Abstract
Cytological and simple sequence repeat (SSR) diversity of eight bananas and four plantains (Musa spp.) of the section Eumusa were analyzed. The chromosome characteristics and karyotype asymmetry reconfirmed triploid chromosome number of 2n = 3x = 33 in all varieties of Musa acuminata besides first reports in var. Desi Daskini Sagar and var. Shankara. Smaller chromosomes were observed in cooking varieties as compared to dessert bananas with a variation of total chromosome length from 39.75 µm in var. Shankara to 81.50 µm in var. Kathia. Metacentric chromosomes were recorded in all the varieties with total F% ranging from 35.65% in var. Amritapani to 45.31% in var. Desi Dakhini Sagara. Sub-median constricted chromosomes were prevalent in all studied varieties. The total chromosome volume and the interphase nuclear volume varied from 10.78 µm3 in var. Robusta to 32.24 µm3 in var. Desi Dakhini Sagara and 625.13 µm3 in var. Gaja Bantala to 2048.37 µm3 in var. Patakpura respectively. No correlation was found between interphase nuclear volume and chromosome length among the genotypes. The dendrogram created on the basis of five karyotype asymmetry indices [degree of asymmetry (A), intrachromosomal asymmetry index (A1), karyotype asymmetry index (AsK%), index of karyotype symmetry (Syi), and total form percentage (TF %)] exhibited different clusters for banana and plantain. A total of 89 amplicons were obtained with 22 SSR markers with an average of 1.68 polymorphic bands having a size range of 120–300 bp. SSR markers analysis in all 12 Musa varieties showed no polymorphism between var. Robusta and var. Grand Naine. Maximum polymorphism (72%) was found among cooking types besides var. Chini Champa of tabletop type. The mean polymorphism among SSR primers was 41.89% in the studied varieties. Out of the two major clusters formed in the dendrogram, Cluster-I showed with all the tabletop varieties and Cluster-II with all cooking varieties. Cluster analysis confirmed more genetic homogeneity among tabletop varieties as compared to the cooking varieties where var. Kathia showed distinct differences in their genetic makeup forming sub-cluster-II with genetic heterogeneity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana belongs to the family Musaceae is one of the principal food for about 400 million people across the globe [34]. Plant parts are used as alternative sources of aesthetics, fiber, and medicine [30] besides, ornamental species like Musa laterita, M. ornata, fiber yielding species like M. textilis. The production and marketing of bananas largely contribute to the livelihood of many peoples in tropical and sub-tropical countries. India is the largest producer of bananas and has widespread biodiversity [21]. Genetically important wild and cultivated Musa varieties found across the country are useful in heterosis breeding programs. The seed sterility of cultivated clones remains a concern in producing new and resistant cultivars through breeding [40]. The challenges faced in banana production due to the biotic and abiotic stresses can be dealt with by conserving the vast genetic resources and its genetic characterization. Moreover, the management of properly characterized Musa germplasm is important because of the high expense and time consumption in maintaining field gene banks for their vegetative mode of propagation. The Indian banana genome is not completely understood and remains a domain of exploration and deep digging.
The genera of Musa and Ensete belong to the family Musaceae. Out of this the wild and domesticated plantains and bananas formed the Musa group represented by five sections namely Eumusa (x = 11), Callimusa (x = 9, 10), Australimusa (x = 10), Rhodochlamys (x = 11), and Ingentimusa (x = 14) represented by only species M. ingens [2, 6]. Eumusa and Rhodochlamys have merged into new sections Musa in the recent classification, and Australimusa which were placed under Callimusa [25]. The primitive genus Ensete is present in tropical Africa and Asia, represented by a few numbers of existing species. Most of the edible bananas having chromosome number x = 11 belong to Eumusa that consists of about eleven species. Two wild species i.e. M. acuminata having AA genome and M. balbisiana having BB genome of section Eumusa crossed naturally and the majority of domesticated varieties thus originated. It resulted in hybrids with different ploidy levels and genomic constituents like diploid (AA, BB, AB), triploid (AAA, AAB, ABB) and also tetraploid (AAAB, AABB, ABBB) [49]. Cultivated banana (Musa spp.) can be divided into either plantain (starchy even after ripening and generally made palatable by boiling or frying and roasting) or tabletop banana (sweet in taste, easily digestible, and consumed raw when ripened). Karyotype variations among varieties provide useful information regarding their apparent origin and evolution. Due to random and repeated translocation, insertion, deletion, and transposition events, the chromosomal characteristics are subsequently altered which can be detected in their metaphase chromosome morphology and karyotype. The evolution of important variations can be analyzed by statistical indices that measure the degree of deviation from a standard karyotype that has all metacentric chromosomes of equal length [51]. A number of intra-chromosomal asymmetries (asymmetry increases with increase in chromosomes with the sub-telomeric or telomeric position of the centromere) and inter-chromosomal asymmetries (asymmetry increases with increase in variation of chromosome length in a chromosome complement) indices have been proposed since Lewitsky [33] to Peruzzi et al. [42]. Afterward, notable studies on karyotype asymmetry in various plant species were carried out [33, 54] including banana [18].
