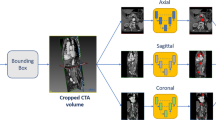
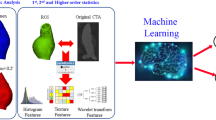
Abstract
Purpose
Although segmentation of Abdominal Aortic Aneurysms (AAA) thrombus is a crucial step for both the planning of endovascular treatment and the monitoring of the intervention’s outcome, it is still performed manually implying time consuming operations as well as operator dependency. The present paper proposes a fully automatic pipeline to segment the intraluminal thrombus in AAA from contrast-enhanced Computed Tomography Angiography (CTA) images and to subsequently analyze AAA geometry.
Methods
A deep-learning-based pipeline is developed to localize and segment the thrombus from the CTA scans. The thrombus is first identified in the whole sub-sampled CTA, then multi-view U-Nets are combined together to segment the thrombus from the identified region of interest. Polygonal models are generated for the thrombus and the lumen. The lumen centerline is automatically extracted from the lumen mesh and used to compute the aneurysm and lumen diameters.
Results
The proposed multi-view integration approach returns an improvement in thrombus segmentation with respect to the single-view prediction. The thrombus segmentation model is trained over a training set of 63 CTA and a validation set of 8 CTA scans. By comparing the thrombus segmentation predicted by the model with the ground truth data, a Dice Similarity Coefficient (DSC) of 0.89 ± 0.04 is achieved. The AAA geometry analysis provided an Intraclass Correlation Coefficient (ICC) of 0.92 and a mean-absolute difference of 3.2 ± 2.4 mm, for the measurements of the total diameter of the aneurysm. Validation of both thrombus segmentation and aneurysm geometry analysis is performed over a test set of 14 CTA scans.
Conclusion
The developed deep learning models can effectively segment the thrombus from patients affected by AAA. Moreover, the diameters automatically extracted from the AAA show high correlation with those manually measured by experts.
Graphical abstract

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
An aortic aneurysm can be defined as a permanent localized dilatation of the aorta with at least a 50% diameter increase compared with the expected normal diameter.4
An Abdominal Aortic Aneurysm (AAA) is an aortic aneurysm that has a diameter of 3 cm or greater and it is located in the abdominal aortic section, which extends from the infrarenal aorta to the common iliac arteries.4 AAA is a relatively common disease and its rupture is one of the commonest causes of death overall.4 In 75% of the clinically-relevant AAA patients there is an intraluminal thrombus.18
The severity of the disease, determined by AAA characteristic diameters and lengths, is currently assessed by the analysis of Computed Tomography Angiography (CTA) scans.15
While the aortic lumen detection is facilitated by the contrast-medium enhancement, the manual assessment of the thrombus boundaries from the surrounding tissues can be challenging. In particular, when dealing with segmentation of anatomical structures, the segmentation of the thrombus is a time consuming task and should be performed by trained-users.11
An automatic tool performing thrombus segmentation would speed-up and standardize the analysis of AAA anatomy, overcoming the problem of intra- and inter-observer variability. In particular, an automatic segmentation tool would ensure a fast-moving thrombus delineation procedure in large databases, facilitating clinical research, and would cope with situations where segmentation is needed quickly.
Some studies have previously addressed the problem of semi-automatic and automatic AAA segmentation to overcome the above-mentioned difficulties.
Maiora et al.15 proposed a segmentation approach based on a supervised Random Forest classifier applied on a set of features extracted from each voxel and its neighborhood. The adopted approach is semi-automatic and no comparison with ground truth segmentations was reported in the paper.
Lareyre et al.11 developed an automatic pipeline to detect the vascular system and the AAA structures. The lumen is first extracted from the CTA scans, then thrombus segmentation is performed exploiting lumen contour propagation.
In recent years, deep learning approaches have achieved excellent performance in medical image analysis.21 In the following, some deep-learning studies focusing on automatic thrombus segmentation are reported. These methods are fully automatic, thus they do not require neither user interaction, nor a priori knowledge of the thrombus geometry.12,13
Hong and Sheikh9 proposed an automatic approach to perform preoperative AAA detection and segmentation. The method is based on two Deep Belief Networks (DBN) for detection: one network is used to detect large aneurysms, while the other one detects small aneurysms, bones, and air. The detection is done in 2D with samples coming from a unique dataset. The thrombus segmentation consists of another DBN trained over 40 image samples, and the obtained results were not quantitatively compared with ground truth segmentations.
