Abstract
The evolutionary history of TUBBY-like proteins (TLPs), which contain a highly conserved tubby domain, can be traced to the early stages of eukaryote evolution, on account of the identification of this gene family in organisms from single-celled to multicellular eukaryotes. While genome-wide structural and evolutionary analyses of the entire TLP gene family have recently been reported in Arabidopsis and rice, little is known about TLP genes in maize. To gain insight into how TLP genes have evolved in maize, we conducted comprehensive analysis of the molecular evolution of TLP genes in this crop. A total of 15 TLP genes (ZmTLP1–15) were identified in maize by genome-wide screening. This family was classified into four subfamilies based on phylogenetic relationships, protein domains, and motif organization. Gene duplication and chromosomal location analysis indicated that segmental duplication has played a major role in the expansion of the maize TLP family. The ZmTLP genes exhibited differential expression profiles under ABA, NaCl, 42, 4 °C, and PEG stress treatment. We performed microsynteny analysis across three gramineous species based on comparisons of the specific regions containing TLP genes, revealing numerous microsyntenic gene pairs among maize, rice, and sorghum, which suggests that the flanking regions of TLP genes may be derived from a common ancient Gramineae ancestor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
TUBBY-like proteins (TLPs) are present in all eukaryotes, from single-celled to multicellular organisms (Liu 2008). The typical TLP has an approximately 270-amino acid tubby domain at its C-terminus (Yang et al. 2008), and most TLPs in plants also contain highly conserved F-box domains (Gagne et al. 2002). The C-termini of F-box proteins generally contain one or several highly variable protein–protein interaction domains, such as the Leu-rich repeat (LRR), kelch repeat, tetratricopeptide repeat (TPR), and WD40 repeat (Jain et al. 2007). The TLP family shares a common, characteristic tertiary structure that consists of a beta barrel packed around an alpha helix in the central pore.
In mammals, TLP genes play important roles in the maintenance and functioning of neuronal cells during post-differentiation and development (Akihiro et al. 2002). Three members of this gene family (TULP1, TULP2, and TULP3) have also been identified in humans and mice (North et al. 1997). The first TUBBY gene was identified in obese mice through positional cloning (Kleyn et al. 1996; Noben-Trauth et al. 1996). The gene derives its name from its role in metabolism: mice with a mutated tubby gene develop delayed-onset obesity (Coleman and Eicher 1990), sensorineural hearing loss, and retinal degeneration (Ohlemiller et al. 1995). Tubby proteins bind to the small cell signaling molecule phosphatidylinositol, which is typically localized to the cell membrane. A similar structural fold to that of TLPs has been identified in the Scramblase family of proteins (Bateman et al. 2009). To date, members of this family have been identified in various multicellular organisms but not in single-celled organisms (North et al. 1997). The tubby gene, which is highly expressed in the paraventricular nucleus of the hypothalamus and several other brain regions, was identified by isolating the genetic locus that transmits this autosomal recessive obesity syndrome (Kleyn et al. 1996; Noben-Trauth et al. 1996). The presence of a highly conserved tubby domain in different species suggests that these proteins play fundamental biological roles in multicellular organisms.
TLPs are thought to function as transcription factors (Boggon et al. 1999), but the detailed molecular mechanism underlying their activity remains unclear. Members of the TLP family serve as potential signaling factors coupled to G-protein activity (Santagata et al. 2001). TLPs are also thought to participate in the insulin receptor signaling pathway (Kapeller et al. 1999). Insulin receptors, which belong to a large family of transmembrane protein-tyrosine kinases (Ullrich and Schlessinger 1990), are widely distributed in the central nervous system (Baskin et al. 1988). The binding of insulin to its cell surface receptor initiates phosphorylation of downstream targets, including Insulin receptor substrate 1 (IRS-1), a member of a large family of adaptor proteins that link upstream kinases to downstream signaling pathways (Mf. 1998). Therefore, TLPs may function as adaptors, linking insulin receptors to downstream signaling protein cascades (Kapeller et al. 1999; Ullrich and Schlessinger 1990).
Arabidopsis thaliana contains 11 members of the TLP gene family, i.e., AtTLP1–11. Although seven of these genes are located on chromosome 1, no local tandem repeats or gene clusters have been identified. Furthermore, accumulating evidence suggests that AtTLP9 participates in the ABA signaling pathway (Lai et al. 2004). AtTLP9 is an F-box protein that interacts with ASK1 (Arabidopsis Skp1-like 1). Transgenic plants with suppressed expression or overexpression of AtTLP9 exhibit aberrant sensitivity to ABA during seed germination and early seedling development (Lai et al. 2004).