DNA markers or molecular markers like AFLP (Amplified Fragment Length Polymorphism), CAPS (Cleaved Amplified Polymorphic Sequence), RAPD (Random Amplified Polymorphic DNA), RFLP (Restriction Fragment Length Polymorphism), retrotransposons, SCAR (Sequence Characterized Amplified Region), and SSRs (Simple-Sequence Repeats) are being extensively used in the last two decades due to reliability and effectiveness in characterizing various plant species [22]. Due to advancement in sequencing technologies, identification of novel sequence-based DNA markers such as GBS (Genotyping-by-sequencing), NGS (Next-Generation Sequencing) based marker system, diversity array technology (DArTSeq), and SNP (Single Nucleotide Polymorphism) markers have facilitated genetic marker development and characterization of more complex genomes [37]. Simple sequence repeats or microsatellites, one type of repetitive DNA sequence (repeats of motif sequence of 1–6 bp length), are quite abundant in plant genomes. Among other available markers, the SSR marker was widely used in molecular breeding because of its high polymorphism, codominant inheritance, efficiency, ability to detect allelic variation [48]. SSR markers still remain efficient in revealing the genetic diversity in M. acuminata [7, 10, 12, 31, 36, 55] and in M. balbisiana [4, 43, 45]. The availability of developed Musa SSR is still far behind that of other crop plants. Christelová et al. [8] standardized a genotyping procedure with a set of 22 SSR markers for Musa [26, 31]. Later on, Christelová et al. [7] used this set of 22SSR markers to characterize 630 Musa accessions of the ITC (International Transit Centre), Leuven, Belgium, and efficiently detected genetic variation that reinforced the morphological and cytogenetic basis of classifications. These markers were used efficiently in marker-assisted selection on banana genotypes besides genetic diversity study [16, 38]. However, Backiyarani et al. [3] recently developed a database (MusatransSSRDB) having the information on transcriptome-derived SSRs. But information on SSR markers of Indian banana varieties is very scanty which can be explored for the genome-specific marker development. Moreover, the species diversity in India needs to be cytogenetically explored for its productive utilization in the breeding and systematic study. Thus, this study is focused on a combined approach of karyotype analysis and SSR marker study using 22 SSR markers of Christelová et al. [8] in twelve varieties of plantains and tabletop Musa available in India to find out genetic diversity, ploidy level, and evolutionary relationships, if any, for utilization in genome characterization and banana breeding program.
Material and methods
Materials
A living collection of twelve varieties of Musa L. of section Eumusa from different places of Odisha have been kept in the net house of the Botany Department, Utkal University, Bhubaneswar, Odisha (Table 1).
Chromosome preparations
Young fresh root tips having 0.5 cm–1.0 cm length of each Musa variety were collected at10.00 am. The root-tips from five individuals of each variety were pre-treated in para-dichloro benzene (pDB): aesculin (1:1) solution for 3½ h at 18 °C followed by overnight fixation in glacial acetic acid: ethanol (1:3) at room temperature. Root-tip squash preparations were done with 45% glacial acetic acid after cold hydrolysis and 2% orcein staining [18]. Scattered chromosome plates were micro-photographed under Olympus BX-53 microscope with 100 × magnification. Measurements were taken with Q-Capture Pro 7 (Canada) software. At least five well-scattered metaphase chromosome plates were chosen out of 10–15 preparations for each variety for getting measurement accuracy of the karyotypes.
Chromosome measurements and statistical analysis
The total chromosomes length and volume of the karyotype were calculated as per the formulae used in our earlier paper [18]. The total form percentage (TF %) of a variety calculated from the individual Form percentage (F %) which is the average of all F% of individual chromosomes of a karyotype [32]. The Interphase Nuclear Volume (INV) was calculated applying formula 4/3πr3, where ‘r’ is the radius of the chromosome [15].