The work proposed by Lòpez-Linares et al. starts with the paper published in 201812 and expands with the paper published in 2019.13 The initial work exploited a Convolutional Neural Network to detect the thrombus and a modified Holistically-Nested Edge Detection (HED) Network to perform segmentation. The dataset was composed of 13 postoperative CTA. The models were trained on 2D single slices in the axial view, and a Gaussian filter was applied on the z-direction to enforce spatial consistency. In the second paper, the pipeline proposed in Lòpez-Linares et al.12 was extended using 3D networks instead of 2D networks, increasing the segmentation performance working on 3D volumes and taking into account spatial coherence. Since 3D networks are data demanding, the used dataset was increased up to 80 CTA preoperative and postoperative scans.
Finally, the work by Caradu et al.3 assessed the quality of PRAEVAorta, a fully automatic software developed by Nurea (https://www.nurea-soft.com/) used to detect the aortic lumen and thrombus.
Once thrombus segmentation is performed, aneurysm treatment involves its geometric measurements collection. In particular, geometrical analysis of the lumen and thrombus is an important factor when planning an EndoVascular Aneurysm Repair (EVAR) procedure or investigating morphological changes of the vessel during follow-up.
Given the need to perform diameters and volume measurements on the obtained segmentations, it is desirable that the measurement procedure is automatic and fast.
Singh et al.22 carried out a work studying intra- and inter- observer variability in the measurements of abdominal aortic and common iliac artery diameter with CT. Their work pointed out that interobserver variability between expert measurements with CT is not negligible and is higher than intraobserver variability. Moreover, the variability was higher for measurement of patients with aneurysms.
Kaladji et al.10 studied a dataset of 32 patients with AAA and measured maximum and minimum diameter of the aneurysm using Endosize (V. 3.1.25 64 bits Therenva SAS, Rennes, France), a semi-automatic three dimensional sizing software. The software needs an initialization step to perform lumen extraction, and it takes 13.1 ± 4.53 minutes to perform one sizing. An Intraclass Correlation Coefficient (ICC) above 0.9 is computed comparing software results with manual measurements. These results show a good correlation between manual and semi-automatic procedures, thus stressing the effectiveness and low human-effort requirement in semi-automatic approaches.
In Caradu et al.,3 the aortic centerline and oblique cross-sectional planes normal to the centerline are extracted to compute the quantitative measures related to aortic geometry. This computation is performed to limit the interobserver variability affecting the maximum cross-section diameter measurements.
In order to segment and quantify the abdominal aortic aneurysm from CTAs, the present study proposes a fully automatic deep learning-based approach. More in detail, following the approach proposed in a previous study which deals with aortic lumen segmentation,7 in this work we make use of a first Convolutional Neural Network (CNN) to localize the thrombus from the whole CTA scans and other three multi-view CNNs to perform a finer segmentation. The novelty of our segmentation approach lies in the integration of 2D networks trained on orthogonal views. In contrast to 2D approaches, the segmentations obtained with our method consider the 3D spatial context. Compared to 3D CNNs, computational and memory requirements are reduced and the dataset available for training is larger. Finally, our pipeline also includes automatic extraction of thrombus measurements, which allows faster geometric analysis of AAA.
Automatic aneurysm geometrical analysis is performed using Vascular Modeling ToolKit (VMTK) open source library.1
Materials and Methods
The proposed approach for automatic thrombus segmentation and geometric evaluation is illustrated in Fig 1. In a first phase, both the aortic lumen and the thrombus are automatically segmented from the input CTA scans using two different deep learning-based procedures. Secondly, the centerline is automatically extracted from the lumen segmentation mask and exploited to perform geometrical evaluations on the thrombus and lumen.
Proposed pipeline for multi-view thrombus segmentation. On the left, the low resolution CTA scan is processed through a first U-Net to identify the thrombus region of interest (ROI). On the right, the ROI is processed with three U-Nets networks trained on orthogonal views. The single network predictions are then combined into a final segmentation.
Dataset
The dataset used in this study has been provided by IRCCS Ospedale Policlinico San Martino (Genoa, Italy) and consists of 85 preoperative CTA scans of patients affected by AAA as primary pathology selected by an expert vascular surgeon to represent a large variability of cases.
The IRCCS Ospedale Policlinico San Martino is the regional reference center for aortic pathologies highly experienced in open and endovascular procedures. The CTA images have been acquired in four different hospital departments using four different scanners (General Electric Medical Systems Optima CT660, General Electric Medical Systems LightSpeed 16, Siemens SOMATOM Flash, Siemens Sensation). For the collected dataset, pixel spacing varies from 0.57 mm to 0.98mm, with an average value of 0.76 ± 0.08 mm. Slice thickness varies in the range 0.5-5 mm, with a mean value of 0.82 ± 0.56 mm.