Maize is an important cereal crop that has become a model plant for investigating genetics, evolution, and basic biological processes. Although TLP genes have been extensively characterized in Arabidopsis, obese mice, human, and other species, their functions remain poorly characterized in maize. Recently, The Maize Genome Sequence Project released the full maize genome assembly (Zea mays L. B73) (Sekhon et al. 2013), which provides the opportunity to perform genome-wide analysis of the TLP gene family to elucidate the evolutionary history and functional mechanisms of these genes in this important species. In this study, we identified and characterized 15 putative TLP genes in the maize genome. The results of this study provide a foundation for further elucidating the functional and evolutionary history of the TLP gene family in maize.
Materials and methods
Identification of non-redundant TLPs in maize
Maize (Zea mays L. B73) genome sequences were downloaded from http://www.maizesequence.org/index.html, and DNATOOLS software was used to construct a local database from the nucleotide and protein sequences of the latest complete maize genome. The Hidden Markov Model profile of the TLP domain (PF01167) was downloaded from the Pfam database (http://pfam.sanger.ac.uk/Software/Pfam) (Punta et al. 2012), which was used as a standard sequence to isolate all possible homologs in maize via BLASTp searches (P value = 0.001). This step was crucial for identifying as many similar sequences as possible. Furthermore, all candidate sequences that met the standards were analyzed in the Pfam database using the SMART program (Letunic et al. 2012) to eliminate any sequences not containing the TLP domain. Information regarding the number of amino acids, the chromosome location, the number of exons and introns, and ORF lengths of ZmTLP genes was obtained from the B73 maize sequence database. The molecular weight (kDa) and isoelectric point (PI) of each gene were calculated by ExPASy (http://www.expasy.org/tools/). All candidate TLP sequences were aligned using MEGA6.0 (Tamura et al. 2013) and checked manually to exclude potentially redundant genes, and all of the non-redundant TLP genes were subjected to further analysis.
Mapping ZmTLPs on chromosomes and gene duplication
To determine the physical locations of the ZmTLP genes, the starting positions of all ZmTLP genes identified from the B73 maize sequence database were determined. A diagram of the chromosome locations of ZmTLP genes was generated by MapInspect (http://www.plantbreeding.wur.nl/uk/software_mapinspect.html) according to their starting positions on the maize chromosomes. ZmTLP gene duplication events were also investigated. ClustalW in MEGA6.0 was used to align TLP amino acid sequences (Thompson et al. 1994) and to compute their evolutionary distances.
Phylogenetic analysis
Analyses of the phylogenetic relationships of ZmTLPs were conducted using the neighbor-joining (NJ) method in MEGA6.0. A phylogenetic tree was initially constructed using the complete TLP sequences of maize with default parameters. Bootstrap analysis was performed using 1000 replicates with the pairwise deletion option.
To compare the phylogenetic relationships of TLP genes from different species, a phylogenetic tree of maize, sorghum, and rice sequences was constructed, and the sequences of the TLP domain-containing proteins were aligned using ClustalX 1.83 (Thompson et al. 1997). Phylogenetic analysis using the NJ method in MEGA6.0, and the Max parsimony method was also used, with 1000 bootstrap replicates, to create a phylogenetic tree and to validate the results from the NJ method.
Sequence analysis of ZmTLPs
The conserved motifs encoded by each ZmTLP gene were also investigated. Protein sequences were analyzed using the online MEME (Multiple Expectation Maximization for Motif Elicitation) tool (http://meme.sdsc.edu/meme4_3_0/intro.html) (Bailey et al. 2006) with the following parameters: (1) optimum motif width was set to ≥6 and ≤50; (2) the maximum number of motifs was set to identify 10 motifs. The SMART program (http://smart.embl-heidelberg.de) and the Pfam database were used to annotate the MEME motifs. Exon–intron structural analysis of the ZmTLP genes was performed using GSDS (http://gsds.cbi.pku.edu.cn/) (Guo et al. 2007).