Idiograms based on mean values of the relative lengths and arm ratios of the chromosomes were analyzed and presented in bar form. Chromosomes of karyotype were classified in terms of the position of its constrictions and length of the chromosome. Type A = median constricted chromosomes with nearly median to sub-median secondary constriction (1.66–3.93 µm); Type B = sub-median constricted chromosomes having nearly sub-median secondary constriction (2.33–2.98 µm); Type C = nearly median primary constricted chromosomes (0.91–3.18 µm); Type D = nearly sub-median primary constricted chromosomes (0.74–3.69 µm). Besides the karyotype asymmetry, other karyotype parameters were calculated from five pairs of karyotype parameters to obtain scatter plots [19, 41, 54]. The dendrogram was constructed with the help of NTSysPC software version 2.20 [44] using five intra-chromosomal asymmetry indices like degree of asymmetry (A), intrachromosomal asymmetry index (A1), karyotype asymmetry index (AsK%), index of karyotype symmetry (Syi), and total form percentage (TF %).
DNA isolation and purification
Genomic DNA of the 12 Musa varieties was isolated following CTAB (cetyl trimethyl ammonium bromide) method with some modifications [47]. Young cigar leaf (2.0 g) was taken in a mortar pestle with a pinch of PVP (Poly Vinyl Pyrrolidone) and crushed into powdered form in liquid nitrogen. DNA extraction buffer with the composition of Tris HCl (100 mM, pH 8.2), EDTA (20 mM) NaCl (0.5 M, w/v), CTAB (2%, w/v) and beta-mercaptoethanol (2%, v/v) was added in equal volume to it and incubated at 60 °C for an hour in a water bath. The mixture was cooled down to room temperature, then centrifuged after emulsifying it with 24:1 chloroform: isoamyl alcohol solution. The precipitation of genomic DNA was made by adding chilled isopropanol into supernatant following dehydration in 70% ethanol and dissolved in T10E1(Tris EDTA) buffer (pH 8.2).Genomic DNA quality and quantity of all the varieties were verified in 0.8% agarose gel. The presence of traces of RNA impurity was purified with the treatment of 150 µl of RNAase-A treatment (10 mg ml−1 stock) at 37 °C for 2 h. Subsequently, precipitation of DNA was done in chilled ethanol mixing with 3 mM sodium acetate following 24:1 chloroform: isoamyl alcohol separation. Further purification was carried out using a silica-based purification kit (Thermo Fisher Scientific) to obtain purified DNA suitable for use in PCR (Polymerase Chain Reaction). The final dilution (25 ng µl−1) of purified DNA was prepared using T10E1 buffer for PCR reactions.
Polymerase chain reaction
PCR was performed as per the protocol of Willams et al. [56] to generated SSR profiles using the set of 22 Musa-specific primers used earlier [8]. Each PCR reaction mixture (25 µl) was prepared as per our earlier protocol [52]. Briefly, dNTPs, Taq DNA polymerase, 10 × PCR assay buffer, 25 ng of primer, and 25 ng of template DNA were mixed and the PCR reaction was performed in a thermal cycler (Bio-Rad T100, USA) following our earlier protocol [52]. The amplificons were separated in 1.5% agarose gel at a constant 55 V for 1½ h and were stained with ethidium bromide (0.5 μg ml−1) dissolved in TAE buffer (40 mM Tris–acetate, pH 8.0; 2 mM EDTA). DNA bands were visualized using a gel documentation system (Geldoc XR system, Bio-Rad, USA) under UV light and were photographed. Gene ruler (100 bp DNA ladder) was used as the size standard in the gel. The PCR reactions were performed three times for each sample and only constantly obtained amplification products were scored.
Data analysis
The size of the amplified bands was scored for each allele in each variety for each set of primers. The Excel score sheet obtained from the SSR profile was used to prepare a similarity matrix [29]. A dendrogram was created by cluster analysis following the UPGMA (unweighted pair group method with arithmetic mean) using the NTSysPC version 2.20 [43].