The mean age of the patients is 75 years (range: 60-91 years), with a male predominance (81% males). The study protocol has been submitted to the local Ethic Committee and patients’ informed consent has been waived due to the retrospective nature of the data.
The collected dataset has been manually segmented by a trained expert (A.Fi.) and by a trainee (F.B.) using the MITK segmentation toolset23 and the expert operator performed the segmentations twice.
Intra-observer and inter-observer variability returns a Dice Similarity Coefficient of 0.96 ± 0.2 and 0.93 ± 0.2, respectively.
The CTA scans and the corresponding segmentations have been pre-processed following the steps described in our previous work.7 The CTAs have been resampled and image intensity has been rescaled in range 0-255. Details regarding data preprocessing can be found in the Supplementary Material.
To address the segmentation problem, the whole dataset has been divided into three sets: a training set used to train the network in thrombus segmentation (n = 63 scans); a validation set used to prevent overfitting and tune the model (n = 8 scans); and a test set (n = 14 scans) used to assess the model performance.
The geometrical analysis, performed using the Vascular Modeling ToolKit (VMTK),1 has been carried out only on the test set. The VMTK is a collection of C++ classes wrapped by python that enable largely automated geometric characterization and visualization of vascular structures from medical images, requiring low user-interaction. In particular, the geometric analysis heavily depends on centerline definitions from the segmented structures. The library relies on two major open source frameworks: the Visualization Toolkit (VTK) and the Insight Toolkit (ITK).1,17,20
Pipeline
Two different steps have to be performed in order to segment and analyze an AAA from a CTA scan. First, the thrombus and the lumen are segmented from the CTAs, then the geometric analysis is performed. The aortic lumen segmentation is already implemented in Fantazzini et al.,7 while the thrombus segmentation is addressed in this paper as described in the following.
Thrombus Segmentation
Following the pipeline for lumen segmentation described in Fantazzini et al.,7 a first U-Net (Ronneberger et al.19) is trained on low resolution CTA scans to perform a coarse segmentation of the thrombus. The network is trained to segment downsampled CTA scans parsed into axial view (downsample factor = 4). After coarsely segmenting the thrombus, the centroid of the low-resolution segmentation is identified. Then, the centroid is reported in the full resolution CT scan, and a cuboid centered in the thrombus is used to crop the CTA scan and focus on the region of interest.
The cuboid has dimensions of 384x384x384, with pixel size set to 0.73mm and slice thickness of 0.62 mm. To choose the size of the cuboid, a bounding box was calculated for each manual segmentation in the dataset. In order to size the final cuboid, the maximum bounding box dimension on the x, y, and z axis is extracted and rounded up to the nearest power of 2, as the U-Net architectures work correctly only if each downsampling/upsampling layer operates on images/feature maps whose height and width are even numbers.
Thus, the bounding box is much larger than the size of the thrombus in the dataset: this allows greater robustness in the case the centroid obtained with coarse segmentation is less accurate. In fact, the coarse segmentation plays a key role in localizing the bounding box: if the preliminary coarse segmentation presents several errors, the calculated centroid could be in the wrong area and consequently the ROI would not fully enclose the thrombus.
In our case, given that the aneurysms in our dataset are of small-medium size and that the bounding box is quite large in size, the chosen bounding box did not result in loss of accuracy in the final segmentation. In any case, the bounding box value should be verified and eventually updated in case aneurysms of larger diameter were added to the dataset.
Unlike the work performed in,7 since the cuboid is quite small in size, the cropped scans have full resolution.
The bounding box extraction is used to focus the attention on the relevant regions and to exclude the background areas. In addition to the advantage of focusing on the area of interest, cropping a Region of Interest (ROI) from the whole CTA is desirable as the computational requirements are reduced.
Starting from the slices detected in the ROI, each CTA scan is parsed into 2D axial, sagittal, and coronal views and three separate U-Nets are used to segment the three orthogonal views separately. These networks are referred to as single-view networks because they process the orthogonal planes independently.
Finally, the raw probability maps provided by the axial, sagittal, and coronal models are integrated into a single final probability map. This aggregation step aims to regularize voxel prediction by considering the spatial information of the three orthogonal views. In order to integrate all the predictions together, the sagittal and coronal segmentations are resliced into the axial view so that the average segmentation can be coherently calculated as follows:
where \({p}_{\text{ax}}(x)\), \({p}_{\text{sag}}(x)\), \({p}_{\text{cor}}(x)\) are the voxel prediction in axial, sagittal, and coronal views respectively.