Expression profile of ZmTLP genes
Since ZmTLPs are thought to function as transcription factors, all ZmTLP genes were investigated at the transcriptional level. The ZmTLPs expression profiles were analyzed by searching the maize EST database (http://www.maizesequence.org/blast) and examining online expression information. Furthermore, publicly available transcriptome data were obtained from R.S. Sekhon (Sekhon et al. 2011). The maize expression data were obtained through BLAST searches against the maize EST database using the DNATOOLS BLAST program. The search parameters were as follows: maximum identity >95 %, length >200 bp, and E value <10−10. While the specific expression of ZmTLP genes was not identified in the local EST database, this information was obtained through the NCBI EST database. Finally, the consolidated data were used to construct a heatmap using R/Bioconductor (http://www.bioconductor.org/).
Ka/Ks analysis of duplication segments
Ka/Ks ratios were used to analyze the “age” of the duplicated gene pairs within each homologous segments by determining the synonymous (Ks)/nonsynonymous (Ka) substitution ratios using DnaSP (version 5.10) (Librado and Rozas 2009). A sliding window was performed to analyze the Ka/Ks ratios with the following parameters: window size, 150 bp; step size, 9 bp. To date the timing of duplication events, the Ks values of flanking conserved genes for each pair of duplicated regions were calculated. Based on a rate of 6.1 × 10−9 substitutions per site per year in millions of years, the Ks values were then translated into divergence time. The divergence time (T) was calculated as T = Ks/(2 × 6.1 × 10−9) × 10−6 Mya (Lynch and Conery 2000).
Interspecies microsynteny analysis
Microsynteny analysis across the three Gramineae species was performed based on comparisons of the specific regions containing TLP genes. Similarly, the TLP genes of maize, sorghum, and rice were regarded as the anchor points according to their physical locations. Levels of identity and similarity between the flanking genes of each TLP gene in one species and those in the other species were determined with the BLASTp program for pairwise comparison (Deleu et al. 2007). A syntenic block was defined as a region where three or more conserved homologs (BLASTp E value < 10−20) were located within a 100 kb region between genomes (Sato et al. 2008).
Plant material and stress treatments
The maize inbred line B73 was grown in a greenhouse under a 14 h light/10 h dark photoperiod at 28 ± 2 °C. When seedlings reached the three-leaf stage, they were subjected to abiotic stress treatments. Similar seedlings were placed in a temperature-controlled environment and subjected to temperature treatments including heat stress (42 ± 1 °C) and cold stress (4 ± 1 °C). For sodium chloride (NaCl) and polyethylene glycol (PEG) treatments, seedling leaves were sprayed with 20 mM NaCl and 20 % PEG solution, respectively. After each treatment, the leaves were harvested at 1 h, immediately frozen in liquid nitrogen, and stored at −80 °C for RNA extraction.
RNA extraction and qRT-PCR analysis
Total RNA was extracted from the samples using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. The quality of total RNA was assessed on a 1 % agarose gel quantified with a NanoDrop ND-1000 spectrophotometer. Reverse transcription of the open reading frame of the RNA was performed using M-MLV reverse transcriptase. Then, qRT-PCR was conducted on an ABI 7300 Real-Time system (Applied Biosystems). Primer Express 3.0 software (Applied Biosystems) was used to design gene-specific primers to amplify 90–150 bp unique PCR products for every gene (Table S1). Each reaction contained 2.0 µl transcription product, 12.5 µl SYBR Green Master Mix Reagent (Applied Biosystems), and 400 nM primers in a final volume of 25 µl. The thermal cycling conditions were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Each reaction was performed with at least three times under the same conditions. The relative expression level of each gene was calculated by comparing the ΔCT values to those of the control group.
Results
Identification of TLPs in maize
The consensus protein sequences (Pfam PF01167) of the TLP Hidden Markov Model (HMM) profile were employed as a query to search against the maize genome database with the BLASTp program. As a result, 38 candidate TLP sequences were identified in maize. To confirm putative TLP genes in the maize genome, the amino acid sequences of all 38 proteins were searched for the presence of TLP domains by Pfam and InterPro. As results of an extensive search for TLP genes, 15 non-redundant maize TLP genes (designated ZmTLP1–15) were identified (Table 1). Although all ZmTLP genes encode proteins with the conserved TLP domain, their remaining sequences are highly diverse. Unlike AtTLPs, not all ZmTLPs have a conserved F-box (45–54 residues)-containing domain. However, like AtTLPs, all identified ZmTLPs are 255–610 amino acids in length. The molecular weights of these deduced ZmTLPs range from 28.59 kDa to 66.85 kDa (Table 1). As shown in Table 1, all ZmTLPs are basic proteins, as their isoelectric points (pIs) are greater than 7.0 (pI > 9.0).