Results and discussions
Cytological characterization
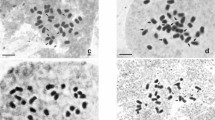
Musa exhibits a range of ploidy levels which was developed in the natural hybridization process of M. acuminata (AA genome) and M. balbisiana (BB genome)—two wild diploid species. The chromosome number in studied varieties is variable in different sections of the family Musaceae with Eumusa having x = 11 chromosomes [49]. In our study, it was found that all varieties of banana and plantain had 2n = 3 × = 33 (Fig. 1, Table 2, Supplementary Table 1). The traditional means of squash preparation and counting of chromosome numbers are still desirable before confirming the ploidy determination by non-counting methods e.g. flow cytometry or by meiotic chromosome study [1]. The chromosome size varied from small to medium in the 12 studied Musa varieties of the present study (Table 2, Supplementary Table 1). The disparity between small and medium chromosomes was quite considerable. Chromosome numbers were found to be the same in all the varieties (Fig. 1, Table 2). The length of the total chromosome varied from 39.75 µm in var. Shankara to 81.50 µm in var. Kathia. Relatively median and nearly median chromosomes were predominant in all twelve varieties. The TF% varied from 35.65% in var. Amritapani to 45.31% in var. Desi Dakhini Sagara. The number of secondary constrictions was 3 in var. Grand Naine, var. Robusta, var. Gaja Bantala to 12 in var. Amritapani, var. Desi Dakhini Sagara. The length of the total chromosome varied from 10.78 µm3 in var. Robusta to 32.24 µm3 in var. Desi Dakhini Sagara. Distinct karyotype variations were observed in different varieties of studied bananas as evident by karyograms and idiograms (Fig. 2). The Interphase Nuclear volume varied from 625.13 µm3 in var. Gaja Bantala to 2048.37 µm3 in var. Patakpura (Table 2). The variation in the number of Type C, Type D suggest the origin of Type-A chromosomes because of structural alteration of chromosome arms through translocation during the micro-evolution [13]. A greater variation and presence of Type C chromosomes rather than Type D chromosomes in most of the species suggested the greater rate of translocation of Type D chromosome to Type C chromosome during the early evolution. The variation in the karyotype formula that indicates structural changes might have led to the diversification of bananas and plantains [14, 23]. The alteration in the TF% might be due to structural alteration of chromosomal arms at an early stage of the natural evolutionary process in Musa as evident by fluorescent in situ hybridization in somatic and meiotic chromosomes [1, 7, 9, 18, 23].
Metaphase plate of the twelve Musa acuminata varieties, a var. Amritapani, b var. Champa, c var. Grand Naine, d var. Patakpura, e var. Dwarf Cavendish, f var. Kathia, g var. Chini Champa, h var. Robusta, i var. Gaja Bantala, j var. Paunsia Bantala, k var. Shankara, l var. Desi Dakhini Sagara. Bar = 10 µm
The karyotype study of all varieties consisted of chromosomes of the nearly median, median, nearly submedian, and sub-median types (Table 2, Supplementary Table 1, Fig. 3). However, the origin of the cultivated varieties was suggested from wild varieties with nearly median and nearly sub-median type chromosomes [18, 23] following the process of reciprocal translocation as evident in M. acuminata ssp. malaccensis [1]. According to Stebbins’s [51] karyotype system, the 2A, 1B, 2B 2C typification of karyotypes were found among the studied varieties (Fig. 3, Table 3).The recorded variation of intra-chromosomes was found mostly the same in all the varieties except var. Paunsia Bantala and var. Gaja Bantala (1B). The inter-chromosomal asymmetry varied among the varieties with the highest value in var. Dwarf Cavendish (2C). The karyotype asymmetry parameters like AI (heterogeneity of chromosome length and/or centromeric index), A1 (intra-chromosomal asymmetry index), A2 (inter-chromosomal asymmetry index), CVCL (relative variation in chromosome length), CVCI (relative variation in centromeric index), DI (Dispersion Index), MAR (Mean Arm Ratio), MCA (Mean Centromeric Asymmetry), Syi (index of karyotype symmetry) varied significantly among the varieties (Table 3). It was observed that most of the intra-chromosomal asymmetry parameters like MAR, AsK% (karyotype asymmetry index), A1, AI, MCA had peak values in var. Dwarf Cavendish and most of the inter-chromosomal asymmetry parameters like A2, DI, CVCL had peak values in var. Desi Dakhini Sagara. Cyto-morphological data create an immense impact on plant evolutionary and diversification studies [35, 50, 51]. With the chromosome number remaining constant, the increasing karyotype asymmetry can be best explained through pericentric inversion and unequal translocation that occurred in the primitive karyotypes leading to advanced ones [51, 54].