Then, the validation set is used to select the best threshold value to binarize the probability map computed with multi-view aggregation and obtain the final segmentation.
We refer to this integration step as a multi-view approach because results obtained with single-view models are merged together, thus considering axial, sagittal, and coronal views.
This integration step regularizes final predictions, avoids the local errors, and improves model accuracy (Fig. 2).
AAA Geometrical Analysis
Once the thrombus has been segmented from the CTAs, both lumen and thrombus segmentations are used as input to the geometric analysis pipeline, without the need for user interaction. Thrombus segmentation spatial extent is used to mask the lumen segmentation, which extends from the aortic root to the common iliac arteries. To perform the geometrical analysis, both lumen and thrombus isosurfaces are reconstructed using Marching Cubes algorithm.14 To analyze only the abdominal aneurysm and exclude the healthy tracts of the aortic lumen, the lumen segmentation is clipped considering the first and last slices where the thrombus is segmented. Then, the clipped lumen segmentation and the thrombus segmentation are paired to obtain the abdominal aneurysm segmentation.
Given the aneurysm segmentation, the first step in the geometric analysis pipeline involves the automatic extraction of the centerline from the 3D model of the aneurysm. Using the VMTK library,1 the centerline can be extracted by specifying its initial and final seed points. In our pipeline, the source seed is automatically computed as the centroid of the first slice of the cropped lumen segmentation, whereas target seeds can be one or two (in the case of iliac involvement) and are calculated as the centroid(s) of the lumen segmentation in the last slice.
A smoothing factor of 0.5 and a resampling step of 2.5 are set as centerline generation parameters (Fig. 3).
Once the centerline is obtained, the cross-sections are generated at every point on the centerline using the vmtkcenterlinesections function. The same centerline is adopted to extract both the lumen and total aneurysm cross-sections. These sections are generated perpendicular to the computed centerline, and for each section the maximum diameter is computed. Finally, the section with the maximum aneurysm diameter value is extracted and both the aneurysm and lumen diameters calculated on this section are compared with manual measurements.
To summarize, the developed pipeline is able to:
-
1.
Reconstruct the lumen and thrombus polygonal meshes from their segmentations.
-
2.
Extract the total aneurysm and lumen diameters.
-
3.
Compute the volumes of the lumen and of the total aneurysm.
Data Analysis
The segmentations predicted by the multi-view segmentation network are compared against manual annotations.
-
Dice Similarity Coefficient (DSC) This metric is used to evaluate the segmentation performance as an overlap measure between the predicted and the manual segmentations. By denoting the manual segmentation as the Ground Truth (GT) and the predicted value with P, the DSC between two binary segmentations is defined as follow:
$$DSC = \frac{{2\left| {GT \cap P} \right|}}{{\left| {GT} \right| + \left| P \right|}}$$ -
Symmetric Surface to Surface Distance This metric is used to evaluate how far the surfaces obtained from the predicted and GT segmentations are from each other. The distances are computed using the distance maps proposed by Maurer et al..16
The results obtained from automatic geometric analysis are compared to manual measurements, performed by an expert surgeon (G.S.), through different approaches.
-
Intraclass Correlation Coefficient 1-1 (ICC 1-1) This coefficient estimates the degree of absolute agreement among any two measurements made on randomly selected objects. It is used to compare model measurements with the measurements made by an expert. To compute the coefficient, the approach described by A. Salarian in2 has been implemented. First of all, the measurement matrix M is created and the number of rows and columns of M are denoted as n and k respectively. Secondly, mean squared error for the rows (MSR) and within-judge variability (MSW) has to be computed.
Hence, ICC 1-1 is defined as:
$$ICC_{1,1} = \frac{MSR - MSW}{{MSR + \left( {k - 1} \right)MSW}}$$ -
Mean absolute difference This index expresses the mean difference between the measured values and their real values. By denoting the real value with GT, the measured value with P, and the number of data with N, the mean absolute difference is computed as follow:
$$\mathop \sum \limits_{i = 1}^{N} \frac{{\left| {GT_{i} - P_{i} } \right|}}{N}$$Both automatic segmentation and geometric analysis results are compared to manual measurements using:
-
Interquartile range (IQR) This metric quantifies the 50% extension of the element distributions located around the median value. It is based on dividing a rank-ordered dataset into four equal portions. The values that separate parts are the first, second, and third quartiles, denoted by Q1, Q2, and Q3 respectively. The IQR range lies between the lower and upper quartiles, Q1 and Q3.