Phylogenetic and structural analysis of ZmTLPs
To detect the evolutionary relationships within the ZmTLP gene family, we constructed an NJ tree based on the alignment of the full-length sequences of the 15 ZmTLPs (Fig. 1). According to the phylogenetic tree, the ZmTLPs were divided into four groups (group A to group D; bootstrap values > 90 %). Fifteen maize ZmTLP genes formed six sister pairs, four of which showed high bootstrap support (99 %). To support the phylogenetic reconstruction, we performed exon–intron analysis by comparing the predicted coding sequence (CDS) with the genomic sequences of the ZmTLP genes (Fig. 1), finding that each ZmTLP CDS is disrupted by one or more introns. Consistent with the phylogenetic analysis results, genes in the same group display similar exon–intron structures, especially in the number of introns, although there are exceptions. For example, ZmTLP13 (in group C) contains the most introns, while ZmTLP4 contains the fewest, and most ZmTLP genes contain three or four introns. Moreover, the intron length is also highly variable, ranging from dozens of bases to approximately 5000 bases. Those observations reflect the high sequence diversity of the TLP family.
Phylogenetic relationships and exon–intron structures of ZmTLPs. The unrooted tree was constructed using the NJ method in MEGA6.0. Bootstrap values (above 50 %) from 1000 replicates are indicated at each node. Exons and introns are indicated by yellow rectangles and thin lines, respectively. The untranslated regions (UTRs) are indicated by blue rectangles
To analyze the phylogenetic relationships of TLP genes from different species, we constructed a combined phylogenetic tree based on alignment of the full-length sequences of maize, sorghum, and rice. The TLP genes were divided into four major classes: class A, B, C, and D, with well-supported bootstrap values, which include representative genes of maize, sorghum, and rice (Fig. 2). Class A genes were further divided into four subclasses according to their bootstrap values and phylogenetic relationship, which were designated A1, A2, A3, and A4. All TLP genes formed 14 sister pairs, 13 of which showed high bootstrap support (>94 %). The maize, rice, and sorghum TLP genes formed 13 sister pairs among the three species, including three pairs between rice and sorghum and 10 pairs between maize and sorghum, which indicates that these genes share a close relationship in terms of origin and evolution. For example, ZmTLP12 and SbTLP12 (A2), ZmTLP5 and SbTLP13 (A3), ZmTLP8 and SbTLP9 (A4), ZmTLP5 and SbTLP8 (A4), ZmTLP14 and SbTLP4 (B), ZmTLP2 and SbTLP7 (C), and ZmTLP1 and SbTLP10 (D) share high homolog (bootstrap values > 99.9 %).
Chromosomal locations and gene duplication
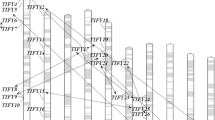
Based on the starting position of each gene on the chromosomes, we determined that the 15 ZmTLP genes are unevenly distributed on chromosomes 1–10 (except for chromosome 7) (Fig. 3). The largest number of TLP genes (three) was detected on chromosome 4, whereas the fewest were found on chromosomes 1, 2, 9, and 10 (one per chromosome). Two genes were found on each of the following chromosomes: chromosome 3 (ZmTLP3 and 14), chromosome 5 (ZmTLP6 and 13), chromosome 6 (ZmTLP7 and 12), and chromosome 8 (ZmTLP4 and 15).
To investigate the expansion mechanism of the ZmTLP gene family during the process of evolution, we investigated gene duplication events, including tandem and segmental duplications (Kong et al. 2007; Moore and Purugganan 2003). Gene duplication between genes on different chromosomes and in the same clade are designated as segmental duplication events. Several rounds of whole-genome duplication events have occurred in both the Arabidopsis and rice genomes (Wang et al. 2005; Yu et al. 2005). In the current study, based on phylogenetic analysis and the chromosomal distribution of the ZmTLP genes, four gene pairs (ZmTLP1/2, ZmTLP3/4, ZmTLP6/9, ZmTLP7/14) were found to be involved in segmental duplication events. However, no tandem duplications or gene clusters were identified.