Scatter plots between a pair of intra- and inter-chromosomal indexes differentiate the varieties according to their asymmetry value. Karyotype with higher intra-chromosomal asymmetry positioned higher on Y-axis and with higher inter-chromosomal asymmetry placed more to the right side of the X-axis. The asymmetry difference found less among the tabletop varieties as compared to cooking varieties (Fig. 3). The karyomorphological representation of the karyotypes by karyogram (Fig. 2) and scatter plots found highly correlated, except the Rec versus Syi plot (Fig. 3d). None of the scatter plot diagrams efficiently described the intra-chromosomal asymmetry evident from the contradictory positions of scattered value according to the intra-chromosomal asymmetry index of varieties. The dendrogram constructed using the five asymmetry parameters (A, A1, AsK%, Syi, and TF%) showed affinity among the dessert banana varieties that placed all the studied varieties under a major cluster due to similar asymmetry values where Cavendish group like var. Dwarf Cavendish, var. Grand Naine and var. Robusta formed a sub-group except for var. Chini Champa and var. Kathia. These two varieties formed a sub-cluster in Cluster-II having AAB genomic constituent, which suggests their ancestry with cooking type. The cooking varieties also got separated from the dessert varieties forming separate clusters with more genetic affinity (Fig. 4).
Molecular characterization
The 22 sets of SSR primers have been tested for polymorphism in 12Musa varieties and produced 89 amplified fragments of ~ 140–230 bp in size (Fig. 5a, b, Table 4). An average polymorphic percentage was recorded at 41.89% among the primers for all studied varieties. A number of 37 alleles found polymorphic out of the total amplified 89 bands that formed 41.89% polymorphism. The dendrogram obtained from SSR markers showed Cluster-I with all the dessert banana varieties and Cluster-II with cooking varieties. Cluster analysis confirmed more genetic homogeneity among banana varieties as compared to the plantain varieties where var. Kathia showed distinct differences in their genetic makeup. Cluster-I differentiates var. Champa, var. Chini Champa, var. Grand Naine, var. Robusta var. Amritpani and var. Patakpura forming single Sub-cluster-I. The Cluster-II also formed Sub-cluster-I and –II with two plantain varieties each (Fig. 6). The genetic relationships of var. Kathia showed high genetic resemblance with Cluster-I (with mostly banana) forming an outgroup (Sub-cluster-II).The involvement of a non-reduced 2n gamete and an n gamete in the natural hybridization process might have resulted in the modern triploid banana [40]. The tabletop banana diversity might be due to their origin from a common ancestor clone and their subsequent vegetative propagation following somatic mutations or involvement of genetically related parents in some subgroups through the sexual reproduction process. The diversity of plantain might be due to somatic mutations of few introduced varieties as reported from time to time [12, 17, 49, 53]. Meiotic restitution in bananas due to unbalanced segregation of chromosome complements through monads, dyads, triads, and tetrad stage of diploid banana might be a cause of genetic variation in the sexual reproduction process [1]. However, the low genetic differences among various accessions of a subgroup cannot elucidate the huge phenotypic diversity observed inside the subgroups [27]. Only a few primers among the tested primers produced amplified fragments in our analyzed varieties. Earlier studies also reported a low fraction of primer amplification using SSR markers [39]. In addition, the primer-anchoring sequences flanking the microsatellite loci might vary adequately to avoid product amplification [11]. So, new SSR primers are required for the genetic analysis of Musa as variations in band intensities were reported in Musa [11] which is in accordance with our finding (Fig. 1). The variations in band intensity could be due to mutations in annealing sequences [28] leading to a null allele or competitive amplification [5]. The alleles of the same length (bp) might have arisen from altered alleles with convergent evolution, and could be the possible elucidation in the evolution of microsatellite markers [20]. In this study, the SSRs used for the phylogenetic analysis of the studied varieties proved an efficient tool in describing the diversity among the varieties that appear consistent with the phylogenetic tree obtained from the karyological data and asymmetry indices. However, capillary-based genotyping could resolve better genotypic information that can contribute more discrete information regarding the genetic variability and systematic position of the varieties [24].
