al. 2013). In present study, glycolysis was only significantly enriched in activation (e.g., the first stage of AR formation of C. bungei) and more DEGs were up-regulated than down-regulated based on KEGG pathway network analysis. Further analysis indicated that the number of DEGs decreased following the AR formative process in C. bungei (Table S5). This observation is in agreement with biochemical results regarding the enzymes involved in glycolysis (Ahkami et al. 2014). The activation stage is the first step of the AR formation. In the stage, the cells were stimulated and become competent to respond to signal factors. Therefore, energy was necessary for the activation stage. Glycolysis generated the energy and carbon skeletons to be used for synthesis of amino acids, proteins and nucleic acids. It might the reason that the glycolysis was only significantly enriched in activation stage.
In this study, 69 unigenes involved in glycolysis were specifically induced during AR formation. Among those DEGs, four common DEGs, including PCKA, FBA2, NCAPP1 and ALDH3F1, were detected. Interestingly, the PCKA and NCAPP1 were simultaneously up-regulated while FBA2 and ALDH3F1 were simultaneously down-regulated during AR formation (Table 6). FBA is a widely distributed enzyme. It is a key constituent of the glycolytic pathway and plays vital roles in carbohydrate metabolism and in the production of triose phosphates and derivatives in signal transduction (Schaeffer et al. 1997). Using a two dimensional polyacrylamide approach, Konishi et al. (2005) revealed that FBA increases root development in rice. In contrast, FBA2 was simultaneously down-regulated during AR formation in our transcriptome. It was indicated that FBA2 was an important negative regulation gene for AR formation in C. bungei. PFK catalyzes the phosphorylation of fructose 6-phosphate to fructose 1,6-bisphosphate. This irreversible reaction is considered to be one of the rate-limiting steps of glycolysis (Hofmann 1976). In present study, PFK3 was specially up-regualted in the activation period (Table S5). Similar results were observed in P. hybrida (Ahkami et al. 2014).
Hormones, hormone-related genes, and AR formation
Plant hormone signaling pathways are important in the control of plant development (Peat et al. 2012). Among plant hormones, auxin is a major factor in the regulation of callus formation and root organogenesis (Lavenus et al. 2013; Legué et al. 2014). Auxin interacts with many plant hormones to mediate developmental processes, such as cell division, elongation and differentiation (Muday et al. 2012). Interestingly, genes associated with auxin homeostasis were the most seriously affected by IBA treatment. However, most induced genes were correlated with decreased free auxin. GH3 can join free auxin to different amino acids and overexpression of GH3 in plants produces severe auxin-deficient phenotypes (Zhang et al. 2009). Here, GH3.1 and GH3.6 were simultaneously up-regulated during AR formation, indicating mechanisms associated with auxin storage were important negative regulation genes for AR formation in C. bungei. Auxin transportation is accomplished by auxin influx and efflux carrier proteins. Numerous auxin influx and efflux carriers have been isolated and characterized (Bennett et al. 1996; Gälweiler et al. 1998; Muday and DeLong 2001; Friml et al. 2002a, b). AtPIN3 is redirected towards one side of columella cells and determines the direction of auxin flux, leading to asymmetric auxin accumulation and differential growth (Friml et al. 2002a, b). The triple mutant pin1 pin3 pin4 in Arabidopsis displays defects in primary root development (Weijers and Jürgens 2005). In our transcriptome, the two common DEGs homologous with AtPIN3 and AtPIN8, respectively, were simultaneously down-regulated in the all AR formative stages. Additionally, the crucial genes for hormone biosynthesis of AUX such as indole-3-pyruvate monooxygenase YUCCA7 (YUC7) were special up-regulated in the AR root formation. These data suggest that not all auxin-induced genes act as positive regulators of AR formation, especially for those involved in auxin transport.
Auxin-BR interactions also are important to AR formation as these hormones modulate both cell expansion and proliferation and their transcriptional responses largely overlap (Nemhauser et al. 2004). Moreover, BR enhances classical auxin growth responses such as lateral root number (Bao et al. 2004) and gravitropic response (Li et al. 2005). In this study, the number of DEGs involved in AUX metabolism and BR metabolism was highest during AR root elongation (Table S7). These results provided the evidence for a functional interaction of BRs with auxin to promote the AR formation. However, the mechanism for this interaction was not clear. Additionally, BR biosynthetic genes, such as CYP72A1, SQMO and CYP85A1 were identified in present study. It might be helpful to research the interaction of BRs with auxin in the future.
