Abstract
Schizophrenia (SZ) is a severe disorder of the human brain which disturbs behavioral characteristics such as interruption in thinking, memory, perception, speech and other living activities. If the patient suffering from SZ is not diagnosed and treated in the early stages, damage to human behavioral abilities in its later stages could become more severe. Therefore, early discovery of SZ may help to cure or limit the effects. Electroencephalogram (EEG) is prominently used to study brain diseases such as SZ due to having high temporal resolution information, and being a noninvasive and inexpensive method. This paper introduces an automatic methodology based on transfer learning with deep convolutional neural networks (CNNs) for the diagnosis of SZ patients from healthy controls. First, EEG signals are converted into images by applying a time–frequency approach called continuous wavelet transform (CWT) method. Then, the images of EEG signals are applied to the four popular pre-trained CNNs: AlexNet, ResNet-18, VGG-19 and Inception-v3. The output of convolutional and pooling layers of these models are used as deep features and are fed into the support vector machine (SVM) classifier. We have tuned the parameters of SVM to classify SZ patients and healthy subjects. The efficiency of the proposed method is evaluated on EEG signals from 14 healthy subjects and 14 SZ patients. The experiments showed that the combination of frontal, central, parietal, and occipital regions applied to the ResNet-18-SVM achieved best results with accuracy, sensitivity and specificity of 98.60% ± 2.29, 99.65% ± 2.35 and 96.92% ± 2.25, respectively. Therefore, the proposed method as a diagnostic tool can help clinicians in detection of the SZ patients for early diagnosis and treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Schizophrenia (SZ) is a severe disorder of the brain which affects the thinking, memory, understanding, speech, and the behavioral characteristics of an individual [1, 2]. This chronic psychiatric disorder affects the employment, marriage and lifestyle of the person [3, 4] and consequently quality of life is then compromised, being unable to function in workplaces, with 20–40% attempting suicide at least once [5]. The World Health Organization (WHO) reports that 20 million people worldwide are affected by this mental disorder [6]. Yet, WHO has also reported that SZ is curable, and precise and timely prognosis is helpful for better treatment and the recovery of the patient.
Currently, there is not a well-known clinical test for SZ, and diagnosis relies on behavioral symptoms such as hallucinations, functional deterioration and disorganized speech observed by experts. Such mentioned assessments are subjective and not very accurate. To overcome the aforementioned limitations, an automatic, reliable and reproducible approach from brain imaging modalities using advanced machine learning method is required. This system can overcome these limitations and can be utilized everywhere with no need to highly-trained experts. For the diagnosis of mental disorders such as SZ, electroencephalogram (EEG) is a powerful tool since it can interpret the brain state so well and widely used in clinical applications [7,8,9]. Moreover, EEG is well accepted due to high temporal resolution, easy setup and, being noninvasive, and portable method. EEG signals have been used in brain source localization for diagnoses of various brain disorders such as epilepsy, schizophrenia and Parkinson [10,11,12,13,14,15,16]. For example, in epilepsy studies, a significant issue is finding activated regions of spikes [15], or in mental disorders such as SZ patients, finding brain source localization is crucial for treatment approaches such as transcranial magnetic stimulation (TMS).
Traditionally, a number of EEG-based machine learning methods have been used for feature extraction, perform feature selection and finally employ conventional classification methods for automated detection of schizophrenia [17,18,19,20,21,22,23,24,25,26,27,28,29,30]. But in recent years, there has been a developing interest in the utilization of deep learning methods as a disruptive alternative to the aforementioned feature based methods [31, 32]. Deep learning algorithms are able to automatically extract significant features and classify them directly from the data. These methods imitate the workings of the human brain in data processing and generating patterns of decision making usage.