SSR band polymorphism among the twelve varieties of M. acuminata amplified with primer a MuS2 and b MuS15. Lane M = Molecular weight marker, 1 = var. Amritapani, 2 = var. Champa, 3 = var. Paunsia Bantala, 4 = var. Desi Dakhini Sagara, 5 = var. Dwarf Cavendish, 6 = var. Grand Naine, 7 = var. Kathia, 8 = var. Chini Champa, 9 = var. Patakpura, 10 = var. Shankara, 11 = var. Robusta, l2 = var. Gaja Bantala
Conclusion
The genetic diversity in cultivated bananas (Musa spp.) has remained a major concern among germplasm conservationists and banana breeders. The detailed karyotype analysis of 12 tabletop bananas and plantains belonging to Eumusa provided structural details of somatic chromosomes of Musa. Our study reconfirms the interrelationship between the karyotype asymmetry and the SSR-based characterization for the phylogenetic analysis of bananas mostly cultivated in Odisha, India. The genetic diversity assessment is able to differentiate the banana and the plantain into separate groups and also demonstrate more genetic variability among plantains rather than the tabletop banana. The narrow genetic variability of tabletop bananas could be increased by the strategic banana breeding programs with a more heterogeneous group of bananas to tackle the disease susceptibility of tabletop bananas.
References
Ahmad F, Poerba YS, Kema GHJ, de Jong H. Male meiosis and pollen morphology in diploid Indonesian wild bananas and cultivars. Nucleus. 2021;64:181–91. https://doi.org/10.1007/s13237-021-00350-7.
Argent G. The wild bananas of Papua New Guinea. Notes R Bot Gard Edinb. 1976;35:77–114.
Backiyarani S, Chandrasekar A, Uma S, Saraswathi MS. MusatransSSRDB (a transcriptome derived SSR database)—An advanced tool for banana improvement. J Biosci. 2019;43:110–6.
Buhariwalla HK, Jarret RL, Jayashree B, Crouch JH, Ortiz R. Isolation and characterization of microsatellite markers from Musa balbisiana. Mol Ecol Notes. 2005;5(2):327–30.
Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–31.
Cheesman EE. Classification of the bananas. The genus Ensete Horan and the genus Musa L. Kew Bull. 1947;2:97–117.
Christelová P, De Langhe E, Hřibová E, Čížková J, Sardos J, Hušáková M, Sutanto A, Kepler AK, Swennen R, Roux N, Doležel J. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers Conserv. 2017;26(4):801–24.
Christelová P, Valárik M, Hřibová E, Channelière S, Roux N, Doležel J. A platform for efficient genotyping in Musa using microsatellite markers. AoB Plants. 2011. https://doi.org/10.1093/aobpla/plr024.
Čížková J, Hřibová E, Humplíková L, Christelová P, Suchánková P, et al. Molecular analysis and genomic organization of major DNA satellites in Banana (Musa spp.). PLoS ONE. 2013;8(1):e54808. https://doi.org/10.1371/journal.pone.0054808.
Creste S, Benatti TR, Orsi MR, Risterucci AM, Figueira A. Isolation and characterization of microsatellite loci from a commercial cultivar of Musa acuminata. Mol Ecol Notes. 2006;6(2):303–6.
Creste S, Neto AT, Vencovsky R, Silva SO, Figueira A. Genetic diversity of Musa diploid and triploid accessions from the Brazilian banana breeding program estimated by microsatellite markers. Genet Resour Crop Ev. 2004;51:723–33.
Crouch HK, Crouch JH, Madsen S, Vuylsteke DR, Ortiz R. Comparative analysis of phenotypic and genotypic diversity among plantain landraces (Musa spp., AAB group). Theor Appl Genet. 2000;101:1056–65.
Das AB, Das P. Estimation of nuclear DNA content and karyotype analysis in nine varieties of Musa acuminata. Cytobios. 1997;90:181–92.
Das AB, Dehery SK, Kiran, Jena SN, Sinha RK. A new diploid Indian accession of Musa laterita of Section Rhodochlamys reported from Gangtok, Sikkim with morphology, chromosome count and 2C DNA content. Cytologia. 2020;85(1):63–9.
Das AB, Mallick R. Varietal difference in 4C DNA content and chromosome characteristic of Coriandrum sativum L. Cytologia. 1989;54:609–16.
de Carvalho Santos TT, de Oliveira Amorim VB, dos Santos-Serejo JA, da Silva Ledo CA, Haddad F, Ferreira CF, Amorim EP. Genetic variability among autotetraploid populations of banana plants derived from wild diploids through chromosome doubling using SSR and molecular markers based on retrotransposons. Mol Breed. 2019;39(7):95. https://doi.org/10.1007/s11032-019-0996-1.
De Langhe E, Swennen R, Vuylsteke D. Plantain in the early Bantu world. Azania Archaeol Res Afr. 1994;29–30(1):147–60. https://doi.org/10.1080/00672709409511667.