Cytokinin and ethylene (ET) were important for AR formation. In this study, the number of DEGs involved in ET metabolism and CTK metabolism was highest during AR callus formation. Moubayidin et al. (2009) reported that organ differentiation largely depends on the cytokinin: auxin ratio. High cytokinin content facilitates shoot formation and high auxin content promotes root formation. In this study, exogenous application of IBA greatly inhibited genes involved in cytokinin biosynthesis, such as cytokinin hydroxylases, but induced genes associated with cytokinin degradation, such as cytokinin oxidases. ET is synthesized as a stress response to wounding and fungus in most plants, and promotes AR development in cut tissues (Kuroha et al. 2002). Ethylene biosynthesis begins at S-adenosylmethionine (SAM) and includes two key steps (Yang and Hoffman 1984). ACS catalyzes the cyclization of SAM to 1-aminocyclopropane-1-carboxylic acid, which is often considered the rate-limiting reaction. The final step of ET biosynthesis, oxygen-dependent conversion of 1-aminocyclopropane-1-carboxylic acid to ET, is catalyzed by ACO. In this study, the expression levels of ethylene biosynthetic genes and ERFs were not coincident. ACO1 and ACO5 were up-regulated while ACS7 and SAMS2 were down-regulated in this study. ERFs control cell dedifferentiation, metabolite biosynthesis, and trait development (Song et al. 2013). ERF3 and ERF110 were up-regulated while ERF098 was down-regulated in our transcriptome. The findings of the present study may facilitate in better understanding the role of cytokinin and ethylene in AR formation.
Abscisic acid regulates plant growth and development such as germination, lateral root development, seedling growth, seed development, seed dormancy, transition from vegetative to reproductive phase and abiotic stress tolerance (Chinnusamy et al. 2008). In the presence of exogenous ABA, lateral root development is inhibited. This occurs at a specific developmental stage, i.e., immediately after the emergence of the lateral root primordium from the parent root and prior to the activation of the lateral root meristem, and is reversible. In this study, the number of DEGs involved in ABA metabolism was highest during AR root formation. In present study, the expression patterns of most unigenes encoding abscisic acid receptor and abscisic acid 8’-hydroxylase were positive in the AR formative stage. The lower levels of ABA might promote the AR formation in the AR formative stage.
Therefore, a complex, dynamic, and possibly interconnected signaling network is involved in AR formation in C. bungei. Our results may help determine the role of hormones in AR formation at the molecular level.
Phenylpropanoid biosynthesis genes associated with AR formation
Phenylpropanoid biosynthesis is one of the most important secondary metabolism pathways in plants, and the most prominent product of this pathway is probably lignin (Hu et al. 2010). Cho et al. (2011) found that IBA-treated Cinnamomum kanehirae cuttings had lower amounts of lignin and exhibited greater induction of ARs. In this study, the number of DEGs increased following the AR formative process. These results indicated that the lignin content was associated with AR formation.
4-coumarate–CoA ligase and caffeoyl-CoA O-methyltransferase are two important enzymes in the lignin biosynthesis (Schmitt et al. 1991). 4CL expression is developmentally regulated and is activated by externa1 stimuli such as pathogen infection and wounding, methyl jasmonate treatment, and UV light irradiation (Chen et al. 2000; Peat et al. 2012; Lavenus et al. 2013). CCoAMT expression is activated in the disease resistance response and the wounding response (Schmitt et al. 1991; Chen et al. 2000). In this study, two CCoAMT unigenes are up-regulated while four 4CL unigenes are down-regulated at all of the different AR formative stages. Peroxidases are heme-containing enzymes that catalyze oxidation of a diverse group of organic compounds (Dawson 1988). Peroxidases are involved in various metabolic steps, such as the oxidation of cinnamyl alcohol prior to polymerization during the lignin formation process (Imberty et al. 1985). One isoform of peroxidase is reported as a predictive marker of AR formation in Betula pendula (Mcdonald and Wynne 2003). In our transcriptome, six common DEGs encoding peroxidases, four unigenes up-regulated and two unigenes down-regulated, were detected across all comparisons. These findings suggest that these genes might regulate the lignification of softwood cuttings to promote AR formation in C. bungei.