Recent developments in neural network architecture design and training have enabled researchers to solve previously intractable learning tasks of deep learning methods. As a result, several research works have focused on the application of deep learning as the state-of-the-art in machine learning especially the Convolutional neural network (CNN) in a wide range of computer vision studies especially in medical applications [33,34,35,36,37,38,39,40] and also for processing EEG signals with very success.[41,42,43,44,45]. Also, there are some works in detection of SZ patients that used CNNs and EEG signals. Recently, in [46] an automatic method for diagnosis SZ patients using CNN from EEG signals is proposed. They have used CNN for discrimination of 14 normal and 14 SZ patients. In another study [47], multi-domain connectome CNN using different fusion strategies for detection of SZ patients from EEG signals are proposed. Moreover, from functional magnetic resonance images (fMRI), a 3-dimensional CNN in combination of autoencoders [48] and 3 dimensional CNN for identification of SZ disorder [49] are presented.
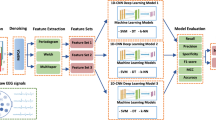
The main novelty of this paper is to provide a more generalized approach to model the brain dysfunction by combination of continuous wavelet transform (CWT), transfer learning with four popular pre-trained deep CNNs (AlexNet, ResNet-18, VGG-19 and Inception-v3) and support vector machine (SVM) as a novel approach for automated diagnosis of the SZ patients from EEG signals. Also, discriminant brain regions for recognition of 14 patients suffering from SZ and 14 healthy subjects are considered and determined by the proposed method. Finding these distinct brain sources is crucial to treat SZ patients with TMS.
Material and methods
Participant and EEG recording
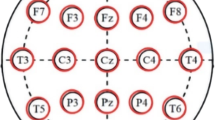
The data used in this study which is publicly available was collected from 14 patients suffering from SZ and 14 healthy subjects [23]. There are seven males with average ages of 28.3 + 4.1 years of and seven females with average ages of 27.9 + 3.3 years in the patient’s group and the same number of males and females in control group with age ranges of 26.8 ± 2.9 for males and 28.7 ± 3.4 years for females. The patients must meet International Classification of Diseases (ICD)–10 criteria for paranoid SZ. Study protocol is approved by the Ethics Committee of the Institute of Psychiatry and Neurology in Warsaw, Poland and written consent are provided from all participants. A minimum age of 18, and a minimum of seven days medication washout period are Inclusion criteria. Pregnancy, organic brain pathology, severe neurological diseases (e.g. epilepsy, Alzheimer’s, or Parkinson disease), and presence of a general medical condition are Exclusion criteria. The signal was recorded for 12 min with participants having their eyes closed and in a relaxed state at sampling rate of 250 Hz. EEG signals passed through the low pass and high pass Butterworth filters with cut off frequencies of 0.5 and 45 Hz, respectively. The signal was divided into 5 s segments before the analysis, therefore each channel of each subject yields 144 segments. The standard 10–20 International system was used to record the data and therefor 19 channels are resulted per subject: Fp1, Fp2, F7, F3, Fz, F4, F8, C3, Cz, C4, P3, Pz, P4, T3, T4, T5, T6, O1, O2. These channels of EEG signals are divided into 5 brain regions (Table 1).
EEG signal to image conversion
Wavelet transform is a tool that provides a two-dimensional time–frequency representation of EEG signal as an image. This image is able to efficiently capture the variation of the spectral content of a signal over time and can represent discriminant properties of normal and SZ subjects. The resultant image represents EEG power changes in frequency and time and is used to feed CNNs. It represents signal as a linear combination of basic functions called wavelets [45]:
where \(a\) is scale (real and positive integer), b is the translational value (real integer), \(\upomega\) is a window and \(\Psi (\mathrm{t})\) is the mother wavelet. In this study, Morse (3,60) mother wavelet which yields better localization in frequency domain compared to other mother wavelets is used.
CNN
CNN is one of the most powerful and popular tools of deep learning methods in the field of medical imaging. It is the state-of-the-art deep learning methodology consisting of many stacked convolutional layers. This network contains a convolutional layer, pooling layer, batch normalization, fully connected (FC) layers and finally a softmax layer [31, 32]. Feature maps are extracted at the convolutional layers. Pooling layers lessen feature maps using maximum or average operators and the most significant features are extracted. Finally, FC layers prepare extracted features to be classified by softmax layer. Nonlinear layers (mostly ReLU function) are used to strengthen the network for solving nonlinear problems. ReLU as activation function are used after each convolutional and fully connected layer. Also, drop out and batch normalization techniques are introduced to overcome the overfitting problem in this neural network.