Dehery SK, Panda E, Saha PR, Sinha RK, Das AB. Chromosome diversity and karyotype asymmetry analysis in four cultivated triploid and three diploid wild genotypes of Musa from North-East India. Nucleus. 2021;64:167–79. https://doi.org/10.1007/s13237-020-00334-z.
Eroğlu HE. Which chromosomes are sub-telocentric or acrocentric? A new karyotype symmetry/asymmetry index. Caryologia. 2015;68:239–45.
Estoup A, Jarne P, Cornuet JM. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol Ecol. 2002;11:1591–604.
FAO. Banana market review: preliminary results for 2018. Rome. Licence: CC BY-NC-SA 3.0 IGO. 2018. pp. 12.
Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F. Trends in plant research using molecular markers. Planta. 2018;247:543–57.
Ghosh S, Das A, Ghorai A, Jha TB. Comparative kayomorphology of edible Musa varieties of West Bengal. Caryologia. 2013;66:243–50.
Guichoux E, Lagache L, Wagner S, Chaumeil P, Léger P, Lepais O, Lepoittevin C, Malausa T, Revardel E, Salin F, Petit RJ. Current trends in microsatellite genotyping. Mol Ecol Resour. 2011;11:591–611.
Häkkinen M. Epitypification of some Musa sect. Callimusa Cheesman and Musa L. sect. Musa names from Vietnam (Musaceae). NeBIO. 2013;4:7–8.
Hippolyte I, Bakry F, Seguin M, Gardes L, Rivallan R, Risterucci AM, Jenny C, Perrier X, Carreel F, Argout X, Piffanelli P, Khan IA, Miller RNG, Pappas GJ, Mbéguié-A-Mbéguié D, Matsumoto T, De Bernardinis V, Huttner E, Kilian A, Baurens FC, D’Hont A, Cotel F, Courtois B, Glaszmann JC. A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol. 2010;10:65. https://doi.org/10.1186/1471-2229-10-65.
IPGRI-INIBAP (Bioversity)/CIRAD. Descriptors for Banana (Musa spp.) Rome IPGRI Montpellier: INIBAP; Montpellier: CIRAD. 1996. pp. 59.
Ishibashi Y, Saitoh T, Abe S, Yoshida MC. Null microsatellite alleles due to nucleotide sequence variation in the grey-sided vole, Clethrionomys rufocanus. Mol Ecol. 1996;5:589–91.
Jaccard P. Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat. 1908;XLIV(163):223–69.
Kennedy J. Bananas and people in the homeland of genus Musa: Not just pretty fruit. Ethnobot Res Appl. 2009;7:179–97.
Lagoda PJL, Noyer JL, Dambier D, Baurens FC, Grapin A, Lanaud C. Sequence tagged microsatellitesite (STMS) markers in the Musaceae. Mol Ecol. 1998;7:659–63.
Levan A, Fredya K, Sandberg A. Nomenclatyure for centromeric position on chromosome. Heridity. 1964;52:201–20.
Lewitsky GA. An essay on cytological analysis of the fixing action of the chrom-acetic formalin and the chromic formalin. Bull Appl Bot Genet Plant Breed. 1931;27:187–240.
Loeillet D. Close-up: banana. Fruitrop. 2008;155:3–39.
Medeiros-Neto E, Nollet F, Moraes AP, Felix LP. Intrachromosomal karyotype asymmetry in Orchidaceae. Genet Mol Biol. 2017;40:610–9.
Miller RN, Passos MA, Menezes NN, Souza MT, Costa MMC, Azevedo VCR, Amorim EP, Pappas GJ, Ciampi AY. Characterization of novel microsatellite markers in Musa acuminata subsp. burmannicoides, var. Calcutta 4. BMC Res Notes. 2010;3:148. https://doi.org/10.1186/1756-0500-3-148.
Nadeem MA, Nawaz MA, Shahid MQ, Doğan Y, Comertpay G, Yıldız M, Hatipoğlu R, Ahmad F, Alsaleh A, Labhane N, Özkan H. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2018;32(2):261–85.
Němečková A, Christelová P, Čížková J, Nyine M, Van den Houwe I, Svačina R, Uwimana B, Swennen R, Doležel J, Hřibová E. Molecular and cytogenetic study of East African highland banana. Front Plant Sci. 2018;9:1371. https://doi.org/10.3389/fpls.2018.01371.
Oriero CE, Odunola OA, Loco Y, Ingelbrecht I. Analysis of B-genome derived simple sequence repeat (SSR) markers in Musa spp. Afr J Biotechnol. 2006;5(2):126–8.