Investigation of conserved motifs in ZmTLP genes
MEME web server is employed to analyze motif distribution and to verify the results of domain prediction. Using the MEME web server, we identified 10 conserved motifs in the ZmTLPs (Fig. 4). Each of these putative motifs was annotated by searching Pfam and SMART, revealing that conserved motifs within the ZmTLP domain-matching motifs are highly similar. Motif 1, 2, 5, 6, and 7 encode the TUB domain, while motif 3, 4, and 8 encode the F-box domain. The same number and types of motif (motif 1, 2, 3, 4, 5, 6, 7, 8, 9, and 10) were identified in nine ZmTLP genes (ZmTLP1, 2, 5, 6, 7, 8, 9, 11, and 14). Meanwhile, two gene pairs (ZmTLP3/4 and ZmTLP6/9) also contain identical motifs, indicating that these genes share high homology, which is similar to the results of phylogenetic tree analysis (Fig. 1). Motif 1, 2, 3, 4, 5, and 6 were found in most ZmTLP genes. Furthermore, conserved motifs 4 and 6 are widely distributed in all 15 ZmTLP genes. Motif 9 is absent in ZmTLP3, ZmTLP4, and ZmTLP12. Unlike ZmTLP10, ZmTLP15 lacks motif 8 and 9. In addition, motif 7, 8, 9, and 10 were also found in genes with unknown functions, indicating that they are likely required for specific functions. Detailed information about the conserved amino acid sequences and lengths of the 10 motifs is presented in Table S2.
Sequence analysis of the ZmTLPs
All of the ZmTLP sequences were searched against the Pfam database, revealing 10 putative ZmTLP sequences with a typical F-box domain at their N-termini. Two ZmTLP sequences (ZmTLP7 and ZmTLP11) have two well-conserved TLP domains at their C-termini. The multiple alignments clearly revealed a highly structured TLP domain located at the C-termini of the proteins, which represents the most conserved section of the ZmTLPs (Fig S1). Furthermore, highly conserved amino acid residues (R, L, L, A, G) were identified in the TLP domains of the 15 ZmTLPs. Meanwhile, according to the multiple sequence alignments, the 10 putative F-box regions were consistently observed (Fig S2). These F-box domains, containing 45–54 amino acids, are highly conserved.
Digital expression analysis of ZmTLP genes
We examined the expression patterns of the ZmTLPs using publicly available genome-wide transcript profiling data from maize tissues as a resource, resulting in the assignment of the ZmTLPs to 18 groups based on their expression patterns in various tissues and organs. The dataset contains RNAseq reads from various studies during the process of maize growth (Fig. 5). As show in Fig. 5, most of these genes were expressed in many tissues and organs, with distinct tissue-specific expression patterns. However, ZmTLP10, ZmTLP13, and ZmTLP15 were rarely expressed in any tissue and organ. The expression patterns of ZmTLP6 and ZmTLP9 were similar, with relatively high expression levels in the thirteenth leaf and in other tissues. ZmTLP5 and ZmTLP12 were highly expressed in almost all tissues, especially the thirteenth and eighth leaves, and ZmTLP5 was also highly expression in the tips of stage-2 leaves. Interestingly, ZmTLP4 was more highly expressed than other genes in most organs, particularly germinating seeds, which suggests that this gene plays an important role in maize development.
Interspecies microsynteny analysis
Microsynteny is genome information that can be used to predict the locations of homologous genes from different species. Gene segments in which 80 % of closely homologous genes are arranged in the same order and transcriptional orientation are characterized as having conserved microsynteny (Mural et al. 2002). We performed microsynteny analysis to detect the evolutionary origins and orthologous relationships within the TLP genes in the three Gramineae species examined. In general, microsynteny relationships are examined between maize and rice/sorghum but not between rice and sorghum. According to the microsynteny maps (Fig. 6), we detected microsynteny relationships within maize, between maize and sorghum, and between maize and rice. We found that 39 out of 42 genes from maize, rice, and sorghum formed 42 microsynteny gene pairs. Among these gene pairs, 19 pairs were generated from the microsyntenic relationship between 13 maize genes and 13 sorghum genes. In addition, 12 maize genes and 13 rice genes also generated 19 microsynteny gene pairs. However, only four gene pairs were detected between eight maize genes. Thus, there were significantly fewer genome sequences within internal duplications in maize than in synteny blocks between maize and the two other species. The smaller amount of internal microsynteny in maize may be indicative of ancient whole-genome duplications followed by gene loss and rearrangement. Ancient large-scale replication provides the basis for the establishment of high levels of microsynteny among these three species. Thus, the flanking regions of TLP genes may be derived from a common ancient Gramineae ancestor, further illustrating that these regions are remarkably conserved.