Pre-trained CNNs
Pre-trained CNNs are trained networks on very large amounts of images with many categories. AlexNet [50], VGGNet [51], Inceptions [52] and Residual network (ResNet) [53] are popular pre-trained CNNs that are trained on ImageNet database and were the winner of the ImageNet Large Scale Visual Recognition Competition (ILSVRC) from 2012 till 2015. ImageNet is a known image database for visual object recognition project that starts with 1.2 million of images from 1000 different categories from animals (dogs, cats, lions, ….) to objects (desks, pens, chairs, …).
AlexNet
AlexNet with 61 million parameters is a simple CNN with a few convolutional layers which has been won the ILSVRC2012 [50]. It has 5 convolutional layers for extraction low and high levels features, max pooling layers and 3 fully connected layers for classification. Figure 1a shows name of layer in left column and the number and size of kernels (filters) of the convolutional and pooling layers in right column. For example, ‘Conv1′ layer has 96 kernels with the size of 11 × 11 × 3 with stride and padding of 4 and 0, respectively.
VGGNet-19
VGGNet is the runner-up of ILSVRC2014 and has been introduced by Simonyan and Zisserman [51]. This network has two versions with different stacked convolutional layers, VGG-16 and VGG-19. VGG-16 has three stacked of 3 convolutional layers and VGG-19 has three stacked of four convolutional layers. In this paper, VGG-19 has been used with 19 uniform convolutional layers and 144 million parameters. Figure 1b shows the structures of this method. For example, ‘Conv1_1′ layer has 64 kernels with the size of 3 × 3 × 3 with stride and padding of 1 and 1, respectively.
Inception-v3
Inception-v3 with 23.9 million parameters was the runner up of ILSVRC2015 [52]. Inception-v3 has many stacked inception modules which are parallel convolutional layers. This network reduced the number of connections, without degrading the efficiency of the network. Figure 2a shows the structures of this method. For example, ‘Conv2d_1′ layer has 32 kernels with the size of 3 × 3 × 3 with stride and padding of 2 and 0, respectively.
ResNet-18
ResNet is the winner of ILSVRC2015 [53]. ResNet has many stacked identity shortcut connections that help to solve the vanishing gradient problem of CNNs. CNNs with many layers face with the vanishing gradient problem, i.e. when there are so many layers, repeating multiplication make very low gradient value near zero and it will be vanished in updating procedure. Therefore, the performance will be degraded as each additional layer. ResNet has some versions with various convolutional layers. ResNet-18 is the version comprise of 18 convolutional layers with 11.7 million parameters. Figure 2b shows the structures of this method. For example, ‘Conv_1′ layer has 64 kernels with the size of 7 × 7 × 3 with stride and padding of 2 and 3, respectively.
Transfer learning
The number of parameters in the model increases as networks gets deeper which in turn results in improved learning efficiency. The deeper networks lead to more complicated computations and as well as demanding more training data. Transfer learning employs a reference deep model trained previously on a huge database and adapts it using a smaller insufficient database for a new application [54,55,56,57,58]. It means we transfer the information (the learned parameters such as weights layers and biases) to our problem with an insufficient database. Transfer learning takes advantage of a pre-trained CNN model on a huge database. This procedure has a number of benefits for researchers such as lower training time, weaker and cheaper hardware requirement, lower computational load, and fewer images for training. In the procedure, the images resulted from EEG signal via CWT transform are used as input and convolutional and pooling layers of pre-trained CNN models namely AlexNet, ResNet-18, VGG-19 and Inception-v3 are used as deep features and fed into the SVM classifier. Then, we have tuned the parameters of SVM to classify SZ patients and healthy subjects. In other words, the fully connected layer and softmax layer of pre-trained CNN models are replaced with a SVM as classifier layer and the parameters of SVM are tuned. It should be noted that there are little differences in input of each network (AlexNet is 227*227. VGGNet-19 and Resent are 224*224, Inception-v3 is 229*229). So in the first step of data preparation, according to different sizes of model inputs, all images were resized to proper sizes.