Q1 = median of the n smallest values
Q3 = median of the n largest values
where n depends on the number of elements in the dataset.
Experiments and Results
Experimental Settings
The 85 preoperative CTAs included in the dataset were acquired in the same hospital. The Convolutional Neural Networks employed for multi-view thrombus segmentation are implemented in Keras5 framework based on Tensorflow6 platform supported with GPU. The binary cross-entropy is chosen as loss function and it is minimized with a mini-batch gradient descent method, using Adam optimizer (learning rate = 0.0001). At each iteration, the random mini-batch is augmented through random rotations, shifts, and zooms. To prevent overfitting during the training process, early stopping criteria are adopted and the patience is set to 15 epochs. Training, validation, and testing scripts are developed using Python 3.6 on a NVIDIA GeForce RTX 2080 Ti graphic card with CUDA compute capability = 7.5, under Windows operating system. Table 1 summarizes the hyperparameters used for the four CNNs.
Aneurysm geometry analysis is implemented using functionalities of the Vascular Modeling ToolKit (VMTK).1
Thrombus Segmentation
Table 2 shows the dimension and the number of images, the training time, and the number of epochs in which the coarse segmentation and the three networks achieved the best model configuration on the validation set. The training process is time expensive for all the three views networks. After the single-view training, the validation set has been used to compute the best threshold value (Th= 0.4). This threshold is used to binarize the final segmentation, obtained after the multi-view integration step.
The quantitative results obtained using single-view and multi-view models on the test set composed by 14 CTA patients are reported in Fig 4; in particular, for each patient, the computed DSC, mean and maximum distance are shown.
Figure 4(a) shows that the DSCs obtained with the multi-view integration are higher than the ones provided by an approach based on the single view. The plots of mean and maximum distances (Figs. 4(b) to (c)) show how the integration step reduces the surface to surface distance obtained with the single-view segmentations. In particular, Patients 1 and 3 present large mean and maxima distances in the axial and sagittal views respectively. At the same time, Patient 11 presents a very large peak in maximum surface distance in the sagittal view. These errors occur because patients' single-view segmentations include wrong parts with similar texture and pixel intensities to the abdominal thrombus, but located in another area. For example, axial segmentation of Patient 1 includes part of the thoracic aortic thrombus, although there were no examples of it in the ground truth segmentations because the model is focused on the abdominal aortic section. Because of these extra segmented parts, single-view segmentations return large maxima and mean distances from the ground truth. Despite this, in all the three cases those peaks are not present in the integrated segmentation. Indeed, the integration step allows to exclude the false recognitions from the final segmentation, improving mean and maxima distances. Moreover, it can be noticed that in more than one patient one of the three views returns poor results, and despite this the final segmentation is equal to or better than the view that returns more satisfactory results. It means that the integration step gives enough stability to our method, preventing that a single wrong segmentation strongly affects the final one.
The final results reported in Table 3 confirm the discussed plots.
Figures 5(a) and (b) display the 3D reconstructions of the thrombus segmentation predicted by the network and produced by the experienced operator, respectively. Comparison between prediction and ground truth is displayed in (c), overlapping the surfaces. The agreement between both reconstructed thrombi is satisfactory, despite spurious points.
Aneurysm Geometrical Analysis
Geometric analysis to retrieve information on aneurysm volume, and lumen and total aneurysm diameters is computed on the 14 patients of the test set for thrombus segmentation. Maximum diameters of the total aneurysm and the lumen are computed over the same perpendicular centerline section, by considering the section with the largest total diameter. The automatic pipeline returns a mean maximum total aneurysm diameter of 55.3 \(\pm\) 11.5 mm, and a mean lumen diameter of 32.3 \(\pm\) 10.3 mm.
By coupling the aorta and thrombus 3D reconstructions, the total aneurysm and abdominal lumen volumes can be computed. The mean total volume of the 14 patients is assessed to be \(143.6 \pm\) 104 \({\text{cm}}^{3}\), while the mean abdominal lumen volume is 68.2 \(\pm\) \({58.7 \text{cm}}^{3}\).
The maximum total aneurysm and lumen diameters extracted are compared with the ones manually measured by an expert in order to evaluate the performances obtained with the geometrical analysis. Mean absolute difference, Intraclass Correlation Coefficient, and Interquartile range of absolute difference are reported in Table 4. These metrics are computed separately for the total aneurysm and the lumen.