Extensive microsynteny of TLP regions across maize, sorghum, and rice. The maize chromosomes, labeled Zm, are shown in orange. The sorghum and rice chromosomes, shown in blue and green, are labeled Sb and Os, respectively. The numbers on the boxes indicate the lengths of the chromosomes in megabases. The entire chromosomes of the three species, harboring TLP regions, are arranged in a circle. Red lines represent the syntenic relationships between TLP regions
Expression levels of ZmTLP genes in response to abiotic stress
Plants are frequently threatened by abiotic stresses such as drought, high salinity, and low temperature during their lifecycle. Since gene expression patterns play an important role in the study of gene function (see promoter cis element analysis, finding that almost all ZmTLP genes contain a putative ABA-responsive element (ABRE), low dehydration-responsive element (DRE), and temperature responsive element (LTRE), Fig S3), we investigated the expression patterns of the TLP genes in response to ABA, NaCl, 42, 4 °C, and PEG stress treatment, particularly focusing on genes in Subgroup I (ZmTLP1, 2, 5, 6, 7, 8, 9, 11, and 14) and II (ZmTLP3, 4, and 12). As shown in Fig. 7, all 12 genes were up- or downregulated under stress treatment relative to the controls, although the changes in expression of some genes were small. Under ABA treatment, ZmTLP3, 4, 5, 6, 8, 9, and 12 were obviously upregulated, especially ZmTLP6. Under NaCl treatment, only the expression of ZmTLP8 was clearly altered. Under 42 °C treatment, the changes in expression were greatest for ZmTLP3, 5, 8, 9, and 12. ZmTLP8 and 9 were also induced under low temperature treatment (4 °C), and the expression of ZmTLP14 was only induced by low temperature treatment. Under PEG treatment, the expression of four genes (ZmTLP2, 3, 4, and 11) increased, and two genes (ZmTLP2 and 11) were specifically induced by this treatment. Interestingly, the expression of ZmTLP7 generally decreased in response to all of the abiotic stress treatments. In addition, the duplicate gene pair ZmTLP3/4 exhibited similar expression patterns.
Discussion
TLP genes comprise a conserved gene family that has been identified in many species. Compared to animals, few TLPs have been functionally studied in plants. In addition to a tubby domain, most plant TLPs also have an F-box domain in corresponding regions (Gagne et al. 2002). The TLP family has been studied in some plant species, such as Arabidopsis (Lai et al. 2004) and rice (Liu 2008). However, few studies of TLP genes have been reported in maize. In this study, we identified 15 non-redundant TLP genes in the maize genome. Through genome-wide comparative analysis of the evolutionary relationships between maize and other plants, such as sorghum and rice, we found that maize TLP genes share more homology with those of Graminaceous plants. TLPs in maize share some patterns observed in other species, but some distinct characteristics were also detected.
In this study, we performed comprehensive analysis of the TLP genes family in maize, including analysis of phylogeny and gene structures, chromosomal locations and gene duplication, conserved motifs, and expression profiles. Phylogenetic analysis of various species showed that all TLP genes formed 14 sister pairs, including 13 with high bootstrap support (>94 %). Among these sisters pairs, there are 13 orthologous pairs in the three species, suggesting that they share a close relationship in terms of origin and evolution, especially maize and sorghum. This finding partially accounts for the high level of TLP gene conservation in these two species during the evolutionary process. We determined that the TLP family in maize had mainly expanded through segmental duplications, and no TLP genes in maize were involved in tandem duplication. The number of TLP genes in maize (15) is similar to that in sorghum (13) and rice (14), perhaps because TLP genes in these species have undergone the same evolutionary patterns, such as segmental duplications. Segmental duplication events are thought to occur more often in the more slowly evolving gene families during the process of evolution (Cannon et al. 2004). To analyze the evolutionary constraints of this family, we investigated the Ka/Ks ratios for four duplicated pairs, showing that the TLP gene family has undergone purifying selection (Fig S4 and Fig S5). In addition, the calculated replication events of four paralogous pairs occurred between 17.59 and 71.38 Mya (Table S3). Interspecies microsynteny analysis demonstrated that TLP genes in the three species examined share many relationships, suggesting that they may have arisen from a common whole genome replication in an ancient common ancestor and that these regions are remarkably conserved. The finding that segmental segments and orthologous pairs generally exist in three different species illustrates that the TLP gene family is a conserved, slowly evolving gene family in plants, which would account for the observation that these three species have a similar number of TLPs.