SVM classifier
SVM is a supervised method of classification in machine learning field and can solve classification problem efficiently. It minimizes error iteratively by maximizing marginal hyperplane. This classifier has been successfully used in EEG signal processing studies [59,60,61]. The linear hyperplane for a training set of data \({x}_{i}\) is defined as Eq. (2) [62]:
where \(w\) and \(b\) are n-dimensional vector and bias, respectively. A hyperplane must have the least possible error in separating data and maximum distance to closest data of each class. Then, according to these two special properties a sample belongs to the left (y = 1) or right (y = − 1) sides of the hyperplane. Equation (3) shows relation of two margins that controls the separability of samples:
The distance (d) to find the best hyperplane is computed as Eq. (4):
Maximizing the margin would be equal to minimizing \(w\). Then, the optimal hyperplane is computed such that [62]:
where C and \({\xi }_{i}\) are the margin parameters and slack variable. Margin parameter determines the tradeoff between maximizing the margin and minimizing classification error, respectively. Slack variable penalizes data points which violate the margin requirements. Here, we used L2-SVM classifier that uses the square sum of slack variables (\({\xi }_{i}\)) as the optimization function. This optimization function is computed as below:
Evaluation performance
Independently, 4 versions of the pre-trained CNNs model were fine-tuned on 90% of data and then evaluated from the residual data. Due to the limited dataset, tenfold cross validation was used and the process is repeated 10 times, with each subsample used exactly once as the testing data until all the dataset has been used for testing and evaluation performance. Then, averaging the 10 results is reported. Then, the average and standard deviation of 10 results is reported. The accuracy, sensitivity and specificity measures are computed as follow:
where TP, TN, FP and FN are true positive, true negative, false positive and false negative from confusion matrix, respectively.
Results
19 channels of EEG signals from each subject were preprocessed using the EEGLAB [63] toolbox in MATLAB software (version 2019a). Then, EEG signals were converted to scalogram images by the CWT method by Morse (3,60) wavelet. Scalogram images were built from 19 channels of each subject. Figure 3 shows a sample scalogram of EEG channels for healthy subject and SZ patient. Horizontal and vertical axes represent time (second) and frequency (Hz) contents, respectively. Then, the scalogram images resulted from EEG signal via CWT transform are used as input and convolutional and pooling layers of pre-trained CNN models are used as feature extractor and fed into the SVM classifier (Fig. 3). Independently, four versions of pre-trained CNNs, Inception-v3, VGG-19, ResNet-18 and AlexNet are used. Then, we have tuned the parameters of SVM to classify SZ patients and healthy subjects. In other words, the fully connected layer and softmax layer of pre-trained CNN models are replaced with a SVM as classifier layer and the parameters of SVM are tuned. Tuning was performed on 90% of scalogram images and then the accuracy, specificity and sensitivity are computed on residual scalograms images. This procedure is done 10-times. Finally, mean and standard deviation of these measures were computed. All processing steps were done with the MATLAB software version 2019a. All codes were implemented on a laptop with an Intel (R) Core (TM) i7-6500U CPU @2.50 GHz 2.60 GHz.
Figure 4 shows the average accuracy for 19 EEG channels using the AlexNet-SVM, VGG-19-SVM, ResNet-18-SVM and Inception-v3-SVM for SZ detection from healthy controls. Maximum accuracy was achieved for ResNet-18-SVM in all EEG channels, followed by Inception-v3-SVM, VGG-19-SVM and AlexNet-SVM having the highest accuracy, respectively. Among all EEG channels, P4 and O2 achieved higher accuracies using ResNet-18-SVM with accuracy of 88.05% and 86.25%, respectively. According to psychological studies, the parietal and occipital are discriminant brain regions in SZ disorder. In [64] after analyzing MR images from SZ patients and normal subjects, they found that gray matter (GM) and white matter (WM) in these brain regions had significant differences between the two groups. Also, in [65], they found discriminant regions in parietal and occipital of SZ patients after investigating GM and WM in MRI.