Figure 6 reports an example of the manual measurements extraction of maximum and minimum diameters of the total aneurysm and lumen in the axial view. Figure 7 shows the 3D reconstruction of the same patient, the section extraction made by our model and the representation of the maximum diameter section of the aneurysm and the lumen.
(a) Manual measurements of the total aneurysm and (b) lumen diameters from CTAs. Both measurements are computed using Endosize software.10
(a) 3D reconstruction of lumen and thrombus segmentations. (b) Sections for both lumen and total aneurysm, generated perpendicular to the centerline with the maximum aneurysm diameter section highlighted (purple). (c) Section with maximum aneurysm diameter: thrombus area is in red; lumen area is in green.
Discussions
Starting from our previous work on lumen segmentation,7 the aim of this work is to extend the proposed segmentation model for thrombus segmentation, allowing a fully automatic analysis of AAAs.
Once the multi-view segmentation method has been trained to extract the thrombus from the CTAs, the automatic extraction of aneurysm geometric information has been addressed. In fact, aneurysm treatment involves measurements of diameters and volumes in order to decide the best procedure to follow.
In this work, by coupling the models for lumen and thrombus segmentation, a fully automatic pipeline for aneurysm measurements extraction has been implemented and validated.
The results obtained with thrombus segmentation are promising. The mean DSC achieves 0.89, providing satisfactory results compared to the state-of-the-art works as briefly discussed in the following.
Table 5 summarizes the main aspects of literature works approaches for thrombus segmentation and analysis, considering only the ones that provide comparison with ground truth.
As in our study, the datasets of the four works reported in Table 5 are obtained from different CT scanners. However, the datasets of these studies include fewer CTAs. López et al.13 present a dataset similar to ours in terms of the number of data but uses a 3D approach, while we split each CTA into the three orthogonal views. In this way, the dataset used to train each neural network involved in our pipeline is much larger (Table 2). For what concerns the validation of the manual segmentations, inter-observer variability is performed over three12 or two3 operators and no intra-observer variability is provided.
Among deep learning-based methods, the performance of our model is consistent with the ones reported in both the works of Lòpez et al.,12,13 but unlike them it presents a 2.5D approach by combining three single-view U-Net CNNs with a final multi-view integration step. Since 3D networks require a lot of data and computation power, several studies24 have suggested the use of 2.5D segmentation approaches that fuse volumetric spatial information into 2D CNNs to improve the accuracy while reducing the computational cost. Integrating the segmentations obtained from different planes allows to decrease the computational cost and improve the accuracy of the final segmentation, as will be shown in the results. Moreover, the results obtained by Caradu et al3 using PRAEVAorta software are comparable with those obtained by Lòpez et al.12 exploiting the 2D approach. Finally, our pipeline does not require a priori knowledge of the lumen region, as in the case of the work of Lareyre et al.11 which addressed a feature-based approach strongly dependent on lumen segmentation, and as in the case of Caradu et al.3 which set lumen segmentation as the initial level for segmenting the thrombus.
For what concerns the maximum aneurysm diameter extraction, Lòpez et al.12,13 perform the diameter computation considering the axial segmentation of the aneurysm, without computing the lumen centerline. For each segmented slice the minimum enclosing circle of the aneurysm mask is computed, and finally the largest diameter is selected among those extracted. In their work, aneurysm diameter extraction is performed using this automatic method on both network-predicted and manual segmentations. Therefore, the mean absolute maximum diameter difference is only used to assess the quality of the automatic segmentation method, as the obtained maximum diameter is not compared with the diameter manually measured by the expert.
In contrast, Caradu et al.3 calculate aneurysm diameters starting from centerline extraction and cross-sections computation. The centerline was computed based on the lumen segmentation and defined as the path along the lumen keeping the largest distance to lumen boundaries. The maximum transverse diameter was then defined as the maximum diameter computed in the planes orthogonal to the centerline, including the lumen and the thrombus. In addition, they compare the diameters obtained with the automatic method to those manually measured by a junior and an experienced surgeon.