DNA metabolism genes associated with AR formation
The DNA metabolism module can be divided into replication and regulation functions. In present study, the number of total DEGs/up-regulated DEGs/unique DEGs was all highest in the AR callus formation. Among those unigenes involved in DNA metabolism, 20 DEGs were involved in the cell division and cell cycle. These results indicate an increase in the induction of DNA metabolism, cell cycle and cell division genes starting 15 days after excision. Acyl-CoA-binding proteins (ACBPs) are implicated in acyl-CoA trafficking in many eukaryotes and some prokaryotes (Xiao and Chye 2009). ACBPs can influence plant development including early embryogenesis, as well as plant stress responses including pathogen resistance (Xiao and Chye 2010). In this study, ACBP5 was simultaneously down-regulated during AR formation. The findings of the present study may facilitate in better understanding the role of DNA metabolism in AR formation.
Conclusions
Our data provides an overall view of dynamic transcriptomic changes occurring at different AR formative stages in C. bungei ‘YU-1’ variety. Based on the data presented in the present study, we propose a preliminary overview of the important biological processes occurring during AR formation (Fig. S2). In favorable conditions, including environment factors such as content of IBA, temperature, humidity, and nutrient status, the plants detect these signals and initiate various developmental processes such as AR formation, which involves hormone regulation (including hormone biosynthesis, conjugation, transport, and degradation), lignin biosynthesis, carbohydrate metabolism, and up-regulation of a series of AR formation-related genes. Crosstalks between CTK and ET promote the callus formation and crosstalks between auxin and BRs promote the AR elongation. The lower levels of ABA might promote the AR formation. Once the carbohydrate supply in the whole plant is sufficient, AR formation was induced under hormone stimulated. AR formation is accompanied by the up-regulation of genes in the lignin biosynthesis pathway. The degree of lignification in stem might be a limiting factor of AR formation. Although this analysis cannot completely account for AR formation in C. bungei ‘YU-1’, it serves as a powerful tool to further explore candidate pathways and genes associated with this complex process. We expect our comprehensive transcriptional overview to prove useful in both furthering the understanding of molecular networks that regulate AR formation, and in the exploration of genes that may improve rooting rates of other trees.
References
Abarca D, Pizarro A, Del Amo A, Diaz-Sala D (2011) Screening of genes associated with early stages of adventitious root formation from progenitor adult cells of pine. BMC Proc 5:133
Ahkami AH, Lischewski S, Haensch KT, Porfirova S, Hofmann J, Rolletschek H, Melzer M, Franken P, Hause B, Druege U et al (2009) Molecular physiology of adventitious root formation in Petunia hybrida cuttings: involvement of wound response and primary metabolism. New Phytol 181:613–625
Ahkami AH, Melzer M, Ghaffari MR, Pollmann S, Javid MG, Shahinnia F, Hajirezaei MR, Druege U (2013) Distribution of indole-3-acetic acid in Petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta 238:499–517
Ahkami A, Scholz U, Steuernagel B, Strickert M, Haensch KT, Druege U, Reinhardt D, Nouri E, Wirén NV, Franken F et al (2014) Comprehensive transcriptome analysis unravels the existence of crucial genes regulating primary metabolism during adventitious root formation in Petunia hybrida. PLoS One 9:e100997
Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134:1624–1631
Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA (1996) Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273:948–950
Birnbaum KD, Sanchez Alvarado A (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132:697–710
Brinker M, van Zyl L, Liu W, Craig D, Sederoff RR, Clapham DH, von Arnold S (2004) Microarray analyses of gene expression during adventitious root development in Pinus contorta. Plant Physiol 135:1526–1539
Chen C, Meyermans H, Burggraeve B, De Rycke RM, Inoue K, De Vleesschauwer V, Steenackers M, Van Montagu MC, Engler GJ, Boerjan WA (2000) Cell-specific and conditional expression of caffeoyl-coenzyme A-3-O-methyltransferase in poplar. Plant Physiol 123:853–868
Chinnusamy V, Gong Z, Zhu JK (2008) Abscisic acid-mediated epigenetic processes in plant development and stress responses. J Integr Plant Biol 50:1187–1195