To improve the SZ recognition performance, EEG channel of each region are combined. So, 19 channels of EEG signals are divided into 5 brain regions (Table 1). Because the highest accuracy achieved using ResNet-18-SVM, this network is used for further analysis. Table 2 mentions the average accuracy, sensitivity and specificity values for scalogram images of 5 defined brain regions using the ResNet-18-SVM in classifying the SZ patients from healthy controls. The highest accuracy achieved 94.84% for parietal region. Finally, brain regions were combined to further improve the performance of SZ recognition. All possible combinations of two, three, four and five brain regions are considered here. The highest accuracy achieved was 98.60% ± 2.29 for scalogram images of combination of four regions of frontal, central, parietal, and occipital. Table 3 mentions the average and standard deviation of accuracy, sensitivity and specificity values for scalogram images of combinations of possible four and five brain regions using the ResNet-18-SVM in classifying the SZ patients. As it is observed, when temporal region is combined with other regions the accuracy is decreased and therefore combination of other four possible brain regions and even five regions (95.30%) with temporal have lower accuracy value. So, we can differentiate between SZ patients and healthy controls with accuracy of 98.60% ± 2.29 for combination of frontal, central, parietal, and occipital regions.
Discussion
In this research, we have used transfer learning with deep CNNs, CWT and SVM methods for automated detection of SZ patients and healthy controls. Accuracy value of 98.60% ± 2.29 is achieved for ResNet-18-SVM architecture in scalogram images of combination of frontal, central, parietal, and occipital regions of EEG signals. Screening of SZ patients is momentous for early diagnosis and treatment.
The relatively small database used in this study, limits the procedure of training enormous parameters of a deep CNN model adequately; Thus, the transfer learning concept to compensate this inadequacy have been exploited. In this study, we demonstrated the feasibility of 4 state-of-the-arts pre-trained CNNs architectures and then used these deep architectures on clinical dataset to perform SZ detection from EEG signals. Also, the fully connected layer and softmax layer were replaced with a SVM. The idea of using SVM as classifier layer is reasonable since prior to the deep learning methods popularity, it was one the most efficient classification methods which could perform discrimination with the highest performance.
As seen in the result section, it can be observed that in terms of accuracy, sensitivity and specificity the ResNet-18-SVM architecture is the best model. The highest average accuracy to recognize SZ patients was obtained by the ResNet-18 method than other pre-trained CNNs (Fig. 4). To understand why the aforementioned network performs better compared to others, one should consider the architectures of these pre-trained CNNs. The structure of VGG-19 and AlexNet is relatively similar but VGG-19 has more convolutional layers with higher accuracies than AlexNet. So, it seems the number of layers affect the performance in this situation. But, ResNet-18 has lower convolutional layers (18) than Inception-v3 (48) with higher accuracy. Consequently, the number of layers is not the only effective factor. ResNet-18 has the residual unit containing multiple stacked identity maps and shortcuts, while, Inception-v3 has multiple parallel convolutional layers in its Inception units. So, according to results on accuracy, it seems that the residual unit performs better than Inception module for discriminating this task.
As observed in Table 3, the combination of frontal, central, parietal, and occipital regions had achieved the highest average accuracy among other combinations. It can be deduced that these regions are the most related regions in recognition of SZ patients and healthy controls. Our findings about best regions are consistent with related studies with other methods [23, 25, 27, 28, 30]. In Table 4, results of this study are compared with related studies that used EEG signals of the same database [25,26,27, 46, 47]. As it is observed, accuracy achieved in this study is higher than those studies with the other machine learning methods and proves the preference of the proposed method.
The main limitation of the research can be considered the dataset size to train the networks. By performing regularization terms and simplifying deep models, we were able to overcome this problem. Our aim in the future is to collect more samples and employing the developed methodology on other types of EEG data. Also, applying different methods for converting 1-D EEG signals into 2-D image which represents information flow between different EEG channels to be fed to CNN Architecture are presented in the future.