In the clinical practice, experts visually inspect the CTA slices and take measurements of the diameters in the slice where the thrombus diameter appears larger. The measurements of lumen and aneurysm diameters depend on the choice of the target CTA slice, and the slice identification is subject to inter- and intra-operator variability. In our automatic pipeline, the centerline extracted from the lumen segmentation is used to compute the aneurysm and lumen diameters from the corresponding 3D models. The diameters are extracted from sections that are perpendicular to the lumen centerline, and the section with maximum aneurysm diameter is chosen. Thus, the difference between automatic and manual measurements, reported in Table 4, may depend on the planes selected to perform the measurements. Moreover, Singh et al.22 have recorded an absolute interobserver difference of the maximal infrarenal aortic diameter of 82% between interobserver pairs and a mean interobserver variability in the infrarenal aortic section of 4.2 mm. Caradu et al.3 dealt with this aspect too by computing the difference in the maximum infrarenal aortic diameter obtained by a senior and a junior surgeon. The evaluation returns a mean absolute difference of 2.8 \(\pm\) 3.8 mm. It means that manual measurements are not objective and hardly reproducible, hence model results on measurements extraction strongly depend on the manual measurements taken.
Our measured ICC value agrees with the one provided by Caradu et al.,3 where they show that the semi-automatic manual segmentations present low interobserver variability given intraclass coefficient values greater than 0.90. In addition, our IQR of the mean absolute difference in total aneurysm maximum diameter is smaller than those in Caradu et al.3 It means that the spread of the middle 50% of values is quite smaller and our measurements have less variability. Hence, we can state that the accuracy of segmentation and analysis of AAA proposed in this work performs similar to analysis made by experts and takes considerably less time than manual segmentation and analysis. In particular, Table 3 shows that the proposed automatic thrombus segmentation takes about 1 minute versus the 25-40 minutes required to perform manual segmentation,11 and geometry analysis for each patient is performed in about 2 minutes, as reported in Table 4.
Limitations
Although the proposed segmentation model achieves a very good performance, in future works it might be necessary to increase the training and validation dataset in order to deal with patients morphology variability. In particular, patients with very large and very small aneurysms should be included in the dataset, since our dataset involves mostly medium-sized aneurysms. Moreover, since the current model is trained only on preoperative CTA scans, postoperative CTA scans could be used to extend the dataset and provide more examples for the network.
Conclusions
The developed pipeline could be used for different purposes. First of all, the automatic analysis of AAAs would enable a fast screening of the CTA scans and a consequently faster diagnosis. Secondly, the automatic geometrical analysis would allow a faster selection of the patients suitable for EVAR procedure, and a faster collection of robust and reproducible measurements for the treatment. Moreover, the model might be used to compare the diameter and total volume of the aneurysm before and after the surgery. This would help in tracking the progress of the endovascular treatment in the long term.
Besides the clinical applications, the 3D thrombus reconstruction can be also exploited in the future to perform finite element simulations.8
Data Availability
None.
Code Availability
None.
References
Antiga, L., M. Piccinelli, L. Botti, B. Ene-Iordache, A. Remuzzi, and D. A. Steinman. An image-based modeling framework for patient-specific computational hemodynamics. Med. Biol. Eng. Comput. 46(11):1097–1112, 2008. https://doi.org/10.1007/s11517-008-0420-1.
Arash Salarian, Intraclass Correlation Coefficient (icc). https://www.mathworks.com/matlabcentral/fileexchange/22099-intraclass-correlation-coefficient-icc
Caradu, C., B. Spampinato, A. M. Vrancianu, X. Bérard, and E. Ducasse. Fully automatic volume segmentation of infrarenal abdominal aortic aneurysm computed tomography images with deep learning approaches versus physician controlled manual segmentation. J. Vasc. Surg. 74(1):246-256.e6, 2021. https://doi.org/10.1016/j.jvs.2020.11.036.
Chaikof, E. L., et al. The Society for Vascular Surgery practice guidelines on the care of patients with an abdominal aortic aneurysm. J. Vasc. Surg. 67(1):2-77.e2, 2018. https://doi.org/10.1016/j.jvs.2017.10.044.
Chollet, Francois et al. Keras. 2015. https://keras.io
Developers, TensorFlow. TensorFlow. Zenodo 2021. https://doi.org/10.5281/ZENODO.5043456.
Fantazzini, A., et al. 3D automatic segmentation of aortic computed tomography angiography combining multi-view 2D Convolutional Neural Networks. Cardiovasc. Eng. Technol. 11(5):576–586, 2020. https://doi.org/10.1007/s13239-020-00481-z.
Haller, S. J., et al. Intraluminal thrombus is associated with early rupture of abdominal aortic aneurysm. J. Vasc. Surg. 67(4):1051-1058.e1, 2018. https://doi.org/10.1016/j.jvs.2017.08.069.
Hong, H. A. and U. U. Sheikh. Automatic detection, segmentation and classification of abdominal aortic aneurysm using deep learning. In 2016 IEEE 12th International Colloquium on Signal Processing & Its Applications (CSPA), Melaka, Malaysia, Mar. 2016, pp. 242–246. https://doi.org/10.1109/CSPA.2016.7515839.