Ortiz R, Swennen R. From crossbreeding to biotechnology-facilitated improvement of banana and plantain. Biotechnol Adv. 2014;32:158–69.
Paszko B. A critical review and a proposal of karotype asymmetry indices. Plant Syst Evol. 2006;258:39–48.
Peruzzi L, Eroǧlu HE. Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet. 2013;7:1–9.
Ravishankar KV, Raghavendra KP, Athani V, Rekha A, Sudeepa K, Bhavya D, Srinivas V, Ananad L. Development and characterization of microsatellite markers for wild banana (Musa balbisiana). J Hortic Sci Biotechnol. 2013;88:605–9.
Rohlf FJ. NTSysPC: numerical taxonomy system, ver.2.20. NY: Exeter Publishing Ltd; 2008.
Rotchanapreeda T, Wongniam S, Swangpol SC, Chareonsap PP, Sukkaewmanee N, Somana J. Development of SSR markers from Musa balbisiana for genetic diversity analysis among Thai bananas. Plant Syst E. 2016;302:739–61.
Ruas M, Guignon V, Sempere G, Sardos J, Hueber Y, Duvergey H, et al. MGIS: managing banana (Musa spp.) genetic resources information and high-throughput genotyping data. Database. 2017. https://doi.org/10.1093/database/bax046 (Oxford).
Saghai-Maroof MA, Soliman KM, Jorgensen HA, Allard HA. Ribosomal DNA spacer length polymorphism in Barley. Mendelian inheritance, chromosomal locations and population dynamics. Proc Nat Acad Sci. 1984;81:8014–8 (USA).
Selkoe KA, Toonen RJ. Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett. 2006;9:615–29.
Simmonds NW, Shepherd K. The taxonomy and origin of the cultivated banana. J Linn Soc Bot. 1955;55:302–12.
Stace CA. Cytology and cytogenetics as a fundamental taxonomic resource for the 20th and 21st century. Taxon. 2000;49:451–77.
Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold (Publishers) Ltd; 1971. p. 216.
Tripathy P, Das AB. Morphological and molecular diversity of blackgram germplasm collectedfrom Odisha. Ecol Genet Genom. 2021;20:100088. https://doi.org/10.1016/j.egg.2021.100088.
Ude G, Pillay M, Ogundiwin E, Tenkouano A. Genetic diversity in an African plantain core collection using AFLP and RAPD markers. Theor Appl Genet. 2003;107:248–55.
Vimala Y, Lavania S, Lavania UC. Chromosome change and karyotype differentiation–implications in speciation and plant systematics. Nucleus. 2021;64:33–54. https://doi.org/10.1007/s13237-020-00343-y.
Wang JY, Zheng LS, Huang BZ, Liu WL, Wu YT. Development, characterization, and variability analysis of microsatellites from a commercial cultivar of Musa acuminata. Gen Resour Crop Evol. 2010;57:553–63.
Willams JGK, Kulelik AR, Livak J, Rafalski A, Tingey SV. DNA polymorphism identified by arbitrary primers are useful as genetic markers. Nucl Acid Res. 1990;18:6531–5.
Acknowledgements
The authors are thankful to the Head of the Botany, Utkal University for providing administrative and instrumental facilities developed under DRS-III, University Grant Commission, and FIST programme, Govt. of India to carry out the research. The financial support received by ABD from the Council of Scientific and Industrial Research (CSIR), Human Resource Development Group, Ministry of Science and Technology, Govt. of India (Scheme No. 21 (1107)/20/EMR-II dt. 21.09.2020) and Department of Biotechnology, Government of India [Project No DBT-NER/AGRI/33/2016 (Group-I, Application No. 02)] are highly acknowledged.
Author information
Authors and Affiliations
Contributions
ABD is responsible for designing of experiments, analysis of data and final manuscript preparation and SKD is responsible for collection of data in experimentation and preparation of first draft of the manuscript. Both the authors finalize all the tables, photographs, graphs and approves the manuscript for submission without any conflict of interest.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding Editor: Maumita Bandyopadhyay; Reviewers: Sreetama Bhadra, Seemanti Ghosh, Alongklod Tanomtong.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dehery, S.K., Das, A.B. Genetic diversity of twelve triploid bananas and plantains under section Eumusa as evident by chromosome morphology and SSR markers. Nucleus 65, 35–48 (2022). https://doi.org/10.1007/s13237-021-00364-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-021-00364-1