Conclusion
Transfer learning with one popular version of deep CNN named ResNet-18-SVM and CWT method is used with very success for automated detection of SZ patients from healthy controls using EEG signals. The accuracy, sensitivity and specificity of the mentioned method are 98.60% ± 2.29, 99.65% ± 2.35 and 96.92% ± 2.25 for combination of frontal, central, parietal, and occipital regions, respectively. Relying on the results, newly proposed deep learning model is capable of effectively analyzing the brain function and can help health care professionals to identify the SZ patients for early identification and intervention.
References
Savio A, Charpentier J, Termenón M, Shinn AK, Grana M (2010) Neural classifiers for schizophrenia diagnostic support on diffusion imaging data. Neural Netw World 20(7):935
Chatterjee I, Agarwal M, Rana B, Lakhyani N, Kumar N (2018) Bi-objective approach for computer-aided diagnosis of schizophrenia patients using fMRI data. Multimed Tools Appl 77(20):26991–27015
Bowie CR, Harvey PD (2006) Cognitive deficits and functional outcome in schizophrenia. Neuropsychiatr Dis Treat 2(4):531–536
Joyce EM, Roiser JP (2007) Cognitive heterogeneity in schizophrenia. Curr Opin Psychiatry 20(3):268–272
Tibbetts PE (2013) Principles of cognitive neuroscience. Q Rev Biol 88:139–140
WHO: https://www.who.int/mental_health/management/schizophrenia/en/.
Afshani F, Shalbaf A, Shalbaf R, Sleigh J (2019) Frontal–temporal functional connectivity of EEG signal by standardized permutation mutual information during anesthesia. Cogn Neurodyn 13(6):531–540
Shalbaf A, Saffar M, Sleigh JW, Shalbaf R (2017) Monitoring the depth of anesthesia using a new adaptive neurofuzzy system. IEEE J Biomed Health Inform 22(3):671–677
Saeedi A, Saeedi M, Maghsoudi A, Shalbaf A (2020) Major depressive disorder diagnosis based on effective connectivity in EEG signals: a convolutional neural network and long short-term memory approach. Cogn Neurodyn. https://doi.org/10.1007/s11571-020-09619-0
López JD, Litvak V, Espinosa JJ, Friston K, Barnes GR (2014) Algorithmic procedures for Bayesian MEG/EEG source reconstruction in SPM. NeuroImage 84:476–487
Friston KJ, Frith CD (1995) Schizophrenia: a disconnection syndrome. Clin Neurosci 3(2):89–97
Jatoi MA, Kamel N, Malik AS, Faye I (2014) EEG based brain source localization comparison of sLORETA and eLORETA. Australas Phys Eng Sci Med 37(4):713–721
Jatoi MA, Dharejo FA, Teevino SH (2020) Comparison of machine learning techniques based brain source localization using eeg signals. Curr Med Imaging. https://doi.org/10.2174/1573405616666200226122636
Jatoi MA, Kamel N, López JD (2020) Multiple sparse priors technique with optimized patches for brain source localization. Int J Imaging Syst Technol 30(1):154–167
Jatoi MA, Kamel N, Teevino SH (2020) Trend analysis for brain source localization techniques using EEG signals. In: 2020 3rd International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), pp 1–5
Gaho AA, Jatoi MA, Musavi SHA, Shafiq M (2019) Brain mapping of cortical epileptogenic zones and their EEG source localization. In: 2019 2nd International Conference on Computing, Mathematics and Engineering Technologies (iCoMET), pp 1–6
Dvey-Aharon Z, Fogelson N, Peled A, Intrator N (2015) Schizophrenia detection and classification by advanced analysis of EEG recordings using a single electrode approach. PLoS ONE 10:e0123033
Kim JW, Lee YS, Han DH, Min KJ, Lee J, Lee K (2015) Diagnostic utility of quantitative EEG in un-medicated schizophrenia. Neurosci Lett 589:126–131
Chen C-MA, Jiang R, Kenney JG, Bi J, Johannesen JK (2016) Machine learning identification of EEG features predicting working memory performance in schizophrenia and healthy adults. Neuropsychiatr Electrophysiol 2(1):1–21