Previous studies have shown that the exon/intron structures and motif compositions of TLPs are relatively conserved. The intron numbers of 15 ZmTLP genes vary, but two-thirds of TLP genes contain three introns, and only a few have more than three introns, which indirectly illustrates the conservative nature of the TLP gene family over the course of evolution. Phylogenetic analysis revealed that almost all members of each subfamily have a similar exon/intron structures. Indeed, introns were specifically inserted into plants and retained in the genome during the course of evolution (Rogozin et al. 2003). Among the four subfamilies in the phylogenetic tree, the gain and loss of introns have occurred in a subfamily-specific manner, which helps confirm the validity of the phylogenetic tree. Meanwhile, the configurations of the motifs identified by MEME reflect conservation within ZmTLPs. Based on the motif diagram and phylogenetic tree, members of the same subfamily have similar structures and numbers of motifs.
We investigated the expression patterns of ZmTLP genes based on publicly available genome-wide transcript profiling data of maize tissues to obtain clues about the functions of these genes. To better understand the gene expression patterns, we also analyzed the cis-elements of ZmTLP gene promoters. We performed qRT-PCR analysis of all ZmTLP genes, but almost no expression of ZmTLP10, 13, or 15 was detected after using various primers. Ultimately, we selected 12 ZmTLP genes to study abiotic stress-induced expression patterns, revealing different expression levels under different stress treatments. The expression of the 12 TLP genes was not significantly altered under NaCl treatment, suggesting that the TLP gene family is not involved in NaCl metabolic pathways. Seven TLP genes were strongly upregulated in response to ABA, suggesting that this family plays an important role in ABA resistance. In particular, ZmTLP6 was significantly upregulated (>40-fold) after ABA treatment, which is consistent with the results of promoter sequence analysis, illustrating that ZmTLP6 plays an important role in the response to ABA stress. Moreover, under low and high temperature treatment, we observed obvious changes in expression in six TLP genes. Three genes (ZmTLP3, 8, and 9) responded to both low and high temperature treatment, suggesting that they play a role in improving plant adaptation to temperature changes. Intriguingly, the results of promoter cis-element analysis were not invariably consistent with the results of qRT-PCR. For instance, ZmTLP9 was induced by ABA, 42 and 4 °C treatment, while it contains only one type of stress-responsive cis-element in its promoter sequence. Additionally, four genes were induced by PEG treatment, especially ZmTLP2 and ZmTLP11, suggesting that these genes have a strong effect on the PEG stress response. In addition, we also found that a duplicate genes pair (ZmTLP3/4) exhibited similar expression patterns, suggesting that these duplicated genes may be involved in the same regulatory pathway in the response to abiotic stress. Finally, we found that the expression of ZmTLP7 decreased in response to all abiotic stresses examined, which may be related to its function in plant adaptation to environmental changes. The above results show that the ZmTLP family functions is some stress responses, such as ABA, temperature, and PEG stress. Maize is often threatened by environmental stresses throughout growth. Some TLP genes have been shown to help plants adapt to stress conditions in other species, such as Cicer arietinum L. (Wardhan et al. 2012). The current qRT-PCR results lay the foundation for further analysis of the biological functions of TLP genes in maize.