Cho Y, Chang CY, Huang LC, Tsai JB, Liu ZH (2011) Indole-3-butyric acid suppresses the activity of peroxidase while inducing adventitious roots in Cinnamomum kanehirae. Bot Stud 52:153–160
Consortium GO (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32:D258–D261
da Costa CT, de Almeida MR, Ruedell CM, Schwambach J, Maraschin FS, Fett-Neto AG (2013) When stress and development go hand in hand: main hormonal controls of adventitious rooting in cuttings. Front Plant Sci 4:133
Dawson JH (1988) Probing structure-function relations in heme-containing oxygenases and peroxidases. Science 240:433–439
Fett-Neto AG, Fett JP, Goulart LW, Pasquali G, Termignoni RR, Ferreira AG (2001) Distinct effects of auxin and light on adventitious root development in Eucalyptus saligna and Eucalyptus globulus. Tree Physiol 21:457–464
Friml J, Benková E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jürgens G et al (2002a) AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108:661–673
Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K (2002b) Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 415:806–809
Gälweiler L, Guan C, Müller A, Wisman E, Mendgen K, Yephremov A, Palme K (1998) Regulation of polar auxin transport by AtPIN1 in Arabidopsis vascular tissue. Science 282:2226–2230
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Han H, Sun X, Xie Y, Feng J, Zhang S (2014) Transcriptome and proteome profiling of adventitious root development in hybrid larch (Larix kaempferi × Larix olgensis). BMC Plant Biol 14:305
Hofmann E (1976) The significance of phosphofructokinase to the regulation of carbohydrate metabolism. Rev Physiol Biochem Pharmacol 75:1–68
Hu Y, Gai Y, Yin L, Wang X, Feng C, Feng L, Li D, Jiang XN, Wang DC (2010) Crystal structures of a Populus tomentosa 4-Coumarate: CoA Ligase shed light on its enzymatic mechanisms. Plant Cell 22:3093–3104
Imberty A, Goldberg R, Catesson AM (1985) Isolation and characterization of Populus isoperoxidases involved in the last step of lignin formation. Planta 164:221–226
Konishi H, Kitano H, Komatsu S (2005) Identification of rice root proteins regulated by gibberellin using proteome analysis. Plant Cell Environ 28:328–339
Koyuncu F, Balta F (2004) Adventitious root formation in leaf-bud cuttings of tea (Camellia sinensis L.). Pak J Bot 36:763–768
Kuroha T, Kato H, Asami T, Yoshida S, Kamada H, Satoh S (2002) A trans-zeatin riboside in root xylem sap negatively regulates adventitious root formation on cucumber hypocotyls. J Exp Bot 53:2193–2200
Lavenus J, Goh T, Roberts I, Guyomarc’h S, Lucas M, De Smet I, Fukaki H, Beeckman T, Bennett M, Laplaze L (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci 18:450–458
Legué V, Rigal A, Bhalerao RP (2014) Adventitious root formation in tree species: involvement of transcription factors. Physiol Plant 151:192–198
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Li L, Xu J, Xu ZH, Xue HW (2005) Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17:2738–2753
Li SW, Shi RF, Leng Y (2015) De novo characterization of the mung bean transcriptome and transcriptomic analysis of adventitious rooting in seedlings using RNA-Seq. PLoS One 10:e0132969
Liu JL, Wei H, Su JL (2008) The Catalpa Bungei” Yu Qiu No. 1” cuttage seedling vegetative organ dissection structure with inserts the ear to injury the research which the organization forms. Henan Sci 7:012
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Lohse M, Nagel A, Herter T, May P, Schroda M, Zrenner R, Tohge T, Fernie AR, Stitt M, Usadel B (2014) Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ 37:1250–1258
Ludwig-Müller J, Vertocnik A, Town CD (2005) Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J Exp Bot 56:2095–2105
Ma LL, Wang P, Zhang ZY, Li LF, Yang RT, Li Y (2014) Evaluation on rooting ability of softwood cuttings on catalpa. North Hortic 15:72–77
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 21:3448–3449
Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y (2008) RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18:1509–1517
Mcdonald MS, Wynne J (2003) Adventitious root formation in woody tissue: peroxidase—a predictive marker of root induction in Betula pendula. In Vitro Cell Dev Plant 39:234–235
Moubayidin L, Mambro RD, Sabatini S (2009) Cytokinin–auxin crosstalk. Trends Plant Sci 14:557–562
Muday GK, DeLong A (2001) Polar auxin transport: controlling where and how much. Trends Plant Sci 6:535–542
Muday GK, Rahman A, Binder BM (2012) Auxin and ethylene: collaborators or competitors? Trends Plant Sci 17:181–195
Negi S, Sukumar P, Liu X, Cohen JD, Muday GK (2010) Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J 61:3–15
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2:e258