Santos-Mayo L, San-José-Revuelta LM, Arribas JI (2017) A computer-aided diagnosis system with EEG based on the P3b wave during an auditory odd-ball task in schizophrenia. IEEE Trans Biomed Eng 64(2):395–407
Ibáñez-Molina AJ, Lozano V, Soriano MF (2018) EEG multiscale complexity in schizophrenia during picture naming. Front Physiol 9:1–12
Aharon ZD, Fogelson N, Peled A, Intrator N (2017) Connectivity maps based analysis of EEG for the advanced diagnosis of schizophrenia attributes. PLoS ONE 12(10):e0185852
Olejarczyk E, Jernajczyk W (2017) Graph-based analysis of brain connectivity in schizophrenia. PLoS ONE 12(11):e0188629
Sun J, Tang Y, Lim KO (2014) Abnormal dynamics of eeg oscillations in schizophrenia patients on multiple time scales. IEEE Trans Biomed Eng 61(6):1756–1764
Buettner R; Hirschmiller M; Schlosser K (2019) High-performance exclusion of schizophrenia using a novel machine learning method on EEG data. In: IEEE International Conference on E-health Networking, Application & Services, Bogotá, Colombia, pp 14–19
Jahmunah V, Oh SL, Rajinikanth V, Ciaccio EJ, Cheong KH, Arunkumar N, Acharya UR (2019) Automated detection of schizophrenia using nonlinear signal processing methods. Artif Intell Med 100:101698
Buettner R, Beil D, Scholtz S, Djemai A (2020) Development of a machine learning based algorithm to accurately detect schizophrenia based on one-minute EEG recordings. In: Proceedings: 53rd Hawaii International Conference on System Sciences, Maui, Hawaii, pp 7–10
Liu T, Zhang J, Dong X, Li Z, Shi X, Tong Y et al (2019) Occipital alpha connectivity during resting-state electroencephalography in patients with ultra-high risk for psychosis and schizophrenia. Front Psychiatry 10:553
Boostani R, Sadatnejad K, Sabeti M (2009) An efficient classifier to diagnose of schizophrenia based on EEG signals. Expert Syst Appl 36(3):6492–6499
Li F, Wang J, Liao Y, Yi C, Jiang Y, Si Y et al (2019) Differentiation of schizophrenia by combining the spatial EEG brain network patterns of rest and task P300. IEEE Trans Neural Syst Rehabil Eng 27(4):594–602
Guo Y, Liu Y, Oerlemans A, Lao S, Wu S, Lew MS (2016) Deep learning for visual understanding: a review. Neurocomputing 187:27–48
Bengio Y, Goodfellow I, Courville A (2017) Deep learning. MIT press, Cambridge
Sun W, Tseng TL, Zhang J, Qian W (2017) Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graph 57:4–9
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M et al (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88
Greenspan H, van Ginneken B, Summers RM (2016) Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging 35(5):1153–1159
Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS et al (2018) Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2(3):158
Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S (2017) Dermatologist-level classification of skin cancer with deep neural networks. Nature 542(7639):115–118
Litjens G, Ciompi F, Wolterink JM, de Vos BD, Leiner T, Teuwen J, Išgum I (2019) State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging 12(8):1549–1565
De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S et al (2018) Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 24(9):1342–1350
Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I et al (2016) Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6:26286
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H (2018) Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med 100:270–278
Roy Y, Banville H, Albuquerque I, Gramfort A, Falk TH, Faubert J (2019) Deep learning-based electroencephalography analysis: a systematic review. J Neural Eng 16(5):051001
Faust O, Hagiwara Y, Hong TJ, Lih OS, Acharya UR (2018) Deep learning for healthcare applications based on physiological signals: a review. Comput Methods Programs Biomed 161:1–3