References
Akihiro I, Nishina PM, Naggerten K (2002) The tubby-like proteins, a family with roles in neuronal development and function. J Cell Sci 115:9–14
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Baskin DG, Wilcox BJ, Figlewicz DP, Dm D (1988) Insulin and insulin-like growth factors in the CNS. Trends Neurosci 11:107–111
Bateman A, Finn RD, Sims PJ, Wiedmer T, Biegert A, Söding J (2009) Phospholipid scramblases and Tubby-like proteins belong to a new superfamily of membrane tethered transcription factors. Bioinformatics 25:159–162
Boggon TJ, Shan WS, Santagata S, Myers SC, Shapiro L (1999) Implication of tubby proteins as transcription factors by structure-based functional analysis. Science 286:2119–2125
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Coleman DL, Eicher EM (1990) Fat (fat) and tubby (tub): two autosomal recessive mutations causing obesity syndromes in the mouse. J Hered 81:424–427
Deleu W, González V, Monfort A, Bendahmane A, Puigdomènech P, Arús P, Garcia-Mas J (2007) Structure of two melon regions reveals high microsynteny with sequenced plant species. Mol Genet Genom 278:611–622
Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc Natl Acad Sci USA 99:11519–11524
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Hereditas 29:1023–1026
Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Jp K (2007) F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol 143:1467–1483
Kapeller R, Moriarty A, Strauss A, Stubdal H, Theriault K, Siebert E, Chickering T, Morgenstern JP, Tartaglia LA, Lillie J (1999) Tyrosine phosphorylation of Tub and its association with Src Homology 2 domain-containing proteins implicate Tub in intracellular signaling by insulin. J Biol Chem 274:24980–24986
Kleyn PW, Fan W, Kovats SG, Lee JJ, Pulido JC, Wu Y, Berkemeier LR, Misumi DJ, Holmgren L et al (1996) Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell 2:281–290
Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, Depamphilis CW (2007) Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J 50:873–885
Lai CP, Lee CL, Chen PH, Wu SH, Yang CC, Shaw JF (2004) Molecular analyses of the Arabidopsis TUBBY-like protein gene family. Plant Physiol 134:1586–1597
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 40:D302–D305
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu Q (2008) Identification of rice TUBBY-like genes and their evolution. FEBS J 275:163–171
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Mf W (1998) The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem 182:3–11
Moore RC, Purugganan MD (2003) The early stages of duplicate gene evolution. Proc Natl Acad Sci USA 100:15682–15687
Mural RJ, Adams EWM, Smith HO, Miklos GL, Wides R, Halpern A, Li PW, Sutton GG, Nadeau J et al (2002) A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296:1661–1671
Noben-Trauth K, Naggert JK, North MA, Pm N (1996) A candidate gene for the mouse mutation tubby. Nature 380:534–538
North MA, Naggert JK, Yan Y, Noben-Trauth K, Nishina PM (1997) Molecular characterization of TUB, TULP1, and TULP2, members of the novel tubby gene family and their possible relation to ocular diseases. Proc Natl Acad Sci USA 94:3128–3133
Ohlemiller KK, Hughes RM, Mosinger-Ogilvie J, Speck JD, Grosof DH, Ms S (1995) Cochlear and retinal degeneration in the tubby mouse. NeuroReport 6:845–849
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV (2003) Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol 13:1512–1517
Santagata S, Boggon TJ, Baird CL, Gomez CA, Zhao J, Shan WS, Myszka DG, Shapiro L (2001) G-protein signaling through tubby proteins. Science 292:2041–2050
Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K (2008) Genome structure of the legume, Lotus japonicus. DNA Res 15:227–239
Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, De Leon N, Kaeppler SM (2011) Genome-wide atlas of transcription during maize development. Plant J 66:553–563
Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM (2013) Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLoS One 8:e61005
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Software version 6.0. Mol Biol Evol 30:2725–2729
Thompson JD, Higgins DG, Tj G (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Dg H (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Ullrich A, Schlessinger J (1990) Signal transduction by receptors with tyrosine kinase activity. Cell 61:203–212
Wang X, Shi X, Hao B, Ge S, Luo J (2005) Duplication and DNA segmental loss in the rice genome: implications for diploidization. New Phytol 165:937–946
Wardhan V, Jahan K, Gupta S, Chennareddy S, Datta A, Chakraborty S, Chakraborty N (2012) Overexpression of CaTLP1, a putative transcription factor in chickpea (Cicer arietinum L.), promotes stress tolerance. Plant Mol Biol 79:479–493
Yang Z, Zhou Y, Wang X, Gu S, Yu J, Liang G, Yan C, Xu C (2008) Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 92:246–253
Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38
Acknowledgments
We thank the members of the Key Laboratory of Crop Biology of Anhui Province for their assistance in this study. This work was supported by the Natural Science Foundation of China (31571685).We are grateful to the reviewers for their helpful comments and criticisms.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We declare that no competing interests exist.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yulong, C., Wei, D., Baoming, S. et al. Genome-wide identification and comparative analysis of the TUBBY-like protein gene family in maize. Genes Genom 38, 25–36 (2016). https://doi.org/10.1007/s13258-015-0338-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-015-0338-6