Kaladji, A., A. Lucas, G. Kervio, P. Haigron, and A. Cardon. Sizing for endovascular aneurysm repair: clinical evaluation of a new automated three-dimensional software. Ann. Vasc. Surg. 24(7):912–920, 2010. https://doi.org/10.1016/j.avsg.2010.03.018.
Lareyre, F., C. Adam, M. Carrier, C. Dommerc, C. Mialhe, and J. Raffort. A fully automated pipeline for mining abdominal aortic aneurysm using image segmentation. Sci. Rep. 9(1):13750, 2019. https://doi.org/10.1038/s41598-019-50251-8.
López-Linares, K., et al. Fully automatic detection and segmentation of abdominal aortic thrombus in post-operative CTA images using Deep Convolutional Neural Networks. Med. Image Anal. 46:202–214, 2018. https://doi.org/10.1016/j.media.2018.03.010.
López-Linares, K., I. García, A. García-Familiar, I. Macía, and M. A. G. Ballester. 3D convolutional neural network for abdominal aortic aneurysm segmentation. 2019. https://doi.org/10.1016/j.media.2018.03.010.
Lorensen, W. E., and H. E. Cline. Marching cubes: A high resolution 3D surface construction algorithm. ACM SIGGRAPH Comput. Graph. 21(4):163–169, 1987. https://doi.org/10.1145/37402.37422.
Maiora, J., B. Ayerdi, and M. Graña. Random forest active learning for AAA thrombus segmentation in computed tomography angiography images. Neurocomputing. 126:71–77, 2014. https://doi.org/10.1016/j.neucom.2013.01.051.
Maurer, C. R., R. Qi, and V. Raghavan. A linear time algorithm for computing exact Euclidean distance transforms of binary images in arbitrary dimensions. IEEE Trans. Pattern Anal. Mach. Intell. 25(2):265–270, 2003. https://doi.org/10.1109/TPAMI.2003.1177156.
McCormick, M., X. Liu, J. Jomier, C. Marion, and L. Ibanez. ITK: enabling reproducible research and open science. Front. Neuroinform. 2014. https://doi.org/10.3389/fninf.2014.00013.
Óleary, S. A., E. G. Kavanagh, P. A. Grace, T. M. McGloughlin, and B. J. Doyle. The biaxial mechanical behaviour of abdominal aortic aneurysm intraluminal thrombus: classification of morphology and the determination of layer and region specific properties. J. Biomech. 47(6):1430–1437, 2014. https://doi.org/10.1016/j.jbiomech.2014.01.041.
Ronneberger, O., P. Fischer, and T. Brox, ‘U-Net: Convolutional Networks for Biomedical Image Segmentation’, ArXiv150504597 Cs, May 2015, doi: https://doi.org/10.1007/978-3-319-24574-4_28.
Schroeder, W., K. Martin, and B. Lorensen. The visualization toolkit: an object-oriented approach to 3D graphics; visualize data in 3D—medical, engineering or scientific; build your own applications with C++, Tcl, Java or Python; includes source code for VTK (supports Unix, Windows and Mac), 4th ed. Clifton Park, NY: Kitware Inc, 2006.
Shen, D., G. Wu, and H.-I. Suk. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19(1):221–248, 2017. https://doi.org/10.1146/annurev-bioeng-071516-044442.
Singh, K., et al. Intra- and interobserver variability in the measurements of abdominal aortic and common iliac artery diameter with computed tomography. The Tromsø study. Eur. J. Vasc. Endovasc. Surg. 25(5):399–407, 2003. https://doi.org/10.1053/ejvs.2002.1856.
Wolf, I., et al. The medical imaging interaction toolkit. Med. Image Anal. 9(6):594–604, 2005. https://doi.org/10.1016/j.media.2005.04.005.
Zhang, Y., Q. Liao, and J. Zhang. Exploring efficient volumetric medical image segmentation using 2.5D method: an empirical study. ArXiv201006163 Cs Eess, 2020. http://arxiv.org/abs/2010.06163
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information
Associate Editor Igor Efimov oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brutti, F., Fantazzini, A., Finotello, A. et al. Deep Learning to Automatically Segment and Analyze Abdominal Aortic Aneurysm from Computed Tomography Angiography. Cardiovasc Eng Tech 13, 535–547 (2022). https://doi.org/10.1007/s13239-021-00594-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13239-021-00594-z