Zhang X, Yao L, Wang X, Monaghan J, Mcalpine D (2019) A survey on deep learning based brain computer interface: recent advances and new frontiers. arXiv. https://doi.org/10.1145/1122445.1122456
Chaudhary S, Taran S, Bajaj V, Sengur A (2019) Convolutional neural network-based approach towards motor imagery tasks EEG signals classification. IEEE Sens J 19(12):4494–4500
Oh SL, Vicnesh J, Ciaccio EJ, Yuvaraj R, Acharya UR (2019) Deep convolutional neural network model for automated diagnosis of schizophrenia using EEG signals. Appl Sci 9(14):2870
Phang CR, Noman F, Hussain H, Ting CM, Ombao H (2019) A multi-domain connectome convolutional neural network for identifying schizophrenia from EEG connectivity patterns. IEEE J Biomed Health Inform 24(5):1333–1343
Oh K, Kim W, Shen G, Piao Y, Kang NI, Oh IS, Chung YC (2019) Classification of schizophrenia and normal controls using 3D convolutional neural network and outcome visualization. Schizophr Res 212:186–195
Qureshi MNI, Oh J, Lee B (2019) 3D-CNN based discrimination of schizophrenia using resting-state fMRI. Artif Intell Med 98:10–17
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Khan SanaUllah, Islam N, Jan Z (2019) A novel deep learning based framework for the detection and classification of breast cancer using transfer learning. Pattern Recogn Lett 125:1–6
Shin H-C, Roth HR, Gao M (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35(5):1285–1298
Byra M, Styczynski G, Szmigielski C (2018) Transfer learning with deep convolutional neural network for liver steatosis assessment in ultrasound images. Int J Comput Assist Radiol Surg 13:1895–1903
Acharya UR, Oh SL, Hagiwara Y, Tan JH, Adeli H, Subha DP (2018) Automated EEG-based screening of depression using deep convolutional neural network. Comput Methods Programs Biomed 161:103–113
Craik A, He Y, Contreras-Vidal JL (2019) Deep learning for electroencephalogram (EEG) classification tasks: a review. J Neural Eng 16(3):031001
Shiao HT, Cherkassky V, Lee J, Veber B, Patterson EE, Brinkmann BH, Worrell GA (2016) SVM-based system for prediction of epileptic seizures from iEEG signal. IEEE Trans Biomed Eng 64(5):1011–1022
Rasheed W, Boon T (2019) Anomaly detection of moderate traumatic brain injury using auto-regularized multi-instance one-class SVM. IEEE Trans Neural Syst Rehabil Eng 28(1):83–93
Shalbaf A, Shalbaf R, Saffar M, Sleigh J (2019) Monitoring the level of hypnosis using a hierarchical SVM system. J Clin Monit Comput 15:1–8
Gholami R, Fakhari N (2017) Support vector machine: principles, parameters, and applications. Handbook of neural computation. Academic Press, Cambridge, pp 515–535
Delorme A, Makeig S (2004) EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods 134(1):9–21
Molina V, Reig S, Sanz J, Palomo T, Benito C, Sánchez J, Sarramea F, Pascau J, Desco M (2005) Increase in gray matter and decrease in white matter volumes in the cortex during treatment with atypical neuroleptics in schizophrenia. Schizophr Res 80(1):61–71
Mitelman SA, Brickman AM, Shihabuddin L, Newmark RE, Hazlett EA, Haznedar MM, Buchsbaum MS (2007) A comprehensive assessment of gray and white matter volumes and their relationship to outcome and severity in schizophrenia. Neuroimage 37(2):449–462
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Approval was obtained by data owner from the Ethics Committee of the Institute of Psychiatry and Neurology in Warsaw, Poland.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Shalbaf, A., Bagherzadeh, S. & Maghsoudi, A. Transfer learning with deep convolutional neural network for automated detection of schizophrenia from EEG signals. Phys Eng Sci Med 43, 1229–1239 (2020). https://doi.org/10.1007/s13246-020-00925-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-020-00925-9