Niemi K, Julkunen-Tiitto R, Tegelberg R, Haggman H (2005) Light sources with different spectra affect root and mycorrhiza formation in Scots pine in vitro. Tree Physiol 25:123–128
Peat TS, Böttcher C, Newman J, Lucent D, Cowieson N, Davies C (2012) Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 24:4525–4538
Quan J, Zhang S, Zhang C, Meng S, Zhao Z, Xu X (2014) Molecular cloning, characterization and expression analysis of the SAMS gene during adventitious root development in IBA-induced tetraploid black locust. PLoS One 9:e108709
Romualdi C, Bortoluzzi S, D’Alessi F, Danieli GA (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics 12:159–162
Schaeffer GW, Sharpe FT, Sicher RC (1997) Fructose 1, 6-bisphosphate aldolase activity in leaves of a rice mutant selected for enhanced lysine. Phytochemistry 46:1335–1338
Schmitt D, Pakusch AE, Matern U (1991) Molecular cloning, induction and taxonomic distribution of caffeoyl-CoA 3-O-methyltransferase, an enzyme involved in disease resistance. J Biol Chem 266:17416–17423
Shi X, Li Y, Yang RT, Wang Q, Yao G (2011) ISSR analysis of genetic diversity of Catalpa bungei CA Mey germplasm resources in China. Jiangsu J Agric Sci 3:032
Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27:431–432
Song XM, Li Y, Hou XL (2013) Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 14:573
Sorin C, Negroni L, Balliau T, Corti H, Jacquemot MP, Davanture M, Sandberg G, Zivy M, Bellini C (2006) Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol 140:349–364
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939
Thomas P, Lee MM, Schiefelbein J (2003) Molecular identification of proline-rich protein genes induced during root formation in grape (Vitis vinifera L.) stem cuttings. Plant Cell Environ 26:1497–1504
Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques MC, Steinhauser D et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138:1195–1204
Wang Y, Weathers PJ (2007) Sugars proportionately affect artemisinin production. Plant Cell Rep 26:1073–1081
Wei K, Wang L, Cheng H, Zhang C, Ma C, Zhang L, Gong W, Wu L (2013) Identification of genes involved in indole-3-butyric acid-induced adventitious root formation in nodal cuttings of Camellia sinensis (L.) by suppression subtractive hybridization. Gene 514:91–98
Wei K, Wang LY, Wu LY, Zhang CC, Li HL, Tian LQ, Cao HL, Cheng H (2014) Transcriptome analysis of indole-3-butyric acid-induced adventitious root formation in nodal cuttings of Camellia sinensis (L.). PLoS One 9:e107201
Weijers D, Jürgens G (2005) Auxin and embryo axis formation: the ends in sight? Curr Opin Plant Biol 8:32–37
Xiao S, Chye ML (2009) An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiol Biochem 47:479–484
Xiao S, Chye ML (2010) New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Prog Lipid Res 50:141–151
Yang SF, Hoffman NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol 35:155–189
Zhang SW, Li CH, Cao J, Zhang YC, Zhang SQ, Xia YF, Sun DY, Sun Y (2009) Altered architecture and enhanced drought tolerance in rice via the down-regulation of indole-3-acetic acid by TLD1/OsGH3.13 activation. Plant Physiol 151:1889–1901
Acknowledgments
This work was supported by Public service platform of science and technology of Jiangsu province (BM2012058) and National natural science foundation of China (31200509).
Author contributions
The study was conceived by YL and PW. The plant material preparation was carried out by SW and LM. Bioinformatics analysis was finished by LL, TY, YM and QW. Manuscript was written by PW. All authors had read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
COG function classification of C. bungei transcriptome (TIFF 1145 kb)
Figure S2
Overview for the biological processes during AR formation in C. bungei (TIFF 123 kb)
Table S4
GO enrichment analysis of DEGs in four comparisons (XLS 80 kb)
Table S5
The detailed information of DEGs involved in glycolysis (XLS 123 kb)
Table S6
The detailed information of DEGs involved in phenylpropanoid biosynthesis (XLS 111 kb)
Table S7
The detailed information of DEGs involved in hormone metabolism (XLS 322 kb)
Table S8
The detailed information of DEGs involved in DNA metabolism (XLS 113 kb)
Rights and permissions
About this article
Cite this article
Wang, P., Ma, L., Li, Y. et al. Transcriptome profiling of indole-3-butyric acid-induced adventitious root formation in softwood cuttings of the Catalpa bungei variety ‘YU-1’ at different developmental stages. Genes Genom 38, 145–162 (2016). https://doi.org/10.1007/s13258-015-0352-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-015-